Machine Learning for Enhanced COPD Diagnosis: A Comparative Analysis of Classification Algorithms
Abstract
1. Introduction
- A review of chronic obstructive pulmonary disease diagnosis models is presented.
- Various machine learning techniques are used for the diagnosis of COPD patients.
- The models are evaluated and assessed, highlighting the superior results of certain models.
Up-to-Date Research
2. Materials and Methods
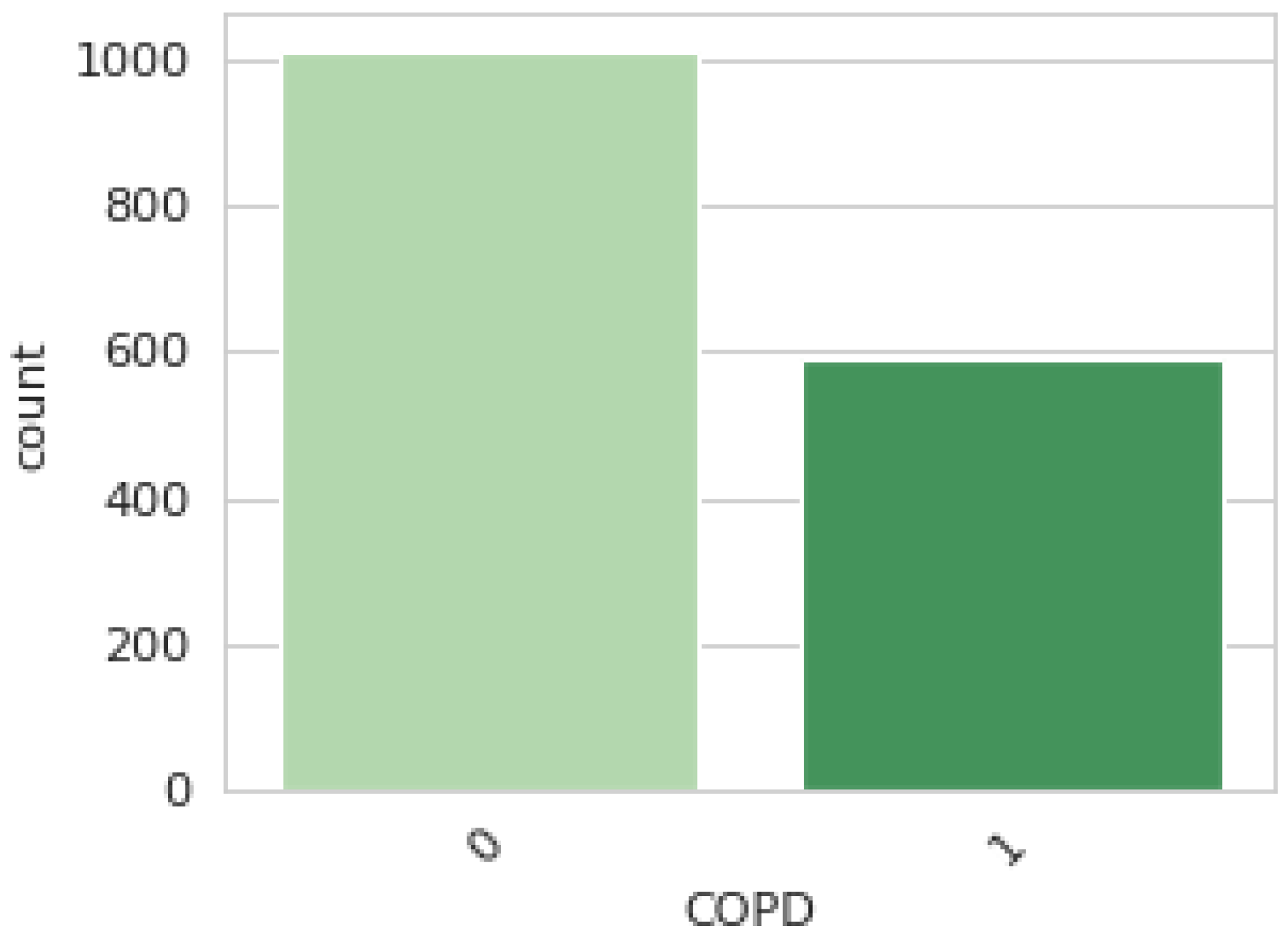
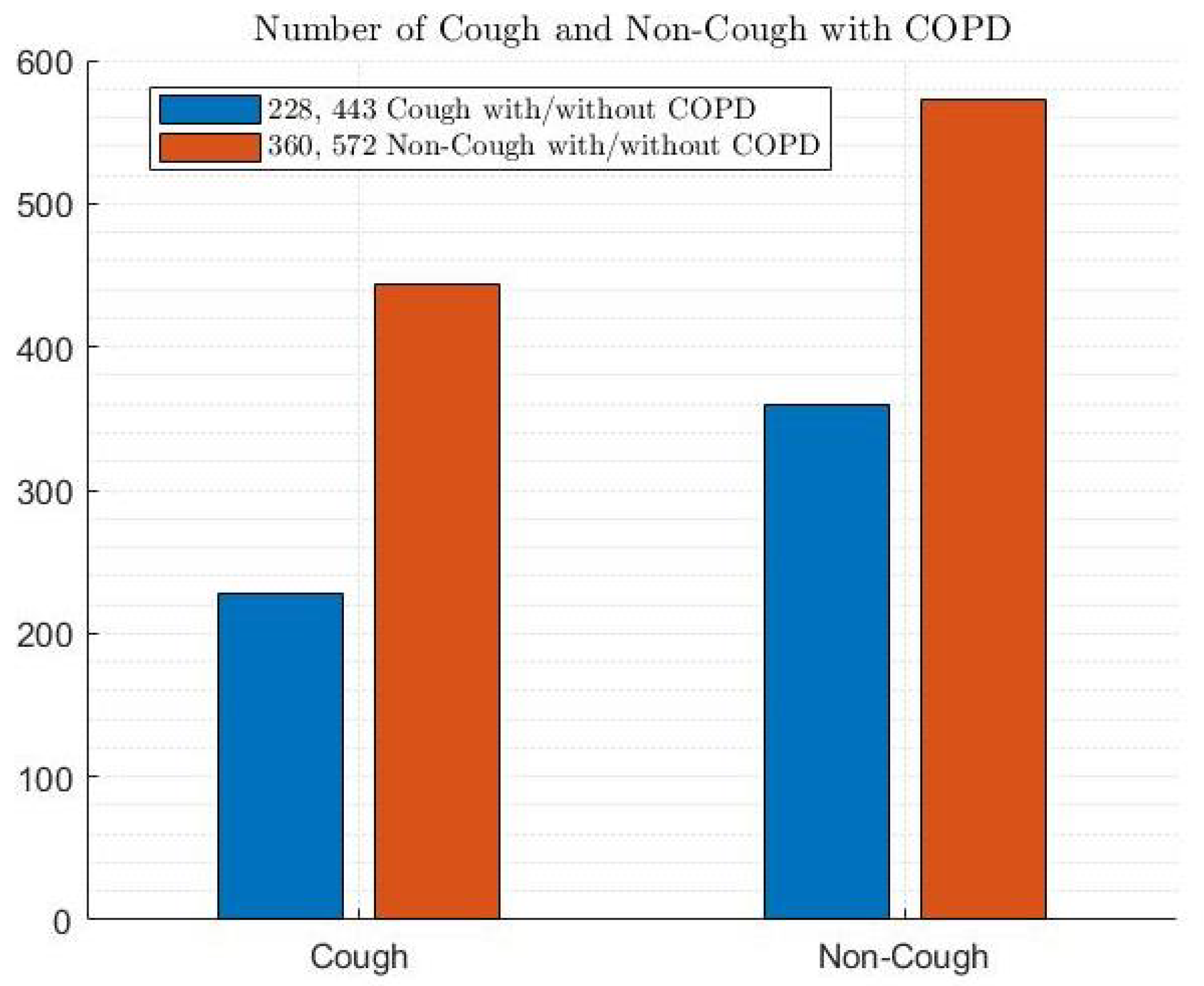
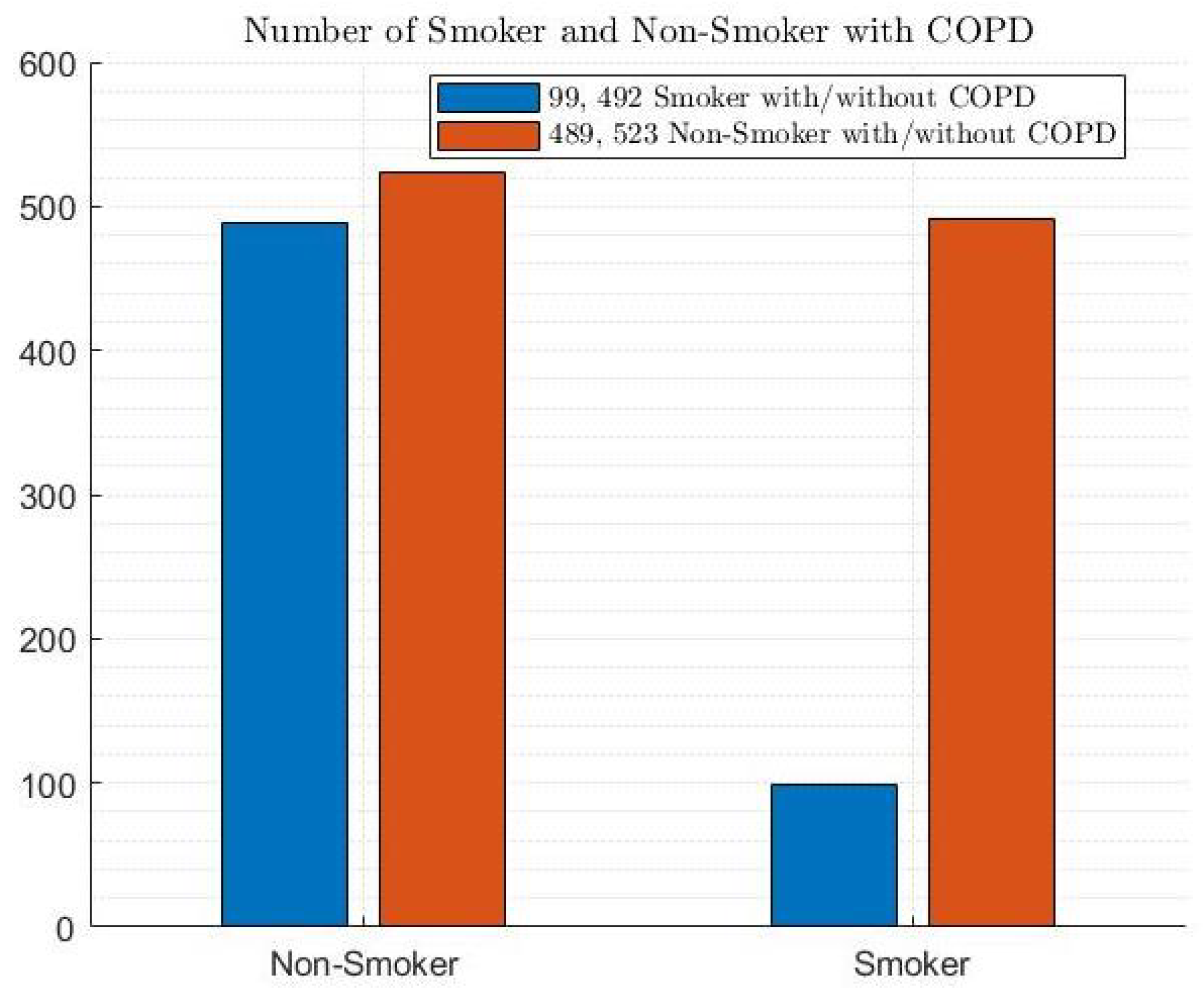
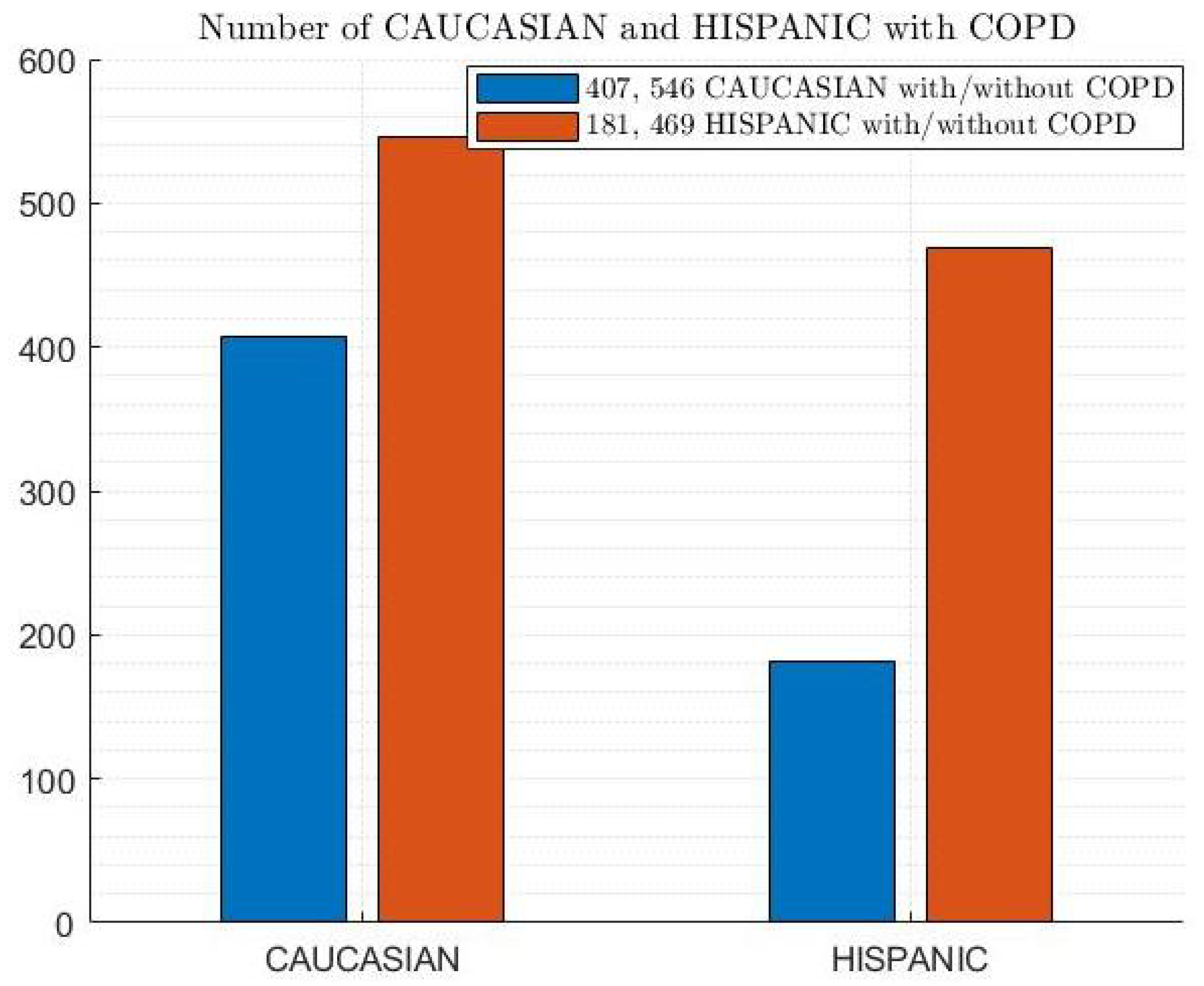
2.1. COPD Dataset
2.2. Machine Learning Models
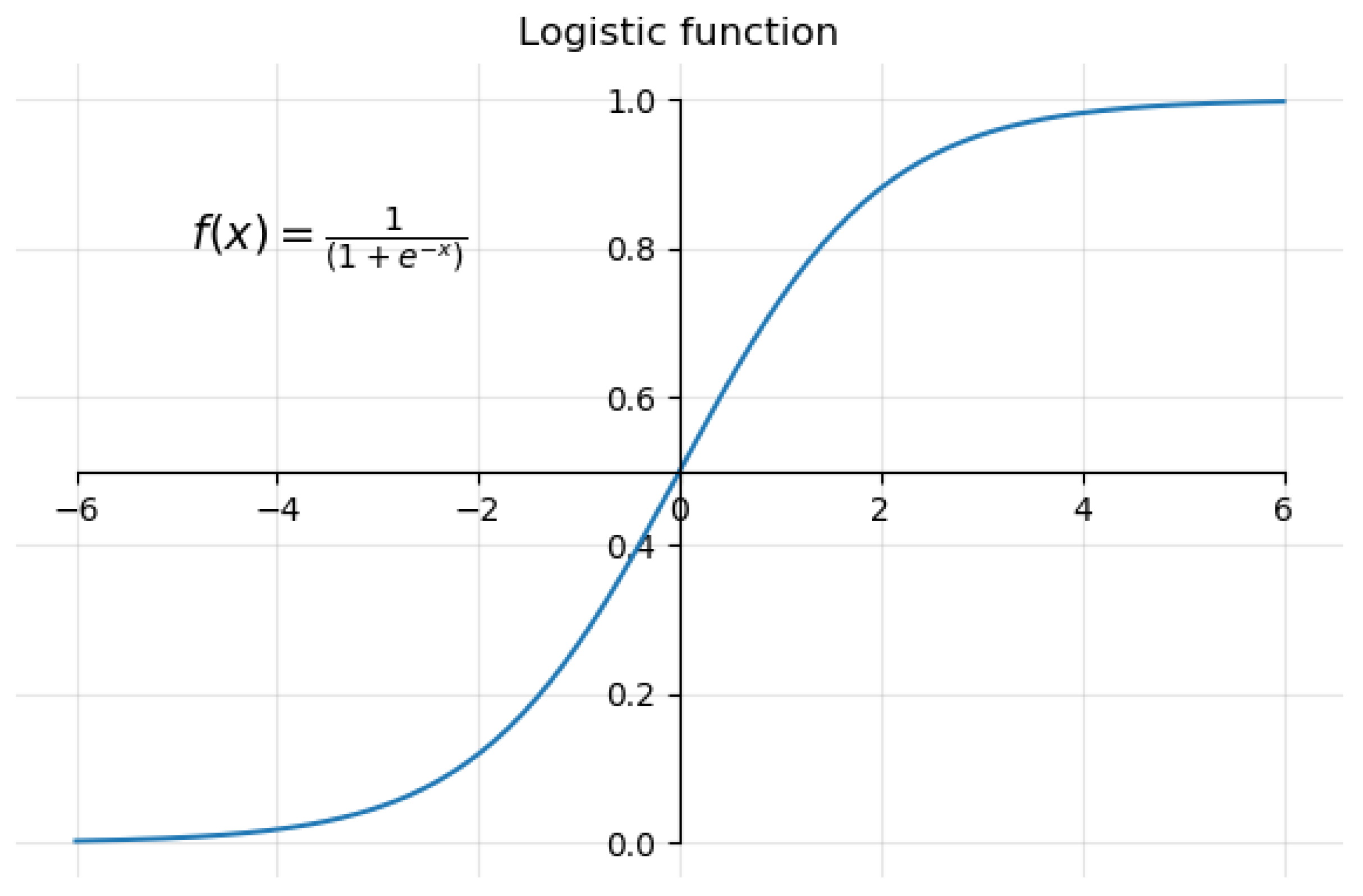
2.2.1. Logistic Regression (LR)
