Object Detection in Laparoscopic Surgery: A Comparative Study of Deep Learning Models on a Custom Endometriosis Dataset
Abstract
1. Introduction
2. Related Work
2.1. Existing AI Applications
2.2. Review of Existing Datasets
2.3. Limitations of Existing Datasets
3. Materials and Methods
3.1. Endometriosis Dataset
3.1.1. Overview
3.1.2. Data Collection and Composition
3.1.3. Annotation Details
3.1.4. Video Characteristics
3.1.5. Data Organization and Accessibility
3.1.6. Considerations and Challenges
3.2. Methods
3.3. Data Preprocessing
3.4. Object Detection Models
- FasterRCNN: We used a FasterRCNN model with a ResNet50 backbone, initialised with pretrained weights from the ImageNet1K_V2 model in PyTorch’s (version 2.3.0) torchvision (version 0.18.0) model zoo. The model has 41.34 M parameters out of which 25.6 M parameters are attributed to the ResNet50 backbone. This model is a two-stage object detection architecture. It combines the aforementioned ResNet50 backbone with the Feature Pyramid Network (FPN) [18] which adds up lateral connections and top-down pathways as well as enhances a multi-scale feature representation. The final stage is represented by a Region Proposal Network (RPN) [19] which takes the feature map and proposes candidate object bounding boxes. FasterRCNN was trained with heavy augmentations to handle a high variability in the dataset.
- YOLOv9: We utilized the largest of the V9 family: the YOLOv9e (https://docs.ultralytics.com/models/yolov9/#performance-on-ms-coco-dataset) model (accessed on 1 December 2024) with 58.1 M parameters, offering the highest accuracy and known for its extensive architecture as well as high performance on object detection tasks. The YOLOv9 model is composed of a backbone: the Generalized Efficient Layer Aggregation Network (GELAN); a neck: Programmable Gradient Information (PGI); and a head: the DualDDetect module. The model was pretrained on the COCO dataset [20] and supports 640 × 640 resolution inputs. It was fine-tuned on our dataset with the default Ultralytics augmentations: https://docs.ultralytics.com (accessed on 1 December 2024).
3.5. Training Strategy
3.5.1. Stratified vs. Non-Stratified Splits
3.5.2. Augmentation Techniques
3.6. Experimental Setup
Evaluation Metrics
