Development of Clinical Radiomics-Based Models to Predict Survival Outcome in Pancreatic Ductal Adenocarcinoma: A Multicenter Retrospective Study
Abstract
Highlights
- This multicenter retrospective study, encompassing two hospital sites for training and leveraging data from forty-seven different hospitals for testing, aimed to develop machine learning models utilising both clinical and radiomics features (individually and in combination) extracted from pre-treatment CT scans. The objective was to predict PFS and OS in patients with PDAC.
- Among the various models developed, the model based on the combination of clinical and CT-derived tumour radiomics features demonstrated superior performance for OS with a C-index of 0.72 in the testing dataset. Conversely, the model based solely on clinical features excelled in predicting PFS, achieving a C-index of 0.70 in the testing dataset.
- The analysis further highlighted the high relevance of clinical features both for OS and PFS outcomes in patients with PDAC.
Abstract
1. Introduction
2. Material and Methods
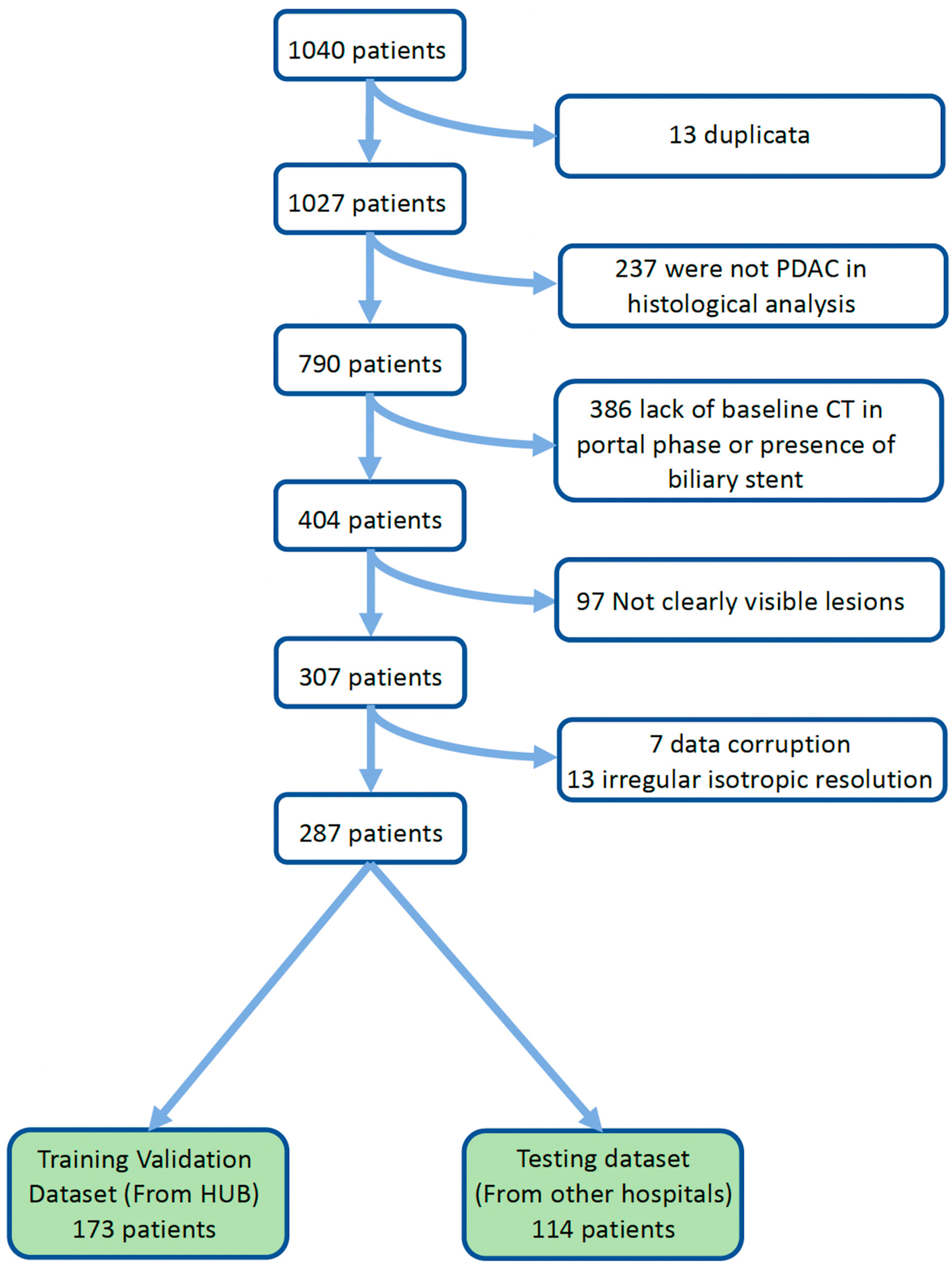
2.1. Study Population
2.2. Patients Stratification
2.3. Data Analysis
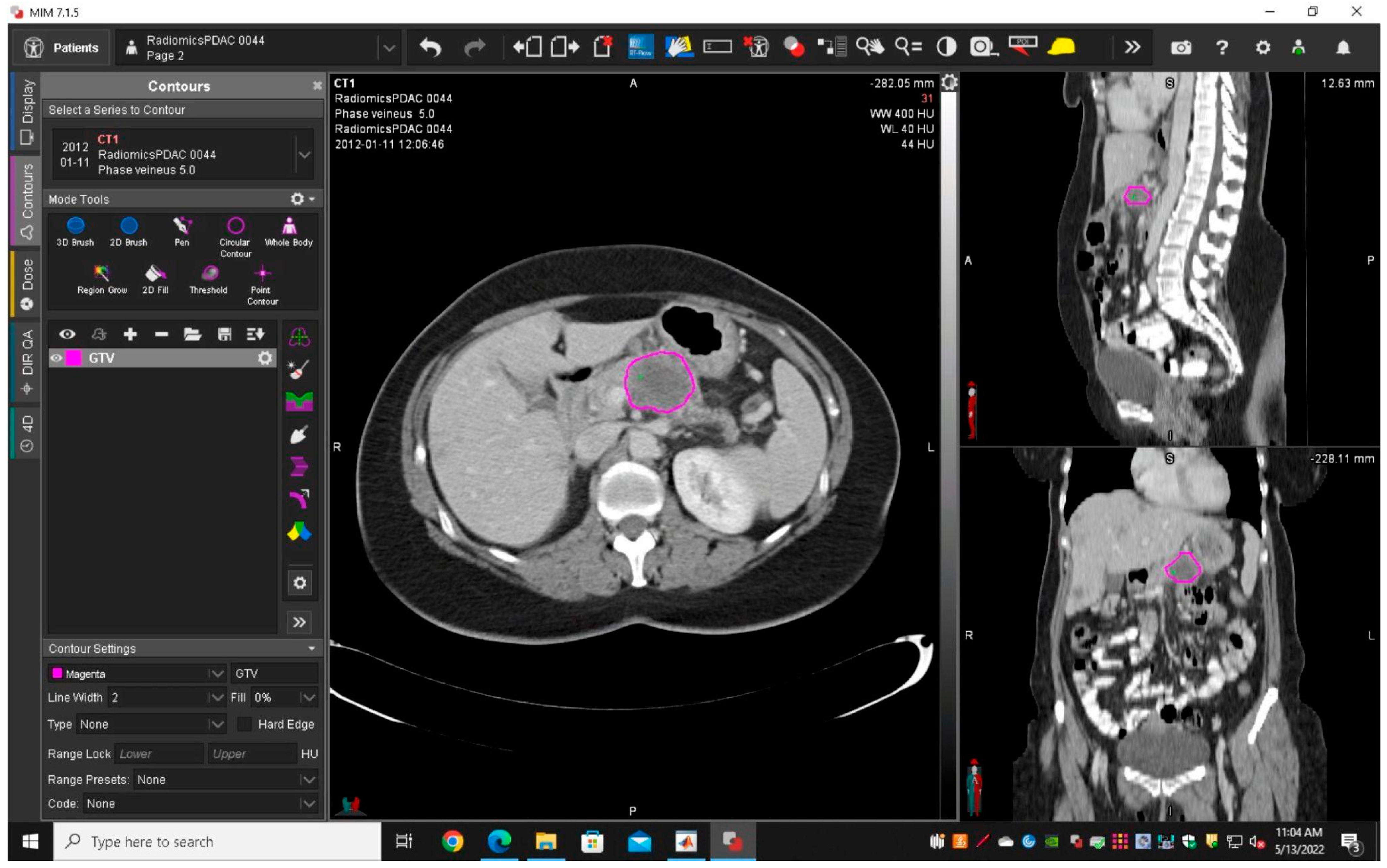
2.3.1. Segmentation and Features Extraction
2.3.2. Feature Selection
2.4. Model Building and Evaluation
2.5. Model Interpretability
2.6. Statistical Analysis
3. Results
3.1. Clinical Data and Patient Stratification
3.2. Image Acquisition and Segmentation
3.3. Features Selection
3.4. Model Performances
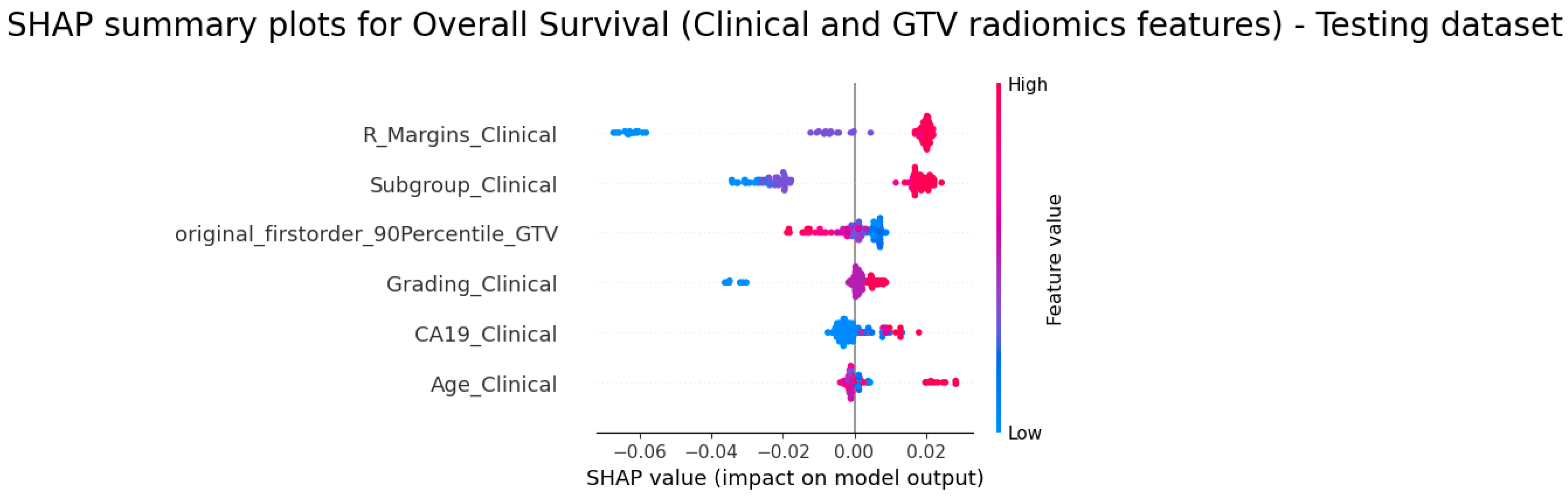
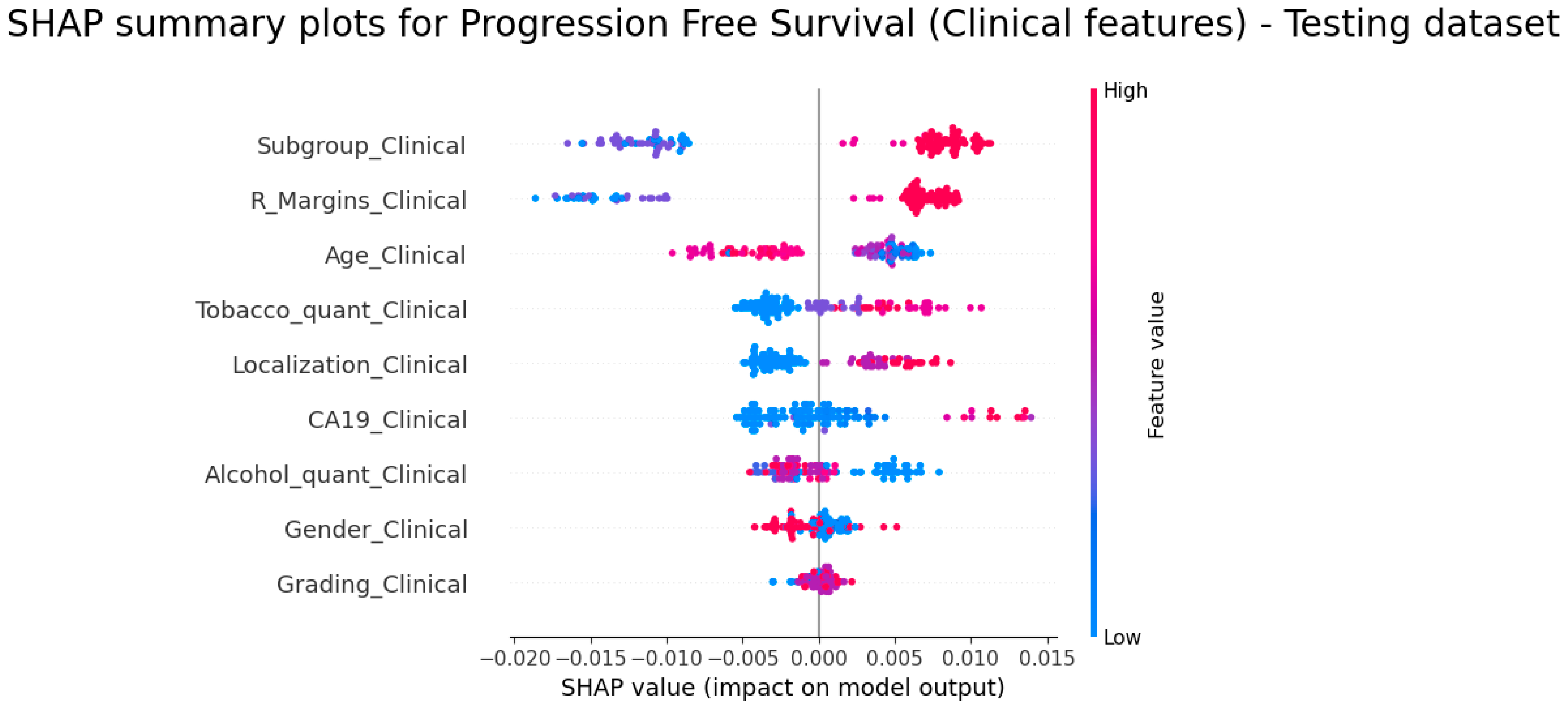
3.5. Explainability of the Models
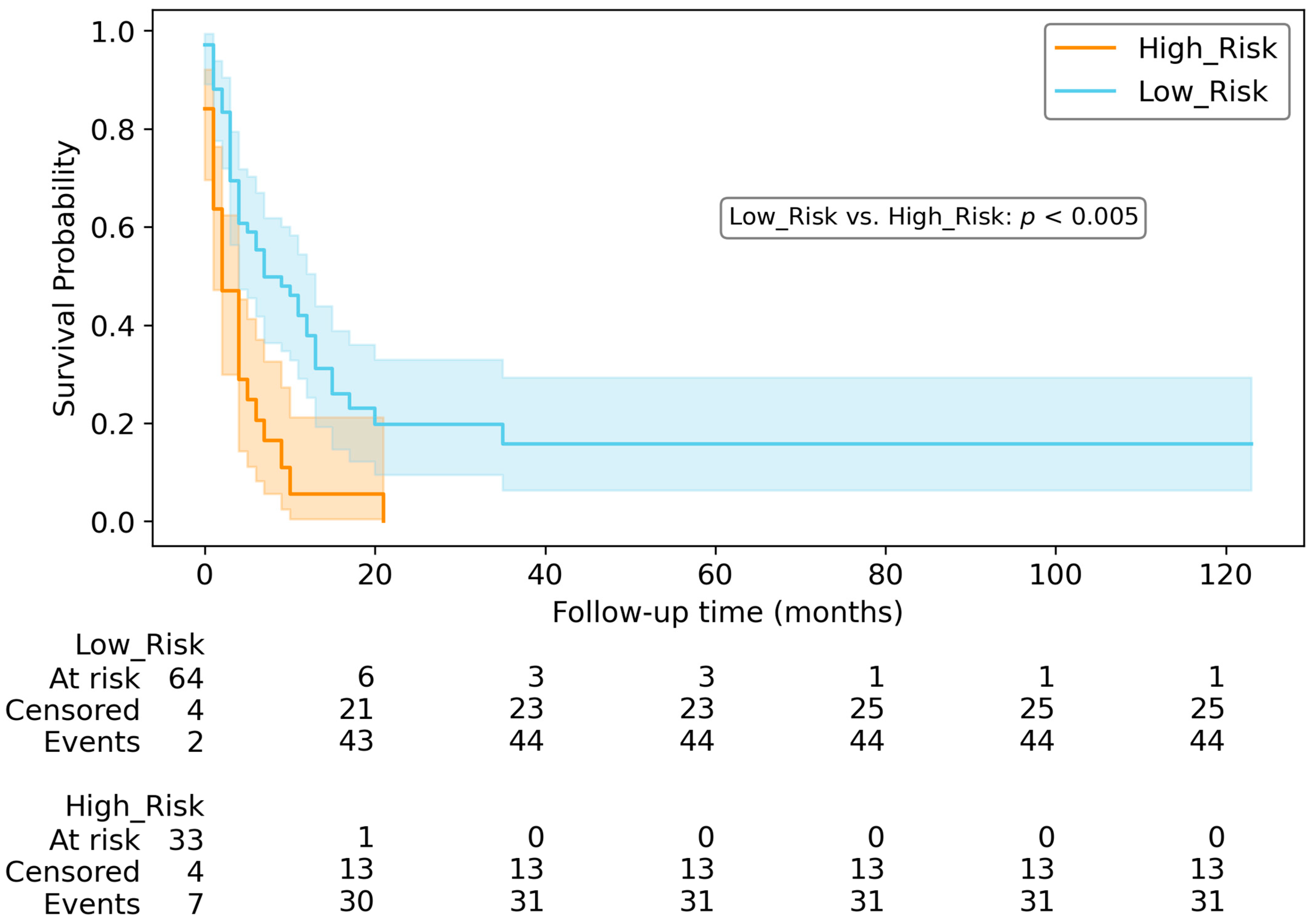
3.6. Survival Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Clinical Relevance Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| AUC | Area Under the Curve |
| BMI | Body Mass Index |
| CT | computed tomography |
| GTV | Gross tumour volume |
| HE | Hopital Erasme |
| HUB | Hopital Universitaire de Bruxelles |
| IJB | Institut Jules Bordet |
| OS | overall survival |
| PDAC | Pancreatic adenocarcinoma |
| PFS | progression-free survival |
| PY | Pack-year |
| RPV | Non tumoral pancreas |
| SHAP | Shapley additive explanations |
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Schlitter, A.M.; Segler, A.; Steiger, K.; Michalski, C.W.; Jäger, C.; Konukiewitz, B.; Pfarr, N.; Endris, V.; Bettstetter, M.; Kong, B.; et al. Molecular, Morphological and Survival Analysis of 177 Resected Pancreatic Ductal Adenocarcinomas (PDACs): Identification of Prognostic Subtypes. Sci. Rep. 2017, 7, 41064. [Google Scholar] [CrossRef] [PubMed]