2.2.2. Support Vector Machine (SVM)
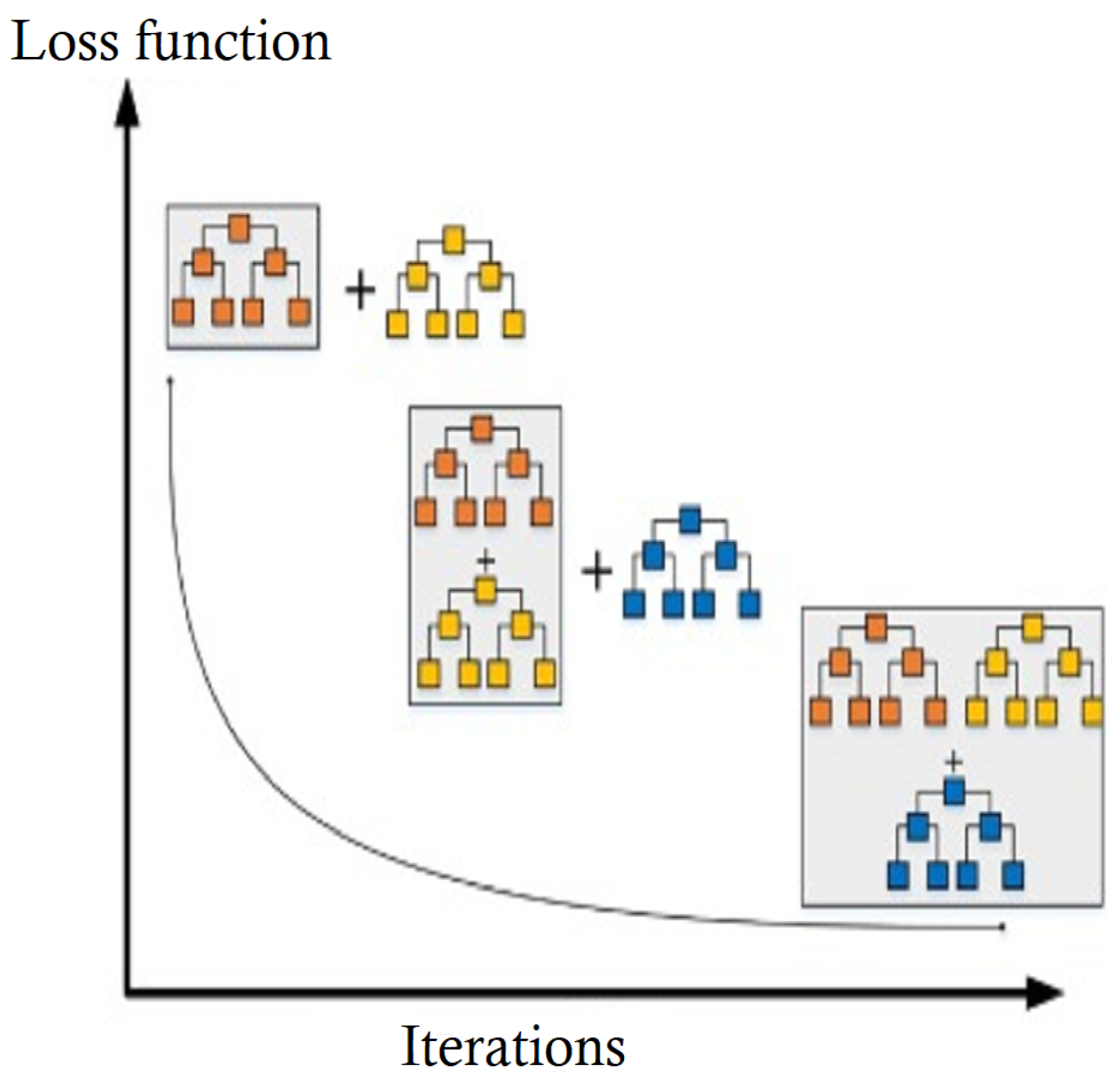
2.2.3. Gradient Boosting Classifier (GBC)
2.2.4. Gaussian Naïve Bayes (GNB)
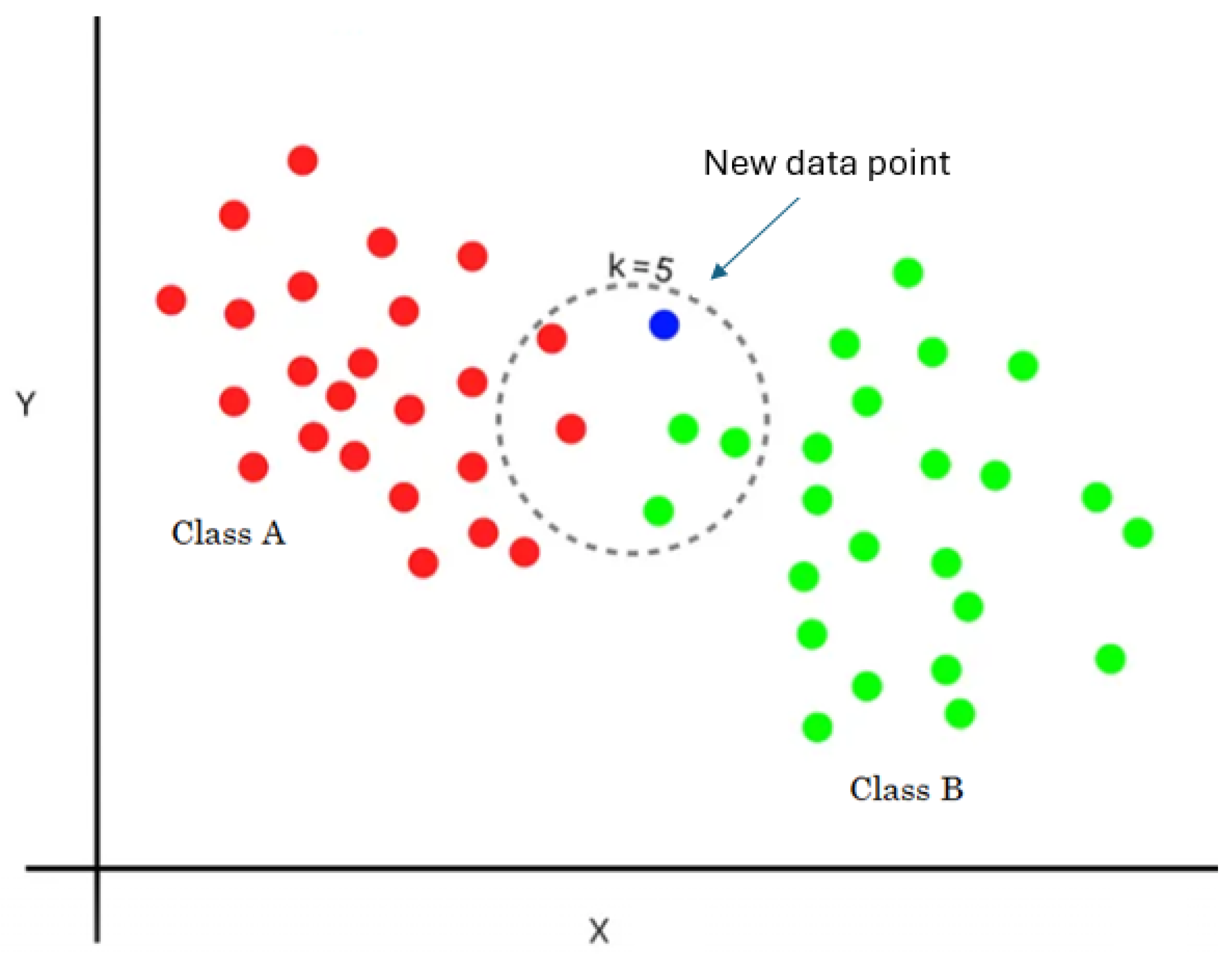
2.2.5. K-Nearest Neighbors Classifier
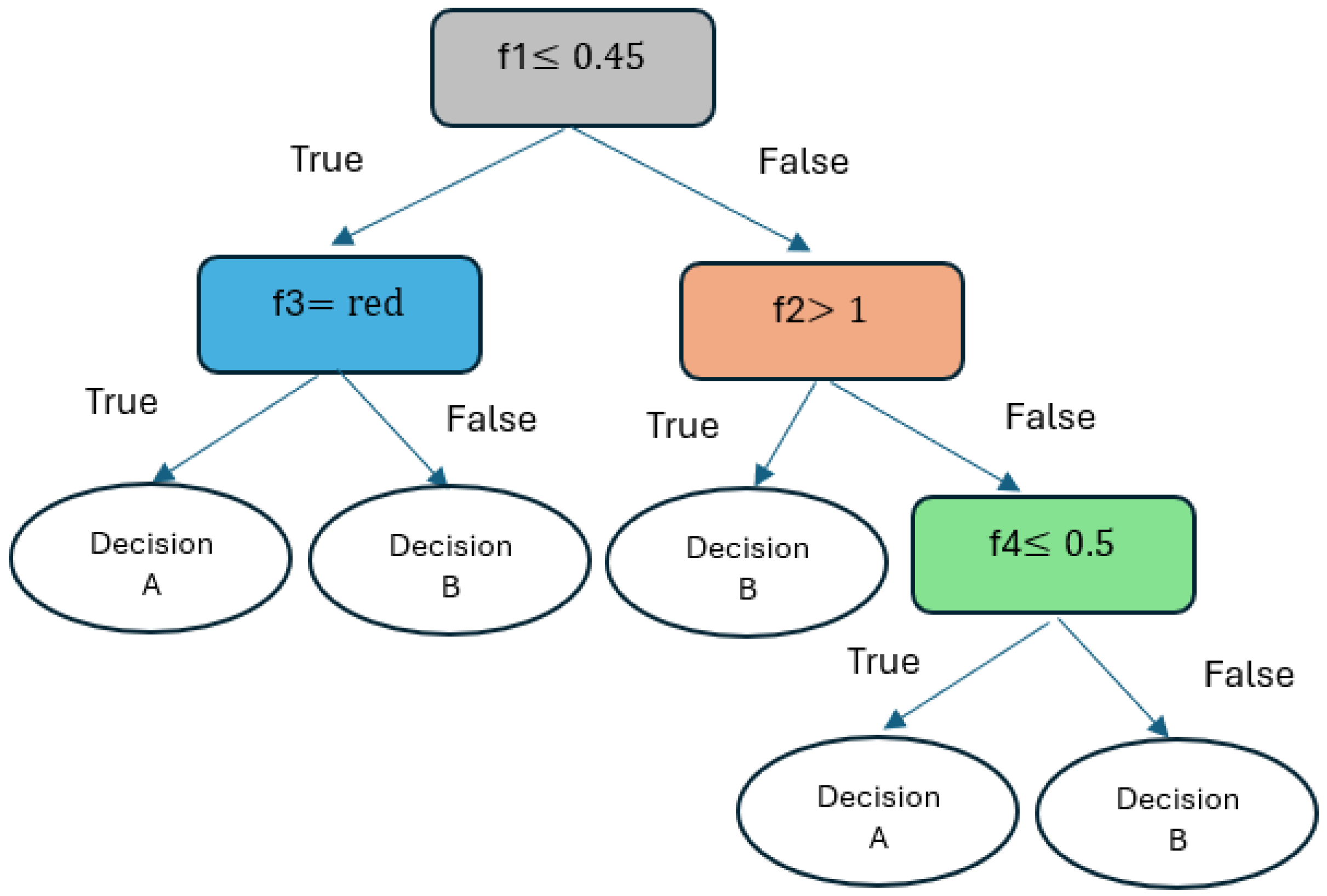
2.2.6. Decision Tree Classifier (DTC)
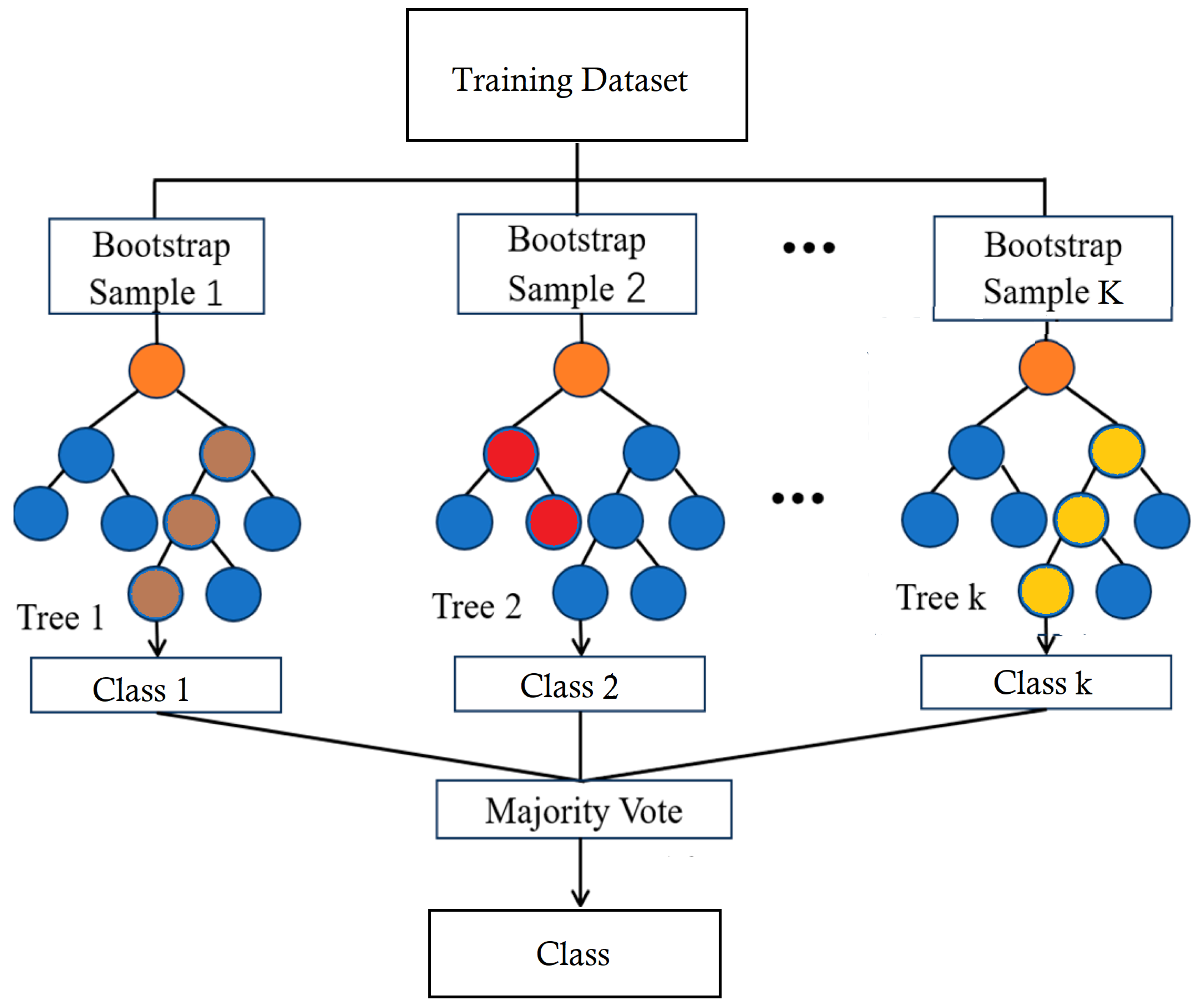
2.2.7. Random Forest Classifier (RFC)
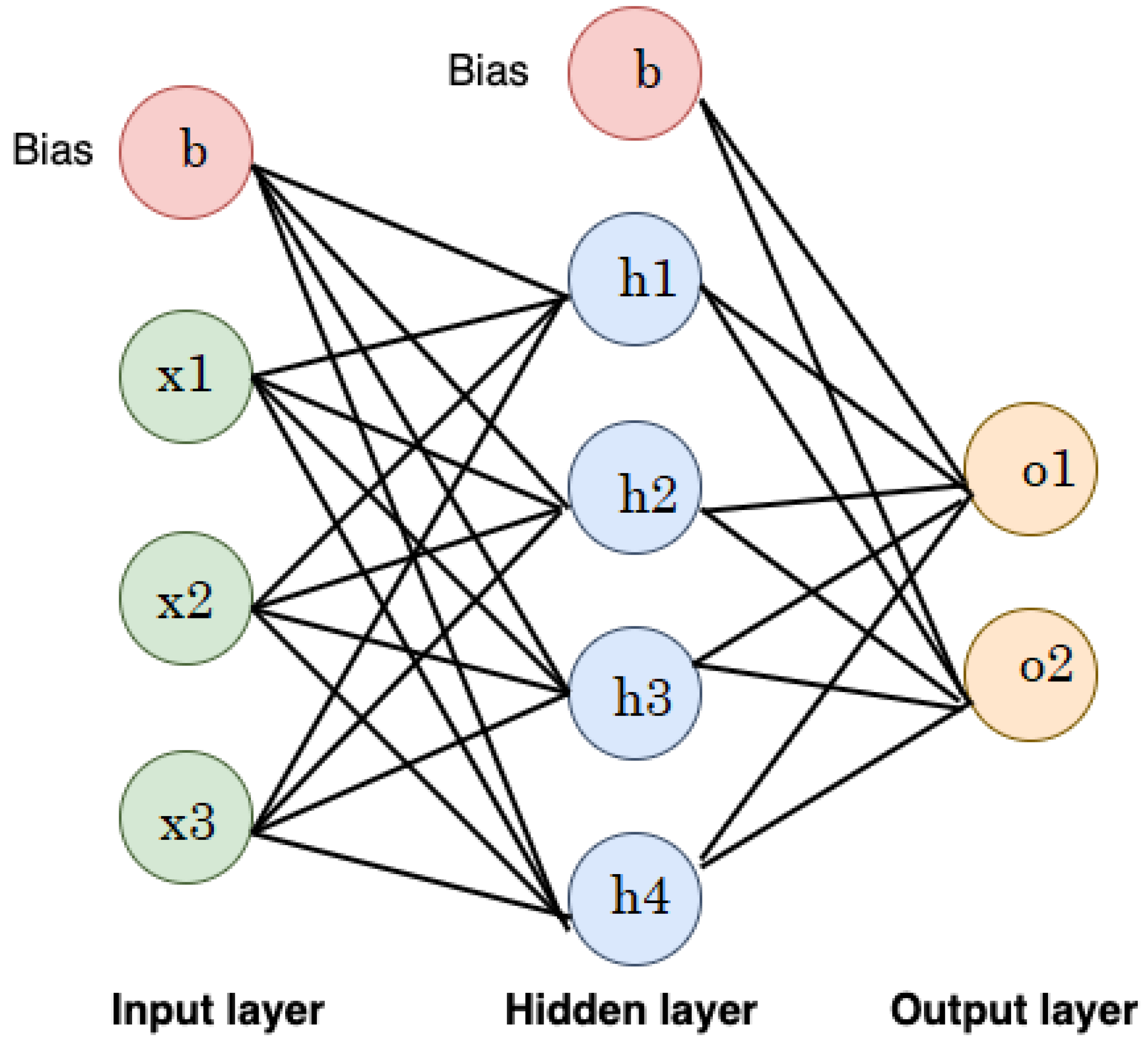
2.2.8. Artificial Neural Networks (ANNs)
2.3. COPD Diagnosis Model and Performance Metrics
- COPD Dataset: This represents the data collection used to train and test the ML models. It likely contains various features extracted from medical records, such as patient demographics, symptoms, and lung function tests.