- Precision: measures the proportion of correctly predicted positive instances among all predicted positives.
- Recall: reflects the model’s ability to identify true positives (actual lesions).
- mAP50: evaluates localization accuracy at an Intersection over Union (IoU) threshold of 0.50.
- mAP50-95: assesses mean Average Precision over a range of IoU thresholds (0.50 to 0.95).
- Fitness: a combined metric that balances precision and recall, often represented by the F-1 score.
4. Main Results
4.1. YOLOv9 Performance
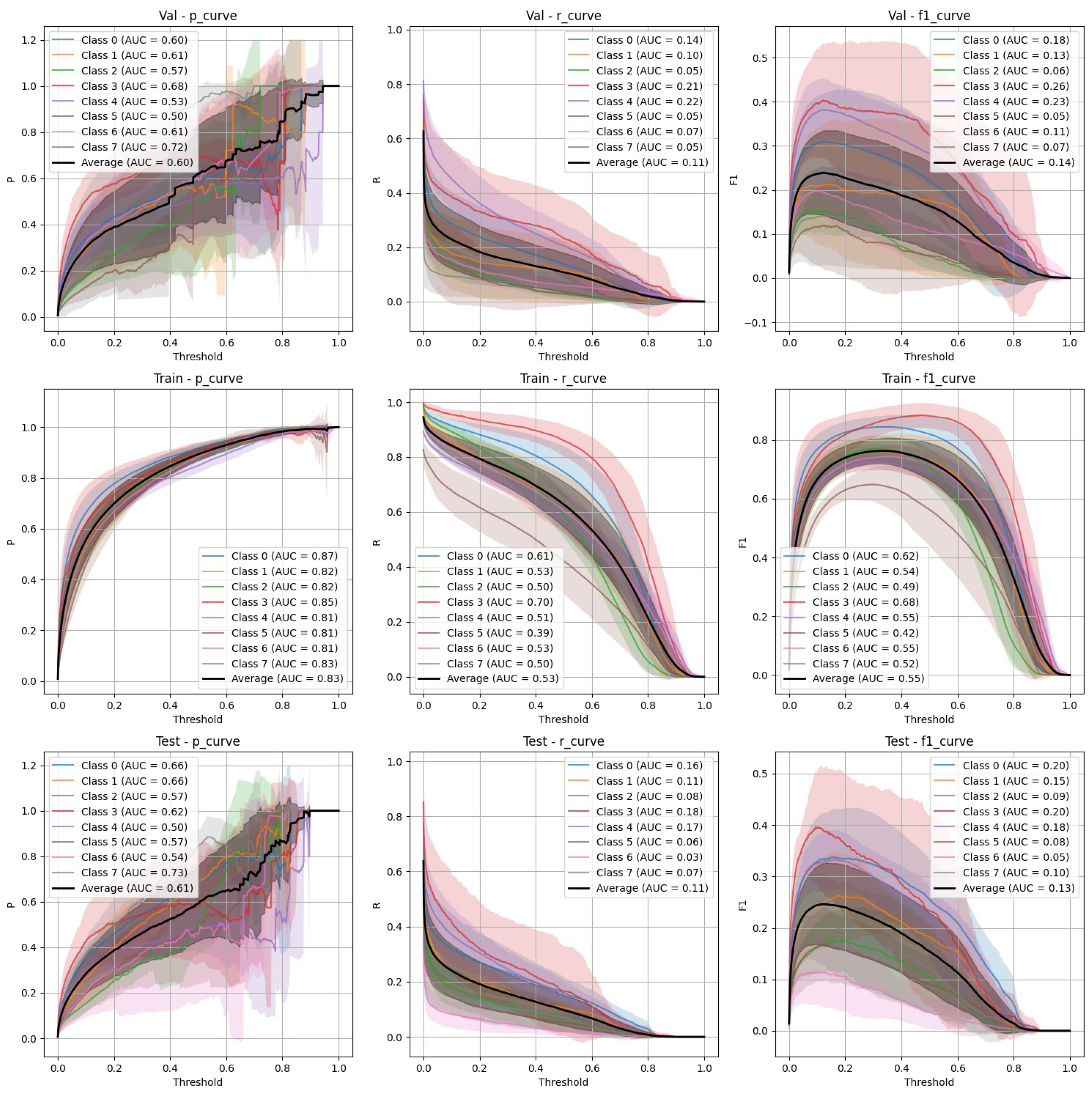
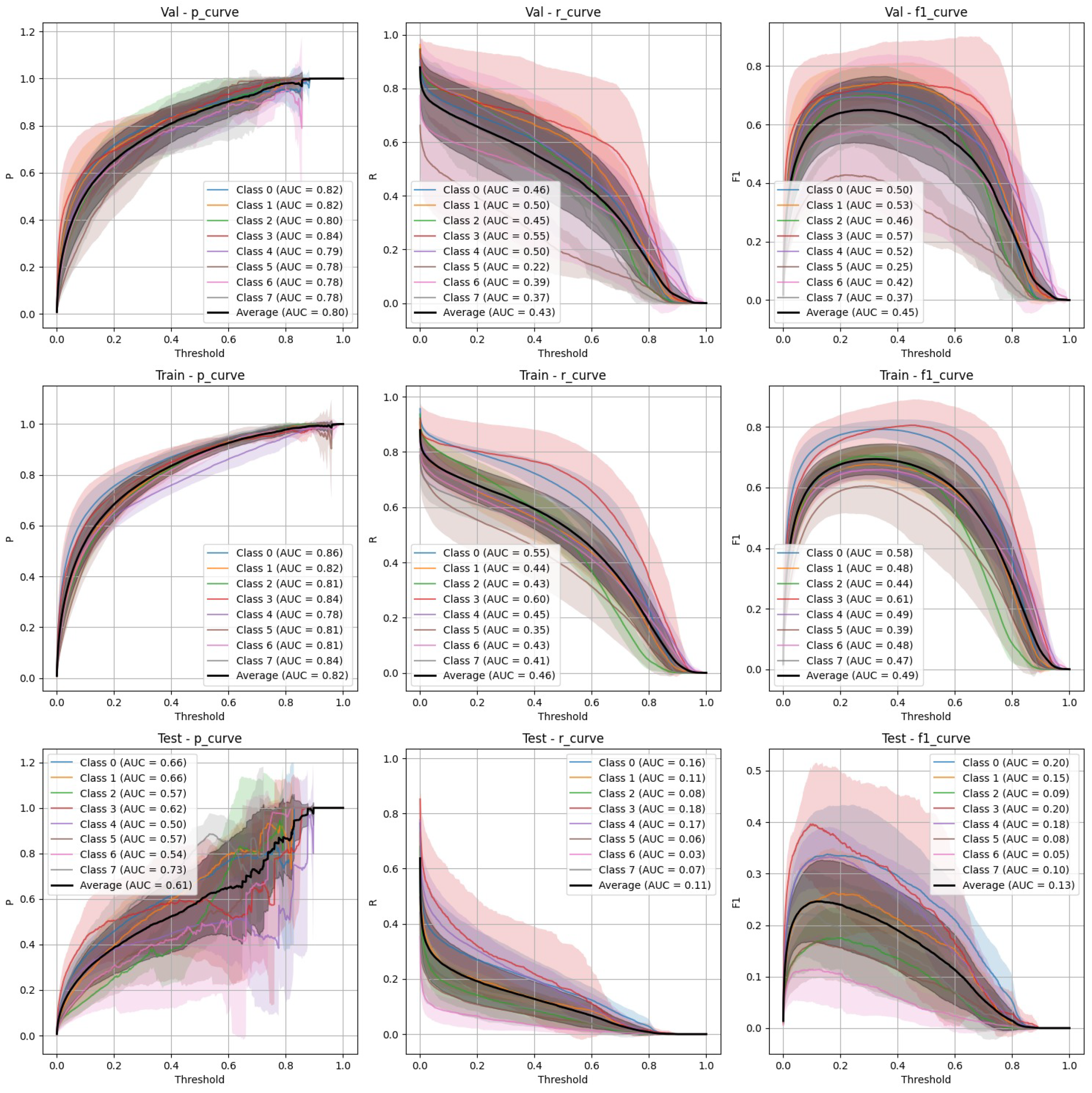
- Stratified split (Figure 3): YOLOv9 showed lower precision and recall compared to FasterRCNN, with substantial variation in the stratified scenario. The F-1 curves demonstrate challenges in maintaining a balance between precision and recall, particularly on the validation and test sets.
- Non-stratified split (Figure 4): YOLOv9 improved its performance in the non-stratified case, as reflected in its higher precision, recall, and mAP scores. The non-stratified F-1 curves also show better consistency across the training and validation sets.