- Neesse, A.; Algül, H.; Tuveson, D.A.; Gress, T.M. Stromal Biology and Therapy in Pancreatic Cancer: A Changing Paradigm. Gut 2015, 64, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Puleo, F.; Nicolle, R.; Blum, Y.; Cros, J.; Marisa, L.; Demetter, P.; Quertinmont, E.; Svrcek, M.; Elarouci, N.; Iovanna, J.; et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology 2018, 155, 1999–2013.e3. [Google Scholar] [CrossRef]
- Bodalal, Z.; Trebeschi, S.; Nguyen-Kim, T.D.L.; Schats, W.; Beets-Tan, R. Radiogenomics: Bridging Imaging and Genomics. Abdom. Radiol. (NY) 2019, 44, 1960–1984. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, M.; Barat, M.; Dohan, A.; Gaujoux, S.; Coriat, R.; Hoeffel, C.; Cassinotto, C.; Chassagnon, G.; Soyer, P. CT and MRI of Pancreatic Tumors: An Update in the Era of Radiomics. Jpn. J. Radiol. 2020, 38, 1111–1124. [Google Scholar] [CrossRef]
- Kuo, M.D.; Jamshidi, N. Behind the Numbers: Decoding Molecular Phenotypes with Radiogenomics--Guiding Principles and Technical Considerations. Radiology 2014, 270, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Barat, M.; Chassagnon, G.; Dohan, A.; Gaujoux, S.; Coriat, R.; Hoeffel, C.; Cassinotto, C.; Soyer, P. Artificial Intelligence: A Critical Review of Current Applications in Pancreatic Imaging. Jpn. J. Radiol. 2021, 39, 514–523. [Google Scholar] [CrossRef]
- Abunahel, B.M.; Pontre, B.; Kumar, H.; Petrov, M.S. Pancreas Image Mining: A Systematic Review of Radiomics. Eur. Radiol. 2021, 31, 3447–3467. [Google Scholar] [CrossRef]
- Qiu, W.; Duan, N.; Chen, X.; Ren, S.; Zhang, Y.; Wang, Z.; Chen, R. Pancreatic Ductal Adenocarcinoma: Machine Learning-Based Quantitative Computed Tomography Texture Analysis For Prediction Of Histopathological Grade. Cancer Manag. Res. 2019, 11, 9253–9264. [Google Scholar] [CrossRef]
- Ahmed, T.M.; Kawamoto, S.; Hruban, R.H.; Fishman, E.K.; Soyer, P.; Chu, L.C. A Primer on Artificial Intelligence in Pancreatic Imaging. Diagn. Interv. Imaging 2023, 104, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, M.A.; Chakraborty, J.; McIntyre, C.A.; Kappagantula, R.; Chou, Y.; Askan, G.; Seier, K.; Gonen, M.; Basturk, O.; Balachandran, V.P.; et al. CT Radiomics Associations with Genotype and Stromal Content in Pancreatic Ductal Adenocarcinoma. Abdom. Radiol. (NY) 2019, 44, 3148–3157. [Google Scholar] [CrossRef] [PubMed]
- Kaissis, G.; Ziegelmayer, S.; Lohöfer, F.; Steiger, K.; Algül, H.; Muckenhuber, A.; Yen, H.-Y.; Rummeny, E.; Friess, H.; Schmid, R.; et al. A Machine Learning Algorithm Predicts Molecular Subtypes in Pancreatic Ductal Adenocarcinoma with Differential Response to Gemcitabine-Based versus FOLFIRINOX Chemotherapy. PLoS ONE 2019, 14, e0218642. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xiao, J.; Yang, J.; Li, M.; Xiong, X.; Nian, Y.; Qiao, L.; Wang, H.; Eresen, A.; Zhang, Z.; et al. Association of Radiomic Imaging Features and Gene Expression Profile as Prognostic Factors in Pancreatic Ductal Adenocarcinoma. Am. J. Transl. Res. 2019, 11, 4491–4499. [Google Scholar] [PubMed]