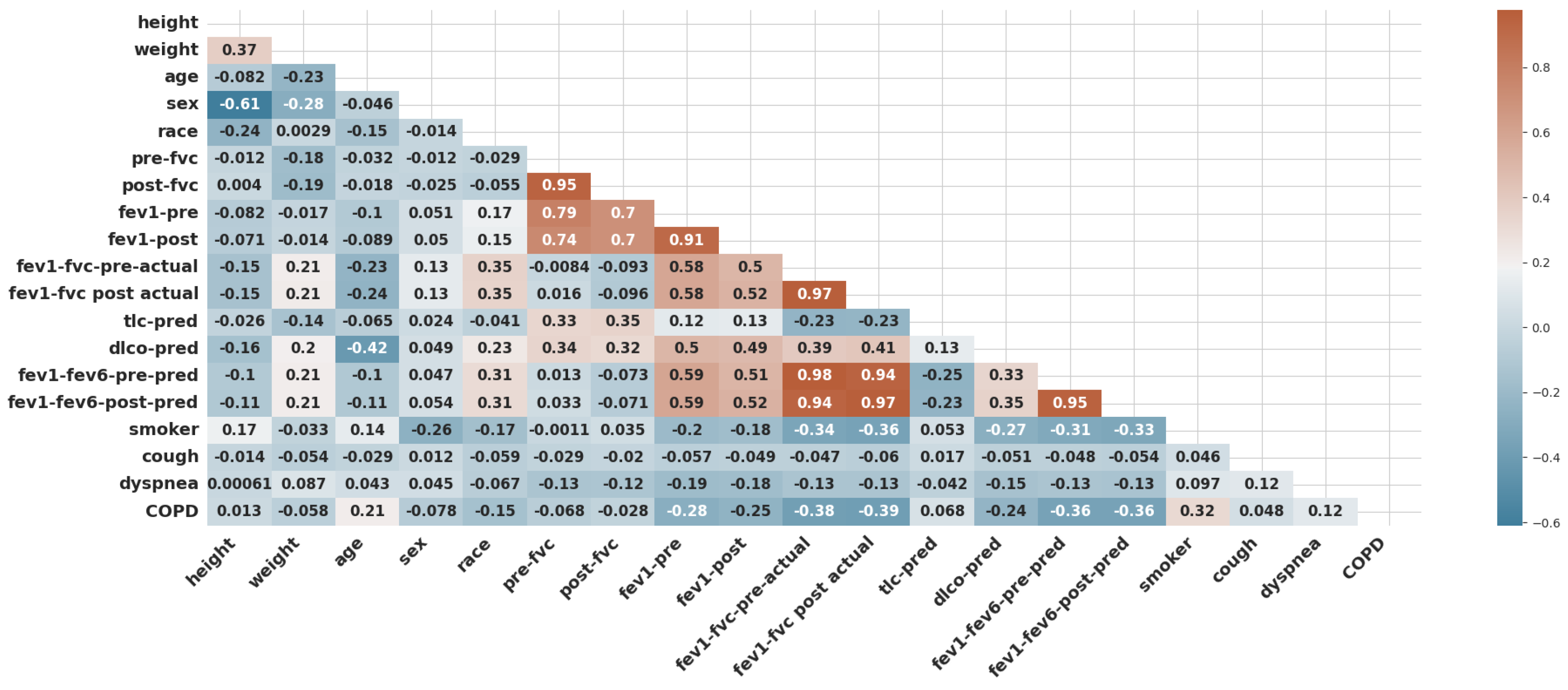
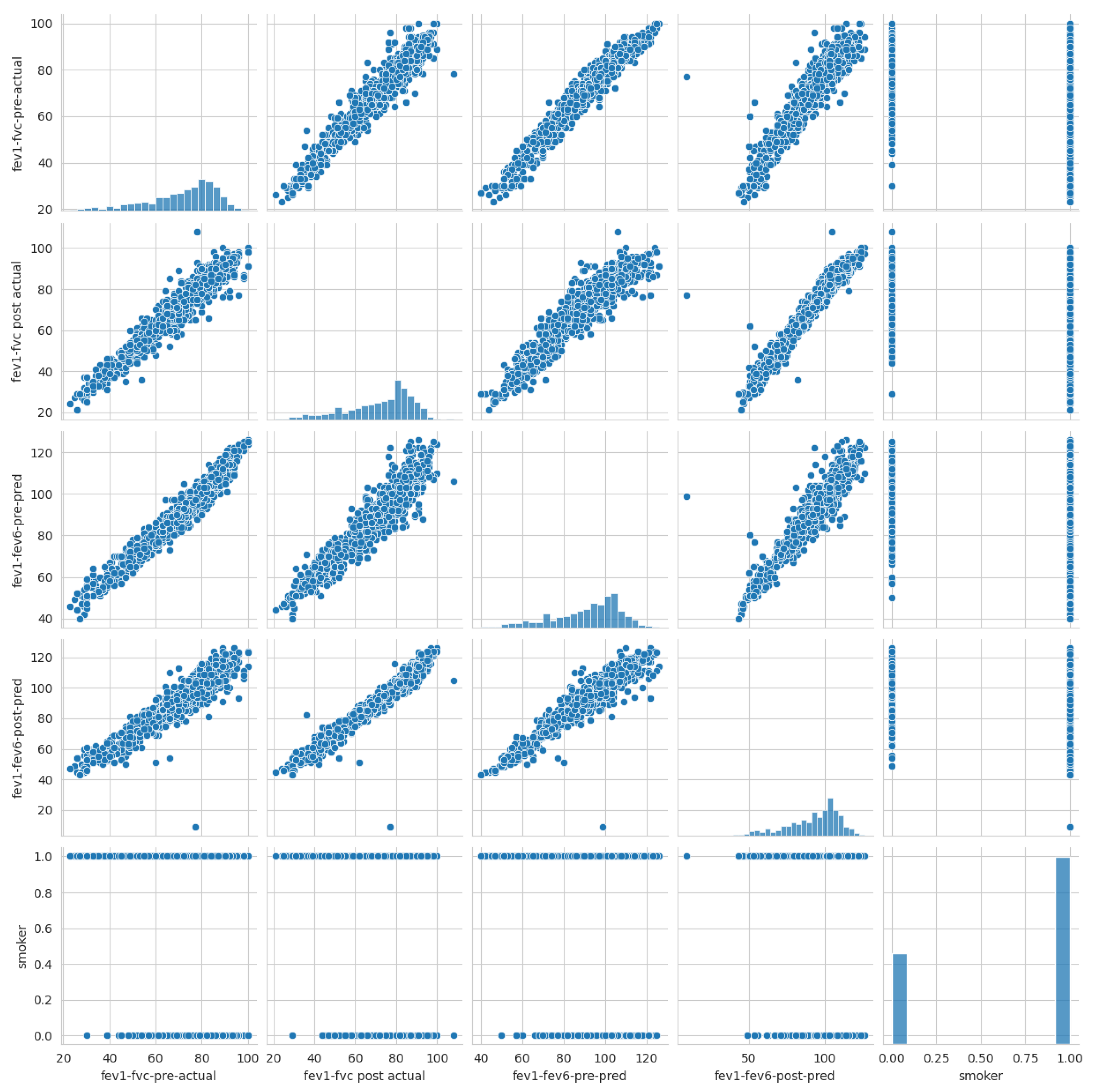
- Data Preprocessing: In this study, data preprocessing involved three key steps to ensure the quality and reliability of the machine learning process. First, one of the simplest and most effective oversampling techniques was applied, which is “RandomOverSampler”, to address class imbalance and improve the representation of minority classes as illustrated in Algorithm 1. As a result of oversampling, the dataset comprised 2030 data points. Second, data scaling was performed using “StandardScaler” to normalize feature values and eliminate biases caused by differing scales. It transforms data by centering around zero (subtracting the mean) and scaling to a standard deviation of one. Finally, feature selection methods were employed to identify and retain the most relevant predictors, reducing model complexity and enhancing efficiency. In this study, five features were selected (fev1-fvc-pre-actual, fev1-fvc post-actual, fev1-fev6-pre-pred, fev1-fev6-post-pred, and smoker) based on correlation coefficients, as shown in Figure 3. The correlation of the selected significant features with the COPD class label is shown in Figure 17. These preprocessing steps were crucial in preparing the dataset for robust analysis and ensuring model performance.