4.2. FasterRCNN Performance
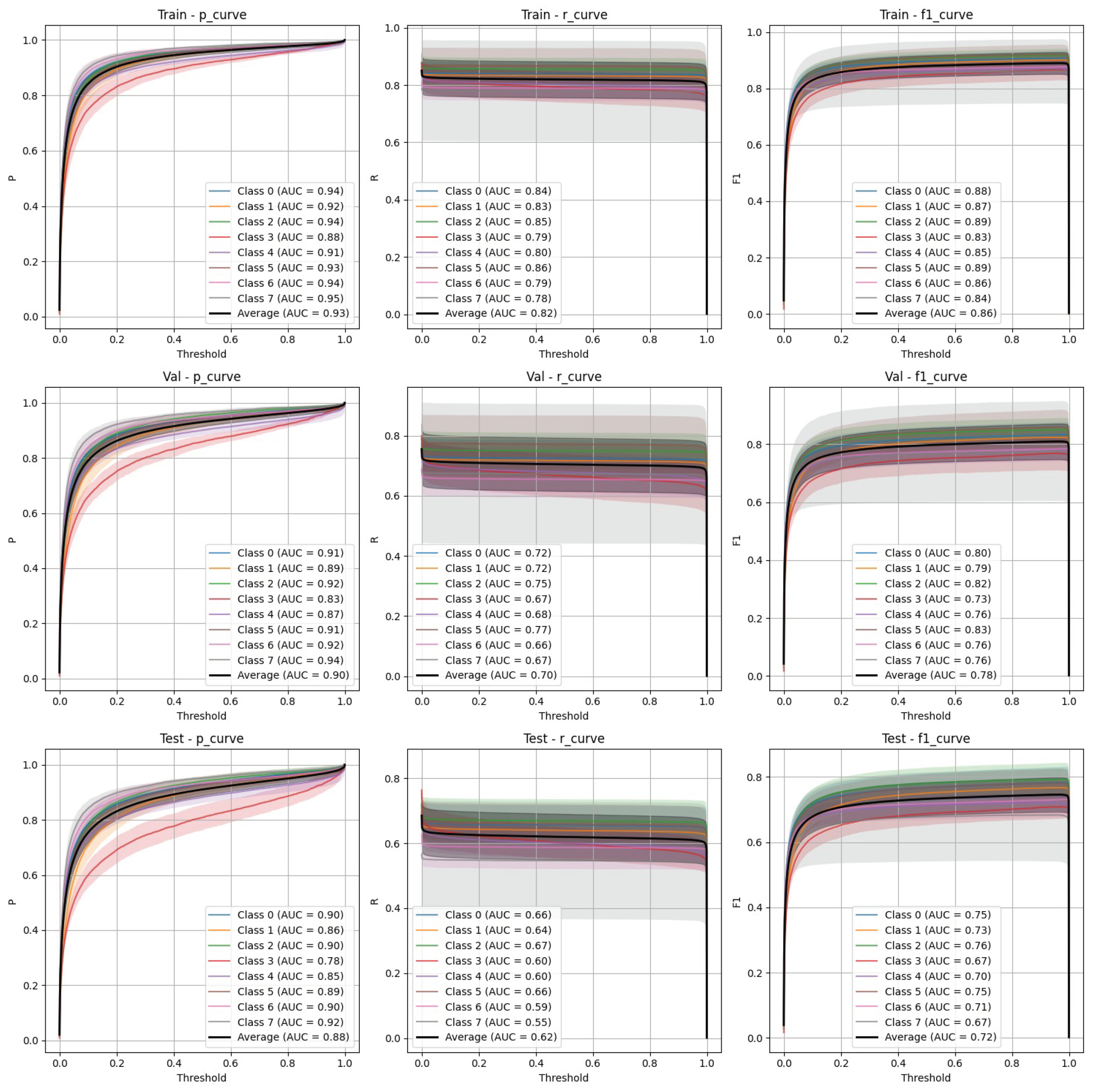
- Stratified split (Figure 5): FasterRCNN demonstrated superior performance across all metrics in the stratified scenario, with precision exceeding 0.97 and relatively stable recall. The mAP50 and mAP50-95 scores indicate that the model was able to detect and segment objects with high accuracy.
- Non-stratified split (Figure 6): While the performance of FasterRCNN remained high in the non-stratified scenario, slight differences were observed in precision and recall across the training, validation, and test datasets. F-1 curves for FasterRCNN consistently showed better generalization and balance across all splits compared to YOLOv9.