- Racu, M.-L.; Bernardi, D.; Chaouche, A.; Zindy, E.; Navez, J.; Loi, P.; Maris, C.; Closset, J.; Van Laethem, J.-L.; Decaestecker, C.; et al. Abstract B043: Impact of SMAD4 Loss in Patients with Pancreatic Ductal Adenocarcinoma Receiving Neoadjuvant Therapy. Cancer Res. 2022, 82, B043. [Google Scholar] [CrossRef]
- Cheng, S.-H.; Cheng, Y.-J.; Jin, Z.-Y.; Xue, H.-D. Unresectable Pancreatic Ductal Adenocarcinoma: Role of CT Quantitative Imaging Biomarkers for Predicting Outcomes of Patients Treated with Chemotherapy. Eur. J. Radiol. 2019, 113, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Broekmans, J.P.E.A.; Boers, T.; Caballo, M.; van den Hurk, M.H.F.; Ozdemir, D.; van Schaik, C.J.; Janse, M.H.A.; van Geenen, E.J.M.; van Laarhoven, C.J.H.M.; et al. Computed Tomography-Based Radiomics Using Tumor and Vessel Features to Assess Resectability in Cancer of the Pancreatic Head. Diagnostics 2023, 13, 3198. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Guo, S.; Jiang, H.; Gao, S.; Shao, C.; Cao, K.; Fang, X.; Li, J.; Wang, L.; Hua, W.; et al. Relationship Between Radiomics and Risk of Lymph Node Metastasis in Pancreatic Ductal Adenocarcinoma. Pancreas 2019, 48, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, V.S.; Karmazanovsky, G.G.; Kondratyev, E.V.; Gruzdev, I.S.; Mikhaylyuk, K.A.; Sinelnikov, M.Y.; Revishvili, A.S. Radiomics Model-Based Algorithm for Preoperative Prediction of Pancreatic Ductal Adenocarcinoma Grade. Eur. Radiol. 2023, 33, 1152–1161. [Google Scholar] [CrossRef]
- Zins, M.; Matos, C.; Cassinotto, C. Pancreatic Adenocarcinoma Staging in the Era of Preoperative Chemotherapy and Radiation Therapy. Radiology 2018, 287, 374–390. [Google Scholar] [CrossRef]
- Tempero, M.A.; Malafa, M.P.; Al-Hawary, M.; Behrman, S.W.; Benson, A.B.; Cardin, D.B.; Chiorean, E.G.; Chung, V.; Czito, B.; Del Chiaro, M.; et al. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.Y.; Lin, C.; Baine, M.; Bennion, N.R.; Yu, L.; Zheng, D.; Vipin, D.; Hollingsworth, M.A.; Zhou, S.M.; Zheng, D. Effect of Interobserver and Interdisciplinary Segmentation Variability on Radiomic Features for Pancreatic Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, e769–e770. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Larue, R.T.H.M.; van Timmeren, J.E.; de Jong, E.E.C.; Feliciani, G.; Leijenaar, R.T.H.; Schreurs, W.M.J.; Sosef, M.N.; Raat, F.H.P.J.; van der Zande, F.H.R.; Das, M.; et al. Influence of Gray Level Discretization on Radiomic Feature Stability for Different CT Scanners, Tube Currents and Slice Thicknesses: A Comprehensive Phantom Study. Acta Oncol. 2017, 56, 1544–1553. [Google Scholar] [CrossRef] [PubMed]