| Algorithm 1: Balancing the Dataset Using Oversampling |
| 1: Input: Dataset D with majority class and minority class 2: Output: Balanced dataset 3: Identify the minority class and majority class 4: While Size() < Size() do 5: Randomly select samples from 6: Duplicate the selected samples 7: Add the duplicated samples to 8: End While 9: Combine and to form 10: Return |
- Data Split: The dataset utilized in this study is divided into two segments: training (75%) and testing (25%). A total 1522 data points were allocated to the training set, and 508 were designated for testing. The training set teaches the ML models to recognize patterns and relationships within the data, while the testing set is used to evaluate the models’ performance on unseen data.
- ML Classifiers: Various ML algorithms were employed, including LR, SVM, GBC, NBC, DT, RFC, KNC, and ANN.
- Trained Models: Each ML classifier was trained on the training dataset, resulting in a set of trained models. These models learned to differentiate between patients with COPD and those without.
- Classification Output: The trained models were applied to the testing dataset to predict whether each patient had COPD or was healthy. The output is a classification label for each patient.
- Accuracy (Acc) refers to the ability to correctly predict whether a patient has the disease. It is calculated as the proportion of correct predictions (both true positives and true negatives, which are identified as the total of +COPD and +Healthy) out of the total number of predictions (i.e., (+COPD) + (-COPD) + (+Healthy) + (-Healthy)).