4.3. Training Challenges
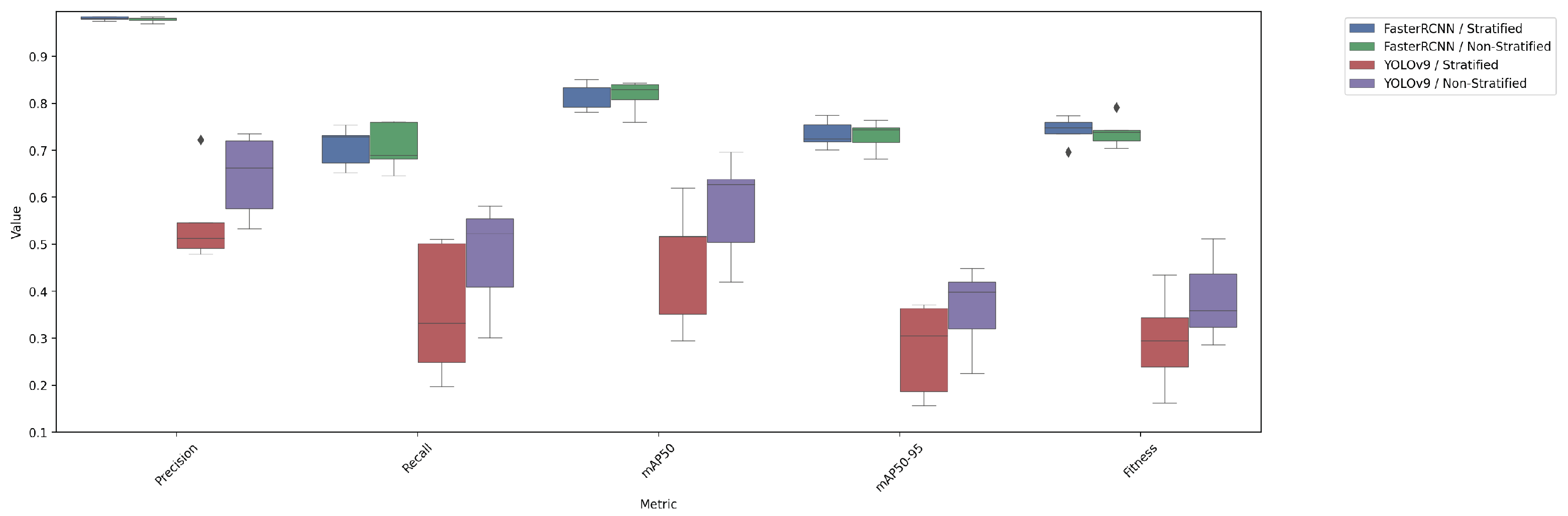
4.4. Comparison of Test Performance Metrics
4.5. Visual Validation and Results
5. Discussion of Challenges and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hummelshoj, L.; Prentice, A.; Groothuis, P. Update on Endometriosis: 9th World Congress on Endometriosis, 14–17 September 2005, Maastricht, the Netherlands. Women’s Health 2006, 2, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Beata, S.; Szyłło, K.; Romanowicz, H. Endometriosis: Epidemiology, Classification, Pathogenesis, Treatment and Genetics (Review of Literature). Int. J. Mol. Sci. 2021, 22, 10554. [Google Scholar] [CrossRef]
- Vobugari, N.; Raja, V.; Sethi, U.; Gandhi, K.; Raja, K.; Surani, S.R. Advancements in oncology with artificial intelligence—A review article. Cancers 2022, 14, 1349. [Google Scholar] [CrossRef] [PubMed]
- Quazi, S. Artificial intelligence and machine learning in precision and genomic medicine. Med. Oncol. 2022, 39, 120. [Google Scholar] [CrossRef] [PubMed]
- Ghaderzadeh, M.; Shalchian, A.; Irajian, G.; Sadeghsalehi, H.; Zahedi bialvaei, A.; Sabet, B. Artificial Intelligence in Drug Discovery and Development Against Antimicrobial Resistance: A Narrative Review. Iran. J. Med. Microbiol. 2024, 18, 135–147. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Yeh, I.H.; Liao, H.Y.M. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. In Proceedings of the 18th European Conference, Milan, Italy, 29 September–4 October 2024; pp. 1–21. [Google Scholar] [CrossRef]
- Foti, P.; Palmucci, S.; Vizzini, I.; Libertini, N.; Coronella, M.; Spadola, S.; Caltabiano, R.; Iraci Sareri, M.; Basile, A.; Milone, P.; et al. Endometriosis: Clinical features, MR imaging findings and pathologic correlation. Insights Imaging 2018, 9, 149–172. [Google Scholar] [CrossRef] [PubMed]
- Sivajohan, B.; Elgendi, M.; Menon, C.; Allaire, C.; Yong, P.; Bedaiwy, M. Clinical use of artificial intelligence in endometriosis: A scoping review. npj Digit. Med. 2022, 5, 109. [Google Scholar] [CrossRef] [PubMed]
- Nifora, C.; Chasapi, M.K.; Chasapi, L.; Koutsojannis, C. Deep Learning Improves Accuracy of Laparoscopic Imaging Classification for Endometriosis Diagnosis. J. Clin. Med. Surg. 2024, 4, 1137–1145. [Google Scholar] [CrossRef]
- Leibetseder, A.; Schoeffmann, K.; Keckstein, J.; Keckstein, S. Endometriosis detection and localization in laparoscopic gynecology. Multimed. Tools Appl. 2022, 81, 6191–6215. [Google Scholar] [CrossRef]
- Hong, W.; Kao, C.; Kuo, Y.; Wang, J.; Chang, W.; Shih, C. CholecSeg8k: A Semantic Segmentation Dataset for Laparoscopic Cholecystectomy Based on Cholec80. arXiv 2020, arXiv:2012.12453. [Google Scholar] [CrossRef]