- Pölsterl, S. Scikit-Survival: A Library for Time-to-Event Analysis Built on Top of Scikit-Learn. J. Mach. Learn. Res. 2020, 21, 1–6. [Google Scholar]
- Baek, E.-T.; Yang, H.J.; Kim, S.H.; Lee, G.S.; Oh, I.-J.; Kang, S.-R.; Min, J.-J. Survival Time Prediction by Integrating Cox Proportional Hazards Network and Distribution Function Network. BMC Bioinform. 2021, 22, 192. [Google Scholar] [CrossRef]
- Park, Y.W.; Eom, J.; Kim, D.; Ahn, S.S.; Kim, E.H.; Kang, S.-G.; Chang, J.H.; Kim, S.H.; Lee, S.-K. A Fully Automatic Multiparametric Radiomics Model for Differentiation of Adult Pilocytic Astrocytomas from High-Grade Gliomas. Eur. Radiol. 2022, 32, 4500–4509. [Google Scholar] [CrossRef]
- Wang, Y.; Lang, J.; Zuo, J.Z.; Dong, Y.; Hu, Z.; Xu, X.; Zhang, Y.; Wang, Q.; Yang, L.; Wong, S.T.C.; et al. The Radiomic-Clinical Model Using the SHAP Method for Assessing the Treatment Response of Whole-Brain Radiotherapy: A Multicentric Study. Eur. Radiol. 2022, 32, 8737–8747. [Google Scholar] [CrossRef] [PubMed]
- Salahuddin, Z.; Chen, Y.; Zhong, X.; Woodruff, H.C.; Rad, N.M.; Mali, S.A.; Lambin, P. From Head and Neck Tumour and Lymph Node Segmentation to Survival Prediction on PET/CT: An End-to-End Framework Featuring Uncertainty, Fairness, and Multi-Region Multi-Modal Radiomics. Cancers 2023, 15, 1932. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, Y.; Liu, D.; Lv, R.; Huang, Y.; Peng, C.; Jiang, S.; Wang, Y.; He, Y.; Lan, X.; et al. Predicting Progression-Free Survival Using MRI-Based Radiomics for Patients with Nonmetastatic Nasopharyngeal Carcinoma. Front. Oncol. 2020, 10, 618. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Park, S.; Sham, J.G.; Kawamoto, S.; Blair, A.B.; Rozich, N.; Fouladi, D.F.; Shayesteh, S.; Hruban, R.H.; He, J.; Wolfgang, C.L.; et al. CT Radiomics-Based Preoperative Survival Prediction in Patients with Pancreatic Ductal Adenocarcinoma. AJR Am. J. Roentgenol. 2021, 217, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Mao, Y.; Xie, H.; Qin, T.; Mai, Z.; Cai, Q.; Wen, H.; Li, Y.; Zhang, R.; Liu, L. Identifying Outcomes of Patients with Advanced Pancreatic Adenocarcinoma and RECIST Stable Disease Using Radiomics Analysis. JCO Precis. Oncol. 2022, 6, e2100362. [Google Scholar] [CrossRef] [PubMed]
- Healy, G.M.; Salinas-Miranda, E.; Jain, R.; Dong, X.; Deniffel, D.; Borgida, A.; Hosni, A.; Ryan, D.T.; Njeze, N.; McGuire, A.; et al. Pre-Operative Radiomics Model for Prognostication in Resectable Pancreatic Adenocarcinoma with External Validation. Eur. Radiol. 2022, 32, 2492–2505. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, M.A.; Chakraborty, J.; Doussot, A.; Langdon-Embry, L.; Mainarich, S.; Gönen, M.; Balachandran, V.P.; D’Angelica, M.I.; DeMatteo, R.P.; Jarnagin, W.R.; et al. Survival Prediction in Pancreatic Ductal Adenocarcinoma by Quantitative Computed Tomography Image Analysis. Ann. Surg. Oncol. 2018, 25, 1034–1042. [Google Scholar] [CrossRef]