- Precision (Pre) indicates how often the model is correct when it predicts a positive outcome.
- Recall (Rec) shows how sensitive the model is to positive instances.
- The F score (Fscore) reflects a model’s ability to both accurately identify positive cases and avoid false positives.
3. Experimental Results
4. Discussion and Analysis of the Results
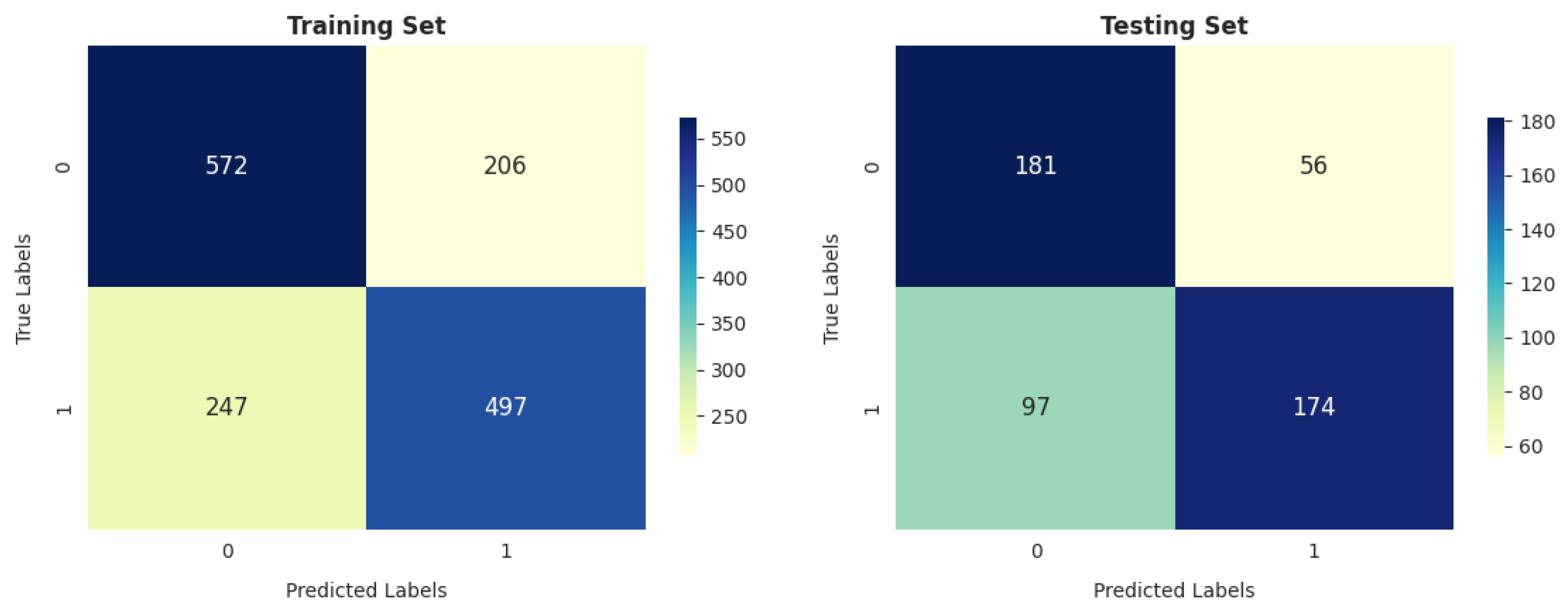
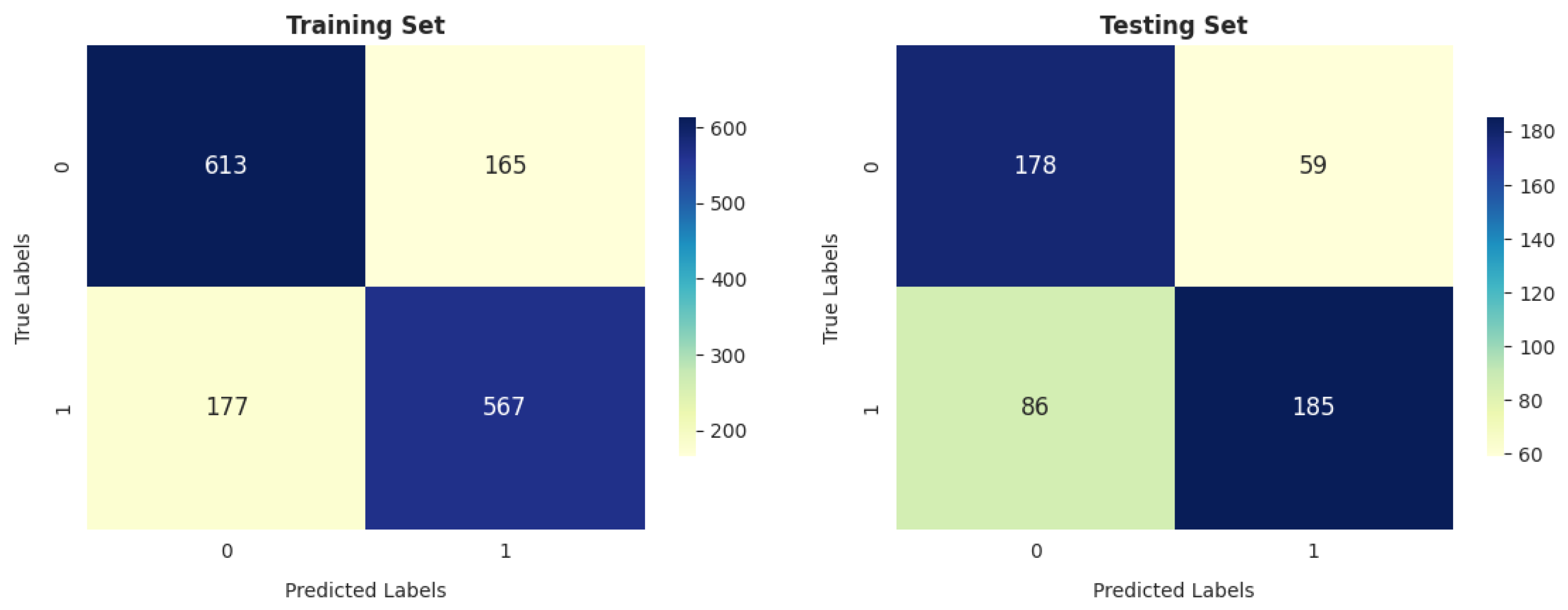
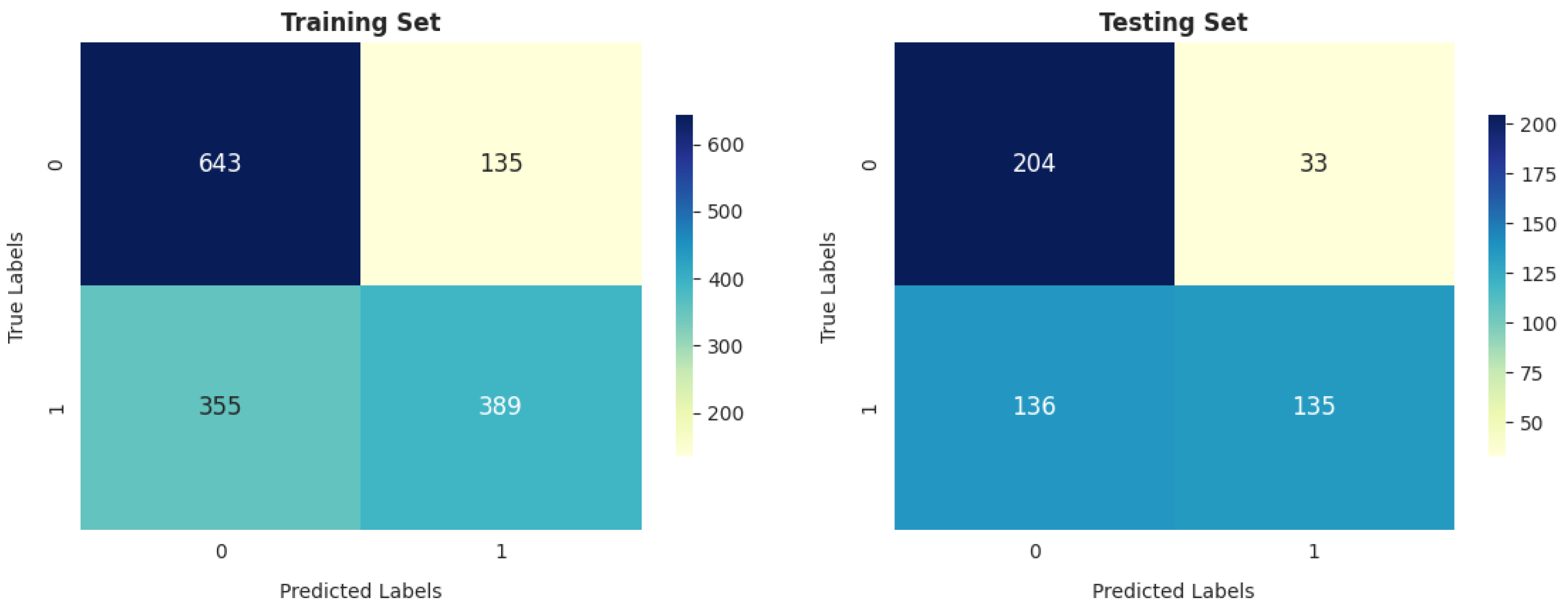
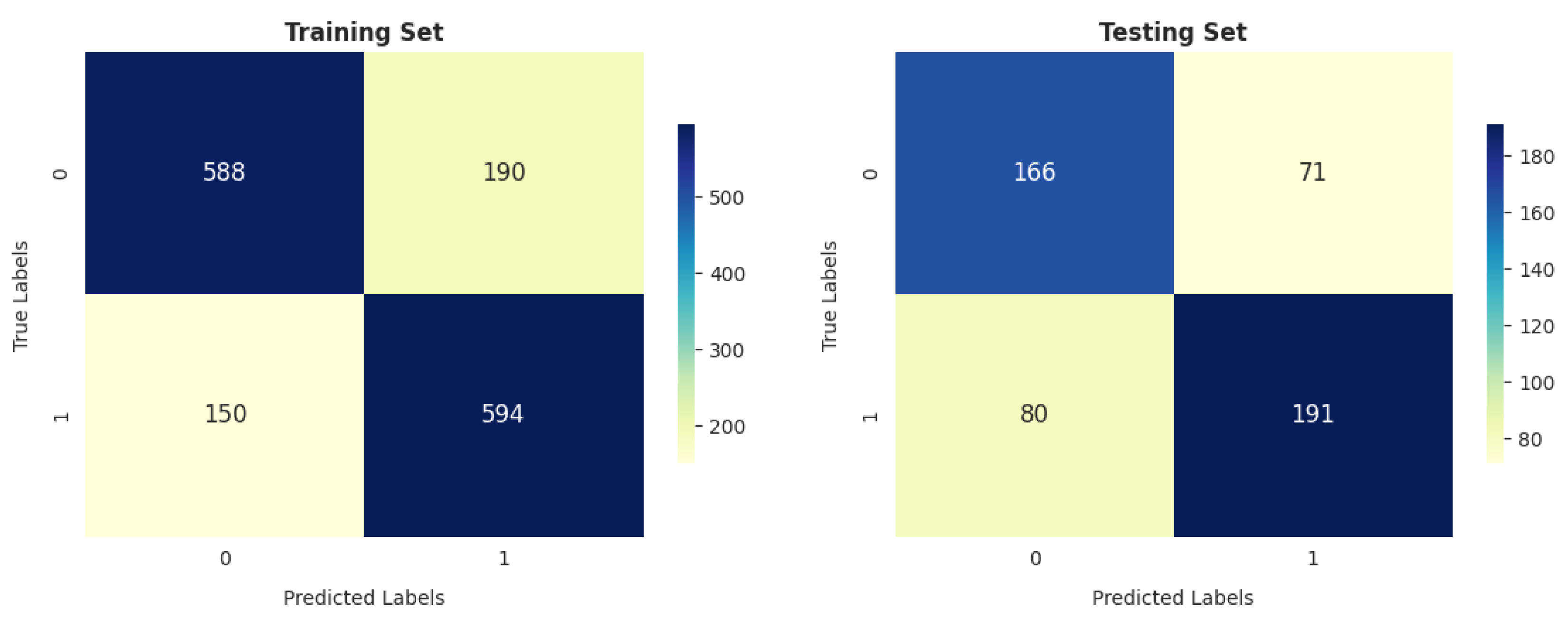
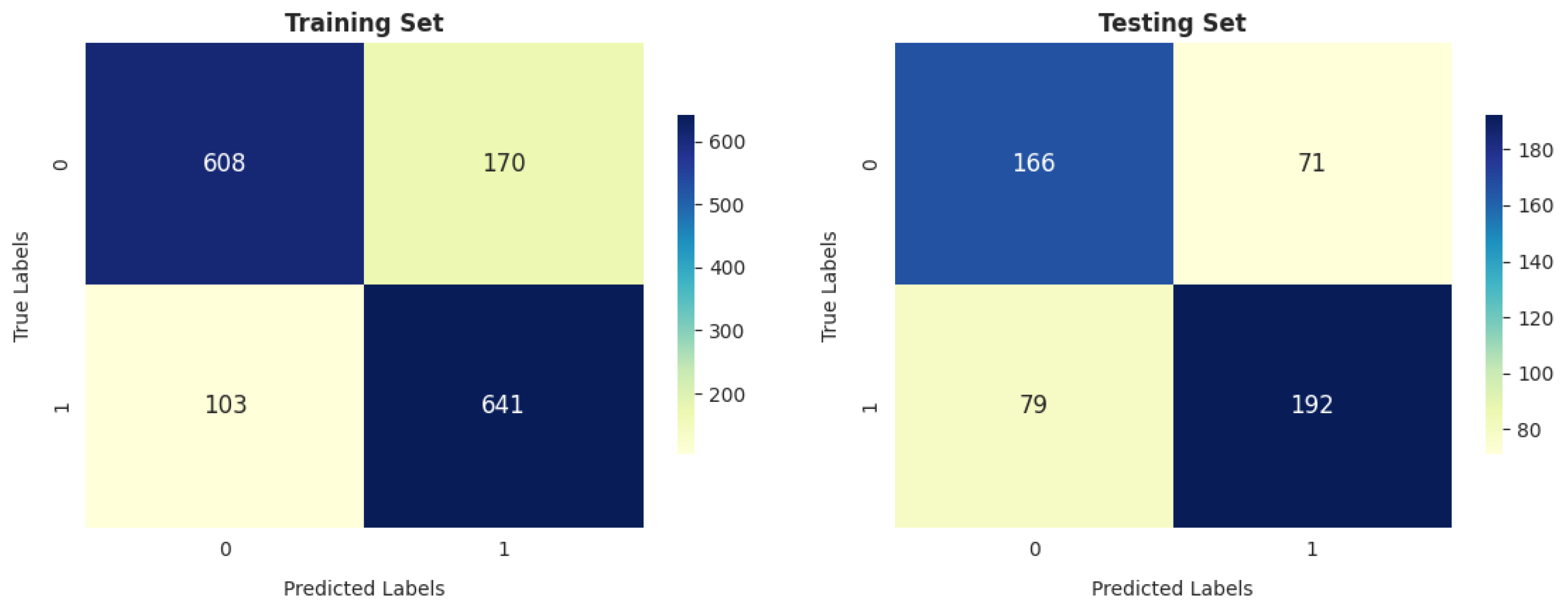
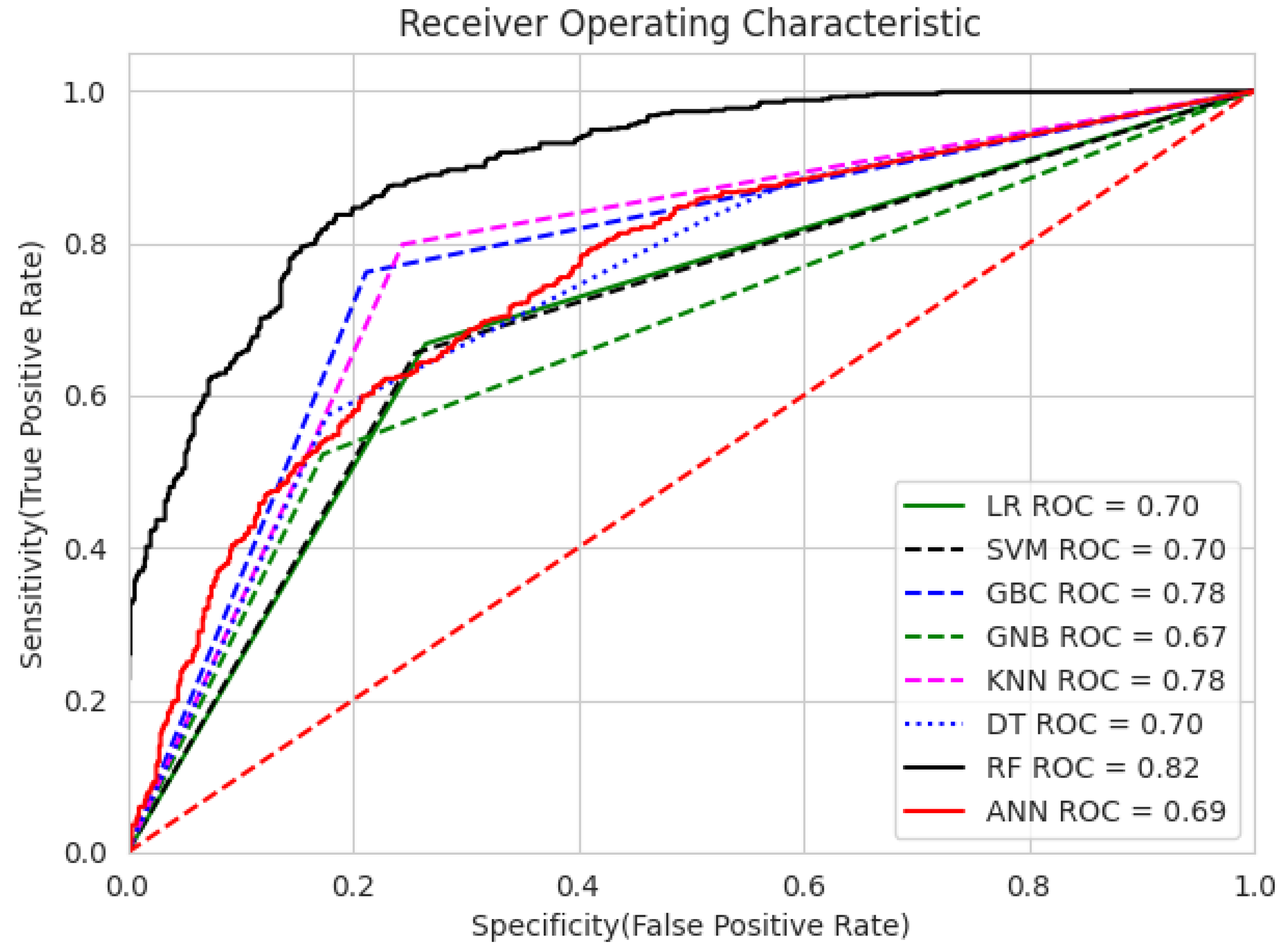
- Our analysis of 1603 patient records revealed that the random forest classifier (RFC) outperformed other models, achieving the highest accuracy, reaching up to 82.06%; precision; recall; and F1 score on the training dataset. It obtained a perfect score and achieved a good balance of false positives and false negatives. This is consistent with the findings of previous research [16], which analyzed 6883 patients from the United Kingdom and the random forest model was found to outperform the others, achieving an impressive accuracy rate of up to 92%. Furthermore, its performance on the testing dataset remained entirely satisfactory, as it achieved an accuracy of 70.47% and the highest F score of 71.91%.