- Carstens, M.; Rinner, F.; Bodenstedt, S.; Jenke, A.; Weitz, J.; Distler, M.; Speidel, S.; Kolbinger, F. The Dresden Surgical Anatomy Dataset for Abdominal Organ Segmentation in Surgical Data Science. Sci. Data 2023, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Leibetseder, A.; Kletz, S.; Schoeffmann, K.; Keckstein, S.; Keckstein, J. GLENDA: Gynecologic Laparoscopy Endometriosis Dataset. In Proceedings of the 26th International Conference, MMM 2020, Daejeon, Republic of Korea, 5–8 January 2020; pp. 439–450. [Google Scholar] [CrossRef]
- Yoon, J.; Hong, S.; Hong, S.; Lee, J.; Shin, S.; Park, B.; Sung, N.; Yu, H.; Kim, S.; Park, S.; et al. Surgical Scene Segmentation Using Semantic Image Synthesis with a Virtual Surgery Environment. In Proceedings of the 25th International Conference, Singapore, 18–22 September 2022; pp. 551–561. [Google Scholar] [CrossRef]
- Figueiredo, R.B.D.; Mendes, H.A. Analyzing Information Leakage on Video Object Detection Datasets by Splitting Images into Clusters with High Spatiotemporal Correlation. IEEE Access 2024, 12, 47646–47655. [Google Scholar] [CrossRef]
- Apicella, A.; Isgrò, F.; Prevete, R. Don’t Push the Button! Exploring Data Leakage Risks in Machine Learning and Transfer Learning. arXiv 2024, arXiv:2401.13796. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Wang, J.; Chen, K.; Yang, S.; Loy, C.C.; Lin, D. Region proposal by guided anchoring. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 2965–2974. [Google Scholar]
- Lin, T.; Maire, M.; Belongie, S.J.; Bourdev, L.D.; Girshick, R.B.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. arXiv 2014, arXiv:1405.0312. [Google Scholar] [CrossRef]
- Ghiasi, G.; Cui, Y.; Srinivas, A.; Qian, R.; Lin, T.Y.; Cubuk, E.D.; Le, Q.V.; Zoph, B. Simple Copy-Paste is a Strong Data Augmentation Method for Instance Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 2917–2927. [Google Scholar]
- Zhang, H.; Cissé, M.; Dauphin, Y.; Lopez-Paz, D. mixup: Beyond Empirical Risk Minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar] [CrossRef]
- Llugsi, R.; El Yacoubi, S.; Fontaine, A.; Lupera, P. Comparison between Adam, AdaMax and Adam W optimizers to implement a Weather Forecast based on Neural Networks for the Andean city of Quito. In Proceedings of the 2021 IEEE Fifth Ecuador Technical Chapters Meeting (ETCM), Cuenca, Ecuador, 12–15 October 2021; pp. 1–6. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929v2. [Google Scholar]
- Nguyen, H. Improving Faster R-CNN Framework for Fast Vehicle Detection. Math. Probl. Eng. 2019, 2019, 3808064. [Google Scholar] [CrossRef]