- Hruban, R.H.; Gaida, M.M.; Thompson, E.; Hong, S.-M.; Noë, M.; Brosens, L.A.; Jongepier, M.; Offerhaus, G.J.A.; Wood, L.D. Why Is Pancreatic Cancer so Deadly? The Pathologist’s View. J. Pathol. 2019, 248, 131–141. [Google Scholar] [CrossRef]
- Sandrasegaran, K.; Lin, Y.; Asare-Sawiri, M.; Taiyini, T.; Tann, M. CT Texture Analysis of Pancreatic Cancer. Eur. Radiol. 2019, 29, 1067–1073. [Google Scholar] [CrossRef]
- Lee, S.J.; Sung, Y.-N.; Kim, S.J.; Shin, S.; Cho, H.; Hruban, R.H.; Hong, S.-M. Comprehensive Histological Evaluation with Clinical Analysis of Venous Invasion in Pancreatic Ductal Adenocarcinoma: From Histology to Clinical Implications. Pancreatology 2020, 20, 1486–1494. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Wood, L.D.; Hruban, R.H.; Hong, S.-M. Desmin and CD31 Immunolabeling for Detecting Venous Invasion of the Pancreatobiliary Tract Cancers. PLoS ONE 2020, 15, e0242571. [Google Scholar] [CrossRef] [PubMed]
- Artinyan, A.; Soriano, P.A.; Prendergast, C.; Low, T.; Ellenhorn, J.D.; Kim, J. The Anatomic Location of Pancreatic Cancer Is a Prognostic Factor for Survival. HPB 2008, 10, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.F.; Moccia, R.D.; Klimstra, D. Management of Adenocarcinoma of the Body and Tail of the Pancreas. Ann. Surg. 1996, 223, 506–511; discussion 511–512. [Google Scholar] [CrossRef] [PubMed]
- Sohn, T.A.; Yeo, C.J.; Cameron, J.L.; Koniaris, L.; Kaushal, S.; Abrams, R.A.; Sauter, P.K.; Coleman, J.; Hruban, R.H.; Lillemoe, K.D. Resected Adenocarcinoma of the Pancreas-616 Patients: Results, Outcomes, and Prognostic Indicators. J. Gastrointest. Surg. 2000, 4, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Lau, M.K.; Davila, J.A.; Shaib, Y.H. Incidence and Survival of Pancreatic Head and Body and Tail Cancers: A Population-Based Study in the United States. Pancreas 2010, 39, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, I.; Sasaki, S.; Konishi, M.; Nakagohri, T.; Inoue, K.; Oda, T.; Kinoshita, T. Onset Symptoms and Tumor Locations as Prognostic Factors of Pancreatic Cancer. Pancreas 2004, 28, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, S.B.; Jamieson, N.B.; Upstill-Goddard, R.; Bailey, P.J.; McKay, C.J.; Australian Pancreatic Cancer Genome Initiative; Biankin, A.V.; Chang, D.K. Defining the Molecular Pathology of Pancreatic Body and Tail Adenocarcinoma. Br. J. Surg. 2018, 105, e183–e191. [Google Scholar]