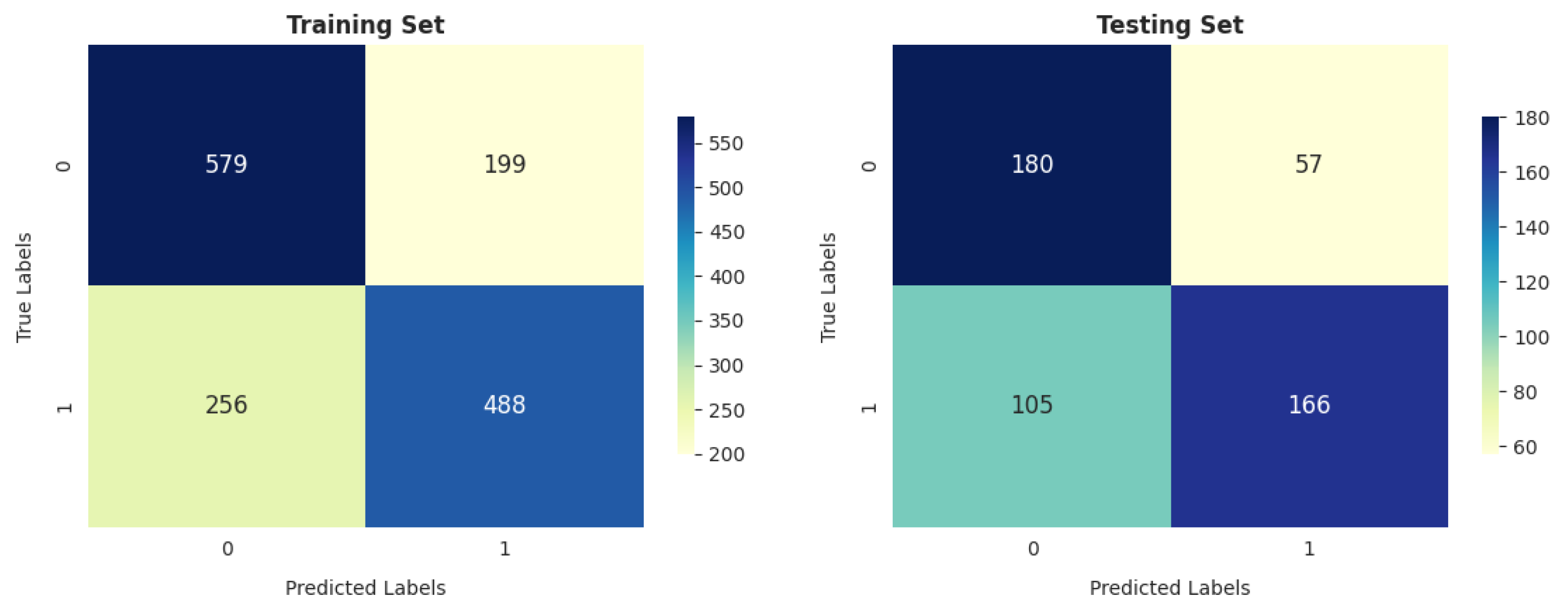
- The gradient boosting classifier (GBC) also demonstrates strong performance on the training dataset, achieving 77.53%, 77.46%, 76.76%, and 76.83% scores for accuracy, precision, recall, and F score, respectively. When tested on the validation dataset, it maintains solid performance, with an accuracy of 71.46%. This accuracy surpasses that of other models.
- The decision tree classifier (DTC) and support vector machine (SVM) exhibit a reasonable accuracy of 70.11% on the training dataset. However, the performance of DT on the testing dataset is somewhat lower, at 67.91%.
- The K-nearest neighbors classifier (KNC) shows strong training results, with an accuracy of 77.66%, precision of 75.77%, recall of 79.84%, and F score of 77.75%. The high recall suggests the model is sensitive to the positive class. While there is a slight drop in testing accuracy compared to training, the model still performs well in generalization.
- Logistic regression (LR) demonstrates decent performance, with a training accuracy of 70.24% and testing accuracy of 69.88%, indicating its ability to generalize reasonably well. It shows good precision on both training (70.70%) and testing (75.65%) datasets, suggesting that it is reliable in avoiding false positives. However, its recall is relatively lower, with 66.80% on the training set and 64.21% on the test set, meaning it misses a significant portion of positive cases. This results in an F score of 68.69% for training and 69.46% for testing, reflecting a moderate balance between precision and recall. While LR is a solid performer overall, it falls short of more complex models like RFC and KNC regarding recall and overall performance.
- Meng et al. [18] employed five common machine learning algorithms (naïve Bayes, KNC, DT, RF, and LR), along with several nonlinear predictors, to characterize PM2.5 pollution-sensitive COPD patients (327 patients), and their results showed that all classifiers yielded similar AUC values. The sensitivities of naïve Bayes, KNC, DT, RF, and LR were 61%, 50%, 46%, 48%, and 39%, respectively. Among these, naïve Bayes outperformed the other algorithms, achieving the highest AUC value of 0.7673. In contrast, our results reveal that the RFC achieved the highest AUC value of 0.82, and the sensitivities of RF, KNC, DT, and LR were 70.85%, 70.48%, 54.61%, and 64.21%, respectively, on the testing data.
- GNB achieves 74.24% precision, which is strong, but its recall is significantly lower (52.28%), leading to a relatively low F score of 61.36% on training data. On the test data, GNB’s performance is even weaker, with 66.73% accuracy and 80.36% precision, as well as a much lower recall of 49.82%. This makes it less suitable compared to the models with more well-balanced performance, such as the RFC and KNC.
- The artificial neural network (ANN) shows moderate performance across both training and testing datasets. Its recall value dropped from 64.92% (in training) to 64.92% (in testing), indicating that it misses a considerable number of positive instances on the test data. The authors of [26] reported that a multilayer ANN achieved better results than SVM when considering 695 patient records.
5. Statistical Test
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Syamlal, G.; Doney, B.; Hendricks, S.; Mazurek, J.M. Chronic Obstructive Pulmonary Disease and U.S. Workers: Prevalence, Trends, and Attributable Cases Associated with Work. Am. J. Prev. Med. 2021, 61, e127–e137. [Google Scholar] [CrossRef]
- Guarascio, A.J.; Ray, S.M.; Finch, C.K.; Self, T.H. The clinical and economic burden of chronic obstructive pulmonary disease in the USA. Clin. Outcomes Res. 2013, 17, 235–245. [Google Scholar]
- Chen, S.; Kuhn, M.; Prettner, K.; Yu, F.; Yang, T.; Bärnighausen, T.; Bloom, D.E.; Wang, C. The global economic burden of chronic obstructive pulmonary disease for 204 countries and territories in 2020–50: A health-augmented macroeconomic modelling study. Lancet Glob. Health 2023, 11, e1183–e1193. [Google Scholar] [CrossRef]
- Ford, E.S.; Mannino, D.M.; Wheaton, A.G.; Giles, W.H.; Presley-Cantrell, L.; Croft, J.B. Trends in the prevalence of obstructive and restrictive lung function among adults in the United States: Findings from the National Health and Nutrition Examination surveys from 1988–1994 to 2007–2010. Chest 2013, 143, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Mannino, D.M.; Gagnon, R.C.; Petty, T.L.; Lydick, E. Obstructive lung disease and low lung function in adults in the United States: Data from the National Health and Nutrition Examination Survey, 1988–1994. Arch. Intern. Med. 2000, 160, 1683–1689. [Google Scholar] [CrossRef]
- Martinez, F.J.; Han, M.K.; Allinson, J.P.; Barr, R.G.; Boucher, R.C.; Calverley, P.M.; Celli, B.R.; Christenson, S.A.; Crystal, R.G.; Fagerås, M.; et al. At the root: Defining and halting progression of early chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2018, 197, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.J.; Agusti, A.; Celli, B.R.; Han, M.K.; Allinson, J.P.; Bhatt, S.P.; Calverley, P.; Chotirmall, S.H.; Chowdhury, B.; Darken, P.; et al. Treatment trials in young patients with chronic obstructive pulmonary disease and pre–chronic obstructive pulmonary disease patients: Time to move forward. Am. J. Respir. Crit. Care Med. 2022, 205, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.P.; Kim, Y.i.; Wells, J.M.; Bailey, W.C.; Ramsdell, J.W.; Foreman, M.G.; Jensen, R.L.; Stinson, D.S.; Wilson, C.G.; Lynch, D.A.; et al. FEV1/FEV6 to diagnose airflow obstruction. Comparisons with computed tomography and morbidity indices. Ann. Am. Thorac. Soc. 2014, 11, 335–341. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Dwivedi, D.P.; Govindraj, V. FEV1/FEV6 is effective as a surrogate for FEV1/FVC in the diagnosis of chronic obstructive pulmonary disease. Indian J. Tuberc. 2021, 68, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Rosa, F.W.; Perez-Padilla, R.; Camelier, A.; Nascimento, O.A.; Menezes, A.M.B.; Jardim, J.R. Efficacy of the FEV1/FEV6 ratio compared to the FEV1/FVC ratio for the diagnosis of airway obstruction in subjects aged 40 years or over. Braz. J. Med Biol. Res. 2007, 40, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, M.; Cataletto, M. Asthma. In StatPearls [Internet]; StatPearls: Treasure Island, FL, USA, 2024. [Google Scholar]
- Amaral, J.L.; Lopes, A.J.; Veiga, J.; Faria, A.C.; Melo, P.L. High-accuracy detection of airway obstruction in asthma using machine learning algorithms and forced oscillation measurements. Comput. Methods Programs Biomed. 2017, 144, 113–125. [Google Scholar] [CrossRef]
- Darabi, H.R.; Tsinis, D.; Zecchini, K.; Whitcomb, W.F.; Liss, A. Forecasting mortality risk for patients admitted to intensive care units using machine learning. Procedia Comput. Sci. 2018, 140, 306–313. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Courteau, J.; Chen, L.; Guo, G.; Vanasse, A. Feature-weighted survival learning machine for COPD failure prediction. Artif. Intell. Med. 2019, 96, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Moll, M.; Qiao, D.; Regan, E.A.; Hunninghake, G.M.; Make, B.J.; Tal-Singer, R.; McGeachie, M.J.; Castaldi, P.J.; Estepar, R.S.J.; Washko, G.R.; et al. Machine learning and prediction of all-cause mortality in COPD. Chest 2020, 158, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, V.; Massaro, S.; Garn, W.; Fakhimi, M.; Stergioulas, L.; Price, D. The cardiovascular phenotype of Chronic Obstructive Pulmonary Disease (COPD): Applying machine learning to the prediction of cardiovascular comorbidities. Respir. Med. 2021, 186, 106528. [Google Scholar] [CrossRef] [PubMed]
- Muro, S.; Ishida, M.; Horie, Y.; Takeuchi, W.; Nakagawa, S.; Ban, H.; Nakagawa, T.; Kitamura, T. Machine Learning Methods for the Diagnosis of Chronic Obstructive Pulmonary Disease in Healthy Subjects: Retrospective Observational Cohort Study. JMIR Med. Inf. 2021, 9, e24796. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Wang, J.; Cui, J.; Li, B.; Wu, S.; Yun, J.; Aschner, M.; Wang, C.; Zhang, L.; Li, X.; et al. Prediction of COPD acute exacerbation in response to air pollution using exosomal circRNA profile and Machine learning. Environ. Int. 2022, 168, 107469. [Google Scholar] [CrossRef] [PubMed]