| Resolution | Number of Videos |
|---|---|
| 1920 × 1080 | 193 |
| 1280 × 720 | 5 |
| 720 × 576 | 1 |
| Class Name | Class ID | Number of Annotated Objects |
|---|---|---|
| Adhesions Dense | 0 | 1424 |
| Adhesions Filmy | 1 | 537 |
| Deep Endometriosis | 2 | 700 |
| Ovarian Chocolate Fluid | 3 | 223 |
| Ovarian Endometrioma | 4 | 302 |
| Ovarian Endometrioma[B] | 4 | 382 |
| Superficial Black | 5 | 835 |
| Superficial Red | 6 | 642 |
| Superficial Subtle | 7 | 509 |
| Superficial White | 8 | 463 |
| Model/Split | Precision | Recall | mAP50 | mAP50-95 | Fitness |
|---|---|---|---|---|---|
| FasterRCNN/stratified | 0.9811 ± 0.0084 | 0.7083 ± 0.0807 | 0.8185 ± 0.0562 | 0.7345 ± 0.0554 | 0.7429 ± 0.0555 |
| FasterRCNN/non-stratified | 0.9787 ± 0.0107 | 0.7076 ± 0.0957 | 0.8162 ± 0.0647 | 0.7309 ± 0.0612 | 0.7395 ± 0.0615 |
| YOLOv9/stratified | 0.5504 ± 0.1864 | 0.3580 ± 0.2701 | 0.4599 ± 0.2503 | 0.2767 ± 0.1877 | 0.2951 ± 0.1939 |
| YOLOv9/non-stratified | 0.6458 ± 0.1662 | 0.4742 ± 0.2193 | 0.5771 ± 0.2113 | 0.3622 ± 0.1656 | 0.3837 ± 0.1701 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bondarenko, A.; Jumutc, V.; Netter, A.; Duchateau, F.; Abrão, H.M.; Noorzadeh, S.; Giacomello, G.; Ferrari, F.; Bourdel, N.; Kirk, U.B.; et al. Object Detection in Laparoscopic Surgery: A Comparative Study of Deep Learning Models on a Custom Endometriosis Dataset. Diagnostics 2025, 15, 1254. https://doi.org/10.3390/diagnostics15101254
Bondarenko A, Jumutc V, Netter A, Duchateau F, Abrão HM, Noorzadeh S, Giacomello G, Ferrari F, Bourdel N, Kirk UB, et al. Object Detection in Laparoscopic Surgery: A Comparative Study of Deep Learning Models on a Custom Endometriosis Dataset. Diagnostics. 2025; 15(10):1254. https://doi.org/10.3390/diagnostics15101254
Chicago/Turabian StyleBondarenko, Andrey, Vilen Jumutc, Antoine Netter, Fanny Duchateau, Henrique Mendonca Abrão, Saman Noorzadeh, Giuseppe Giacomello, Filippo Ferrari, Nicolas Bourdel, Ulrik Bak Kirk, and et al. 2025. "Object Detection in Laparoscopic Surgery: A Comparative Study of Deep Learning Models on a Custom Endometriosis Dataset" Diagnostics 15, no. 10: 1254. https://doi.org/10.3390/diagnostics15101254
APA StyleBondarenko, A., Jumutc, V., Netter, A., Duchateau, F., Abrão, H. M., Noorzadeh, S., Giacomello, G., Ferrari, F., Bourdel, N., Kirk, U. B., & Bļizņuks, D. (2025). Object Detection in Laparoscopic Surgery: A Comparative Study of Deep Learning Models on a Custom Endometriosis Dataset. Diagnostics, 15(10), 1254. https://doi.org/10.3390/diagnostics15101254