- Yu, G.P.; Ostroff, J.S.; Zhang, Z.F.; Tang, J.; Schantz, S.P. Smoking History and Cancer Patient Survival: A Hospital Cancer Registry Study. Cancer Detect. Prev. 1997, 21, 497–509. [Google Scholar]
- Nakamura, K.; Nagata, C.; Wada, K.; Tamai, Y.; Tsuji, M.; Takatsuka, N.; Shimizu, H. Cigarette Smoking and Other Lifestyle Factors in Relation to the Risk of Pancreatic Cancer Death: A Prospective Cohort Study in Japan. Jpn. J. Clin. Oncol. 2011, 41, 225–231. [Google Scholar] [CrossRef]
- Gapstur, S.M.; Jacobs, E.J.; Deka, A.; McCullough, M.L.; Patel, A.V.; Thun, M.J. Association of Alcohol Intake with Pancreatic Cancer Mortality in Never Smokers. Arch. Intern. Med. 2011, 171, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Pelucchi, C.; Galeone, C.; Polesel, J.; Manzari, M.; Zucchetto, A.; Talamini, R.; Franceschi, S.; Negri, E.; La Vecchia, C. Smoking and Body Mass Index and Survival in Pancreatic Cancer Patients. Pancreas 2014, 43, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, C.; Huang, H.; Jiang, Q.; Zhao, D.; Tian, Y.; Ma, J.; Yuan, W.; Sun, Y.; Che, X.; et al. Effects of Alcohol Drinking and Smoking on Pancreatic Ductal Adenocarcinoma Mortality: A Retrospective Cohort Study Consisting of 1783 Patients. Sci. Rep. 2017, 7, 9572. [Google Scholar] [CrossRef] [PubMed]
- Orlhac, F.; Frouin, F.; Nioche, C.; Ayache, N.; Buvat, I. Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiology 2019, 291, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Orlhac, F.; Eertink, J.J.; Cottereau, A.-S.; Zijlstra, J.M.; Thieblemont, C.; Meignan, M.; Boellaard, R.; Buvat, I. A Guide to ComBat Harmonization of Imaging Biomarkers in Multicenter Studies. J. Nucl. Med. 2022, 63, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhou, X.; Zhang, Y.; Sun, X.; Liu, H.; Zhang, Y. Reporting and Methodological Quality of Survival Analysis in Articles Published in Chinese Oncology Journals. Medicine 2017, 96, e9204. [Google Scholar] [CrossRef] [PubMed]
- Lorensen, W.E.; Cline, H.E. Marching Cubes: A High Resolution 3D Surface Construction Algorithm. Comput. Graph. 1987, 21, 163–169. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Sanduleanu, S.; Woodruff, H.C.; de Jong, E.E.C.; van Timmeren, J.E.; Jochems, A.; Dubois, L.; Lambin, P. Tracking Tumor Biology with Radiomics: A Systematic Review Utilizing a Radiomics Quality Score. Radiother. Oncol. 2018, 127, 349–360. [Google Scholar] [CrossRef]


| Datasets 1 and 2 | Dataset 3 | p-Value | |
|---|---|---|---|
| Number of patients | |||
| Total | 173 (60.3%) | 114 (39.7%) | |
| Age (years) | 66 (34–89) | 66.5 (38–89) | 0.65 |
| Gender | 0.77 | ||
| Men | 85 (49.1%) | 54 (47.4%) | |
| Women | 88 (50.9%) | 60 (52.6%) | |
| BMI (kg/m2) | 24.2 (12.84–36) | 24.16 (16.76–39.06) | 0.72 |
| Tobacco history | 0.62 | ||
| No smoking | 79 (45.7%) | 57 (50.0%) | |
| <20 cigarette/day or <15 PY | 34 (19.7%) | 21 (18.4%) | |
| >20 cigarette/day or >15 PY | 27 (15.6%) | 12 (10.5%) | |
| Stopped | 20 (11.6%) | 11 (9.6%) | |
| Alcohol consumption | 0.76 | ||
| No consumption | 70 (40.5%) | 42 (36.8%) | |
| <1 unit/day | 33 (19.1%) | 25 (21.9%) | |
| 1–2 unit/day | 16 (9.2%) | 14 (12.3%) | |
| >3 unit/day | 26 (15.0%) | 14 (12.3%) | |
| Old consumption | 16 (9.2%) | 8 (7.0%) | |
| Ca19.9 (kU/L) | 1100 (0.6–243,000) | 580 (0.6–357,437) | 0.163 |
| Localisation of the lesions | 0.04 | ||
| Head | 77 (44.5%) | 68 (59.6%) | |
| Body | 57 (33.0%) | 26 (22.8%) | |
| Tail | 39 (22.5%) | 20 (17.5%) | |
| Subgroup | 0.01 | ||
| Resectable | 13 (7.5%) | 16 (14.0%) | |
| Borderline Resectable | 30 (17.3%) | 31 (27.2%) | |
| Unresectable | 23 (13.3%) | 6 (5.3%) | |
| Metastatic | 107 (61.8%) | 61 (53.5%) | |
| R_margins | 0.02 | ||
| No surgery | 144 (83.2%) | 79 (69.3%) | |
| Open-closed without resection | 7 (4.0%) | 4 (3.5%) | |
| Resection margin R0 | 10 (5.8%) | 16 (14.0%) | |
| Resection margin R1-R2 | 12 (6.9%) | 15 (13.2%) | |
| Histological grade | 0.35 | ||
| Well differentiated | 11 (6.4%) | 7 (6.1%) | |
| Moderately differentiated | 24 (13.9%) | 30 (26.3%) | |
| Poorly differentiated | 31 (17.9%) | 25 (21.9%) | |
| Treatment type | 0.24 | ||
| FOLFIRINOX | 57 (33.0%) | 46 (40.4%) | |
| Gemcitabine + Abraxane | 85 (49.1%) | 48 (42.1%) | |
| Cisplatin + 5-Fluorouracil | 4 (2.3%) | 4 (3.5%) | |
| Gemcitabine + Cisplatin | 5 (2.9%) | 0 (0.0%) | |
| No treatment | 22 (12.7%) | 16 (14.0%) | |
| Treatment strategy | 0.01 | ||
| Only surgery | 2 (1.2%) | 8 (7.0%) | |
| Surgery + Adjuvant | 10 (5.8%) | 7 (6.1%) | |
| Neoadjuvant | 28 (16.2%) | 29 (25.4%) | |
| Only palliative | 133 (77.0%) | 70 (61.4%) | |
| OS (months) | 9 (0–72) | 10 (0–123) | 0.634 |
| PFS (months) | 3 (0–28) | 3 (0–123) | 0.496 |
| Predicting Models for OS | ||||
|---|---|---|---|---|
| Clinical | GTV1 | RPV | Clinical>V1 | Clinical>V1&RPV |
| Subgroup | original_firstorder_90Percentile_GTV | original_glcm_ClusterShade_RPV | Subgroup | Subgroup |
| CA19 | original_firstorder_Skewness_RPV | CA19 | CA19 | |
| R_Margins | original_shape_Sphericity_RPV | R_Margins | R_Margins | |
| Grading | original_glcm_InverseVariance_RPV | Grading | Grading | |
| Age | original_shape_Flatness_RPV | Age | Age | |
| original_glcm_Contrast_RPV | original_firstorder_90Percentile_GTV | original_glcm_ClusterShade_RPV | ||
| original_shape_LeastAxisLength_RPV | original_firstorder_90Percentile_GTV | |||
| original_firstorder_Skewness_RPV | ||||
| Predicting Models for PFS | ||||
|---|---|---|---|---|
| Clinical | GTV1 | RPV | Clinical>V1 | Clinical>V1&RPV |
| Subgroup | original_glcm_Contrast_GTV | original_glcm_Contrast_RPV | Subgroup | Subgroup |
| CA19 | original_glcm_Correlation_RPV | CA19 | CA19 | |
| R_Margins | R_Margins | original_glcm_Correlation_RPV | ||
| Localization | original_glcm_Contrast_GTV | R_Margins | ||
| Age | original_firstorder_90Percentile_GTV | |||
| Alcohol_quant | original_glcm_Correlation_GTV | |||
| Gender | Localization’ | |||
| Tobacco_quant | original_glcm_Idn_GTV | |||
| Grading | Age | |||
| Models | Training–Validation Set | Testing Set | p-Value |
|---|---|---|---|
| CLINICAL | 0.71 ± 0.07 | 0.7 | - |
| GTV1 | 0.55 ± 0.05 | 0.63 | 0.156 |
| RPV | 0.62 ± 0.06 | 0.56 | 0.001 * |
| CLINICAL>V1 | 0.72 ± 0.08 | 0.72 | 0.212 |
| CLINICAL>V1&RPV | 0.71 ± 0.08 | 0.71 | 0.817 |
| GTV1 volume | 0.5 ± 0.08 | 0.58 | 0.005 * |
| RPV volume | 0.5 ± 0.1 | 0.44 | 0.0002 * |
| Models | Training–Validation Set | Testing Set | p-Value |
|---|---|---|---|
| CLINICAL | 0.67 ± 0.08 | 0.7 | - |
| GTV1 | 0.62 ± 0.08 | 0.51 | 0.004 * |
| RPV | 0.55 ± 0.09 | 0.5 | 0.002 * |
| CLINICAL>V1 | 0.67 ± 0.11 | 0.66 | 0.657 |
| CLINICAL>V1&RPV | 0.63 ± 0.06 | 0.67 | 0.388 |
| GTV1 volume | 0.52 ± 0.08 | 0.51 | 0.001 * |
| RPV volume | 0.48 ± 0.06 | 0.54 | 0.003 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mokhtari, A.; Casale, R.; Salahuddin, Z.; Paquier, Z.; Guiot, T.; Woodruff, H.C.; Lambin, P.; Van Laethem, J.-L.; Hendlisz, A.; Bali, M.A. Development of Clinical Radiomics-Based Models to Predict Survival Outcome in Pancreatic Ductal Adenocarcinoma: A Multicenter Retrospective Study. Diagnostics 2024, 14, 712. https://doi.org/10.3390/diagnostics14070712
Mokhtari A, Casale R, Salahuddin Z, Paquier Z, Guiot T, Woodruff HC, Lambin P, Van Laethem J-L, Hendlisz A, Bali MA. Development of Clinical Radiomics-Based Models to Predict Survival Outcome in Pancreatic Ductal Adenocarcinoma: A Multicenter Retrospective Study. Diagnostics. 2024; 14(7):712. https://doi.org/10.3390/diagnostics14070712
Chicago/Turabian StyleMokhtari, Ayoub, Roberto Casale, Zohaib Salahuddin, Zelda Paquier, Thomas Guiot, Henry C. Woodruff, Philippe Lambin, Jean-Luc Van Laethem, Alain Hendlisz, and Maria Antonietta Bali. 2024. "Development of Clinical Radiomics-Based Models to Predict Survival Outcome in Pancreatic Ductal Adenocarcinoma: A Multicenter Retrospective Study" Diagnostics 14, no. 7: 712. https://doi.org/10.3390/diagnostics14070712
APA StyleMokhtari, A., Casale, R., Salahuddin, Z., Paquier, Z., Guiot, T., Woodruff, H. C., Lambin, P., Van Laethem, J.-L., Hendlisz, A., & Bali, M. A. (2024). Development of Clinical Radiomics-Based Models to Predict Survival Outcome in Pancreatic Ductal Adenocarcinoma: A Multicenter Retrospective Study. Diagnostics, 14(7), 712. https://doi.org/10.3390/diagnostics14070712