- Zafari, H.; Langlois, S.; Zulkernine, F.; Kosowan, L.; Singer, A. AI in predicting COPD in the Canadian population. Biosystems 2022, 211, 104585. [Google Scholar] [CrossRef]
- Wang, X.; Ren, H.; Ren, J.; Song, W.; Qiao, Y.; Ren, Z.; Zhao, Y.; Linghu, L.; Cui, Y.; Zhao, Z.; et al. Machine learning-enabled risk prediction of chronic obstructive pulmonary disease with unbalanced data. Comput. Methods Programs Biomed. 2023, 230, 107340. [Google Scholar] [CrossRef]
- Kocks, J.W.; Cao, H.; Holzhauer, B.; Kaplan, A.; FitzGerald, J.M.; Kostikas, K.; Price, D.; Reddel, H.K.; Tsiligianni, I.; Vogelmeier, C.F.; et al. Diagnostic Performance of a Machine Learning Algorithm (Asthma/Chronic Obstructive Pulmonary Disease [COPD] Differentiation Classification) Tool Versus Primary Care Physicians and Pulmonologists in Asthma, COPD, and Asthma/COPD Overlap. J. Allergy Clin. Immunol. Pract. 2023, 11, 1463–1474. [Google Scholar] [CrossRef]
- Smith, L.A.; Oakden-Rayner, L.; Bird, A.; Zeng, M.; To, M.S.; Mukherjee, S.; Palmer, L.J. Machine learning and deep learning predictive models for long-term prognosis in patients with chronic obstructive pulmonary disease: A systematic review and meta-analysis. Lancet Digit. Health 2023, 5, e872–e881. [Google Scholar] [CrossRef]
- Bian, H.; Zhu, S.; Zhang, Y.; Fei, Q.; Peng, X.; Jin, Z.; Zhou, T.; Zhao, H. Artificial Intelligence in Chronic Obstructive Pulmonary Disease: Research Status, Trends, and Future Directions–A Bibliometric Analysis from 2009 to 2023. Int. J. Chronic Obstr. Pulm. Dis. 2024, 19, 1849–1864. [Google Scholar] [CrossRef]
- Shen, X.; Liu, H. Using machine learning for early detection of chronic obstructive pulmonary disease: A narrative review. Respir. Res. 2024, 25, 336. [Google Scholar] [CrossRef]
- Xu, Y.; Long, Z.A.; Setyohadi, D.B. A Comprehensive Review on the Application of Artificial Intelligence in Chronic Obstructive Pulmonary Disease (COPD) Management. In Proceedings of the 2024 18th International Conference on Ubiquitous Information Management and Communication (IMCOM), Kuala Lumpur, Malaysia, 3–5 January 2024; pp. 1–8. [Google Scholar] [CrossRef]
- Maldonado-Franco, A.; Giraldo-Cadavid, L.; Tuta-Quintero, E.; Cagy, M.; Bastidas Goyes, A.; Botero-Rosas, D. Curve-Modelling and Machine Learning for a Better COPD Diagnosis. Int. J. Chronic Obstr. Pulm. Dis. 2024, 19, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, Y.; Yang, D.; Zhang, W.; Kouritas, V.; Chen, X. Developing and validating machine learning-based prediction models for frailty occurrence in those with chronic obstructive pulmonary disease. J. Thorac. Dis. 2024, 16, 2482. [Google Scholar] [CrossRef]
- Culver, B.H.; Graham, B.L.; Coates, A.L.; Wanger, J.; Berry, C.E.; Clarke, P.K.; Hallstrand, T.S.; Hankinson, J.L.; Kaminsky, D.A.; MacIntyre, N.R.; et al. Recommendations for a Standardized Pulmonary Function Report. An Official American Thoracic Society Technical Statement. Am. J. Respir. Crit. Care Med. 2017, 196, 1463–1472. [Google Scholar] [CrossRef]
- Sheta, A.; El-Ashmawi, W.; Baareh, A. Heart Disease Diagnosis Using Decision Trees with Feature Selection Method. Int. Arab. J. Inf. Technol. (IAJIT) 2024, 21, 427–438. [Google Scholar] [CrossRef]
- Sheta, A.; Elashmawi, W.H.; Al-Qerem, A.; Othman, E.S. Utilizing Various Machine Learning Techniques for Diabetes Mellitus Feature Selection and Classification. Int. J. Adv. Comput. Sci. Appl. 2024, 15, 1372. [Google Scholar] [CrossRef]
- Sheta, A.; Turabieh, H.; Thaher, T.; Too, J.; Mafarja, M.; Hossain, M.S.; Surani, S.R. Diagnosis of Obstructive Sleep Apnea from ECG Signals Using Machine Learning and Deep Learning Classifiers. Appl. Sci. 2021, 11, 6622. [Google Scholar] [CrossRef]
- Sheta, A.; Turabieh, H.; Braik, M.; Surani, S.R. Diagnosis of obstructive sleep apnea using logistic regression and artificial neural networks models. In Proceedings of the Future Technologies Conference (FTC) 2019, San Francisco, CA, USA, 24–25 October 2019; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1, pp. 766–784. [Google Scholar]
- Urso, A.; Fiannaca, A.; La Rosa, M.; Ravì, V.; Rizzo, R. Data Mining: Prediction Methods. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, UK, 2019; pp. 413–430. [Google Scholar]
- Genkin, A.; Lewis, D.D.; Madigan, D. Large-Scale Bayesian Logistic Regression for Text Categorization. Technometrics 2007, 49, 291–304. [Google Scholar] [CrossRef]
- Boateng, E.; Abaye, D. A Review of the Logistic Regression Model with Emphasis on Medical Research. J. Data Anal. Inf. Process. 2019, 7, 190–207. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Kecman, V. Support Vector Machines—An Introduction. In Support Vector Machines: Theory and Applications; Wang, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–47. [Google Scholar] [CrossRef]
- Dada, E.G.; Bassi, J.S.; Chiroma, H.; Abdulhamid, S.M.; Adetunmbi, A.O.; Ajibuwa, O.E. Machine learning for email spam filtering: Review, approaches and open research problems. Heliyon 2019, 5, e01802. [Google Scholar] [CrossRef] [PubMed]
- Nhat-Duc, H.; Van-Duc, T. Comparison of histogram-based gradient boosting classification machine, random Forest, and deep convolutional neural network for pavement raveling severity classification. Autom. Constr. 2023, 148, 104767. [Google Scholar] [CrossRef]
- Rish, I. An empirical study of the naive Bayes classifier. In Proceedings of the IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, WA, USA, 4–6 August 2001; IBM: New York, NY, USA, 2001; Volume 3, pp. 41–46. [Google Scholar]
- Fürnkranz, J. Decision Tree. In Encyclopedia of Machine Learning; Sammut, C., Webb, G.I., Eds.; Springer: Boston, MA, USA, 2010; pp. 263–267. [Google Scholar]
- Saud, A.; Shakya, S.; Neupane, B. Analysis of Depth of Entropy and GINI Index Based Decision Trees for Predicting Diabetes. Indian J. Comput. Sci. 2022, 6, 19–28. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Braik, M.; Al-Zoubi, H.; Al-Hiary, H. Pedestrian detection using multiple feature channels and contour cues with census transform histogram and random forest classifier. Pattern Anal. Appl. 2020, 23, 751–769. [Google Scholar] [CrossRef]







| Model | Train | Test | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc | Pre | Rec | Fscore | Acc | Pre | Rec | Fscore | |
| LR | 0.7024 | 0.7070 | 0.6680 | 0.6869 | 0.6988 | 0.7565 | 0.6421 | 0.6946 |
| SVM | 0.7011 | 0.7103 | 0.6559 | 0.6820 | 0.6811 | 0.7444 | 0.6125 | 0.6721 |
| GBC | 0.7753 | 0.7746 | 0.7621 | 0.7683 | 0.7146 | 0.7582 | 0.6827 | 0.7184 |
| GNB | 0.6781 | 0.7424 | 0.5228 | 0.6136 | 0.6673 | 0.8036 | 0.4982 | 0.6150 |
| KNC | 0.7766 | 0.7577 | 0.7984 | 0.7775 | 0.7028 | 0.7290 | 0.7048 | 0.7167 |
| DT | 0.7011 | 0.7558 | 0.5739 | 0.6524 | 0.6791 | 0.7872 | 0.5461 | 0.6449 |
| RFC | 0.8206 | 0.7904 | 0.8616 | 0.8244 | 0.7047 | 0.7300 | 0.7085 | 0.7191 |
| ANN | 0.6873 | 0.6920 | 0.6492 | 0.6699 | 0.6713 | 0.7261 | 0.6162 | 0.6667 |
| Classifier | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|
| LR | 4.0 | 5.5 | 4.0 | 4.0 |
| SVM | 5.25 | 5.5 | 5.5 | 5.0 |
| GBC | 2.0 | 2.5 | 3.0 | 2.5 |
| GNB | 8.0 | 3.0 | 8.0 | 8.0 |
| kNC | 2.5 | 5.0 | 2.0 | 2.5 |
| DT | 5.75 | 3.0 | 7.0 | 7.0 |
| RFC | 1.5 | 3.5 | 1.0 | 1.0 |
| ANN | 7.0 | 8.0.0 | 5.5 | 6.0 |
| p-value | 0.0634819 | 0.3325939 | 0.0542263 | 0.0542263 |
| Accuracy (RF is the control classifier) | |||||
| i | Algorithm | p-value | ÷ i | Hypothesis | |
| 7 | GNB | 2.65361389 | 0.00796349 | 0.00714286 | Rejected |
| 6 | ANN | 2.24536560 | 0.02474467 | 0.00833333 | Not Rejected |
| 5 | DT | 1.73505523 | 0.08273102 | 0.01 | Not Rejected |
| 4 | SVM | 1.53093109 | 0.12578642 | 0.0125 | Not Rejected |
| 3 | LR | 1.02062073 | 0.30743417 | 0.01666667 | Not Rejected |
| 2 | KNC | 0.40824829 | 0.68309140 | 0.025 | Not Rejected |
| 1 | GBC | 0.20412415 | 0.83825649 | 0.05 | Not Rejected |
| Precision (GBC is the control classifier) | |||||
| i | Algorithm | p-value | ÷ i | Hypothesis | |
| 7 | ANN | 2.24536560 | 0.02474467 | 0.00714286 | Rejected |
| 6 | LR | 1.22474487 | 0.22067136 | 0.00833333 | Not Rejected |
| 5 | SVM | 1.22474487 | 0.22067136 | 0.01 | Not Rejected |
| 4 | KNC | 1.02062073 | 0.30743417 | 0.0125 | Not Rejected |
| 3 | RFC | 0.40824829 | 0.68309140 | 0.01666667 | Not Rejected |
| 2 | GNB | 0.20412415 | 0.83825649 | 0.025 | Not Rejected |
| 1 | DT | 0.20412415 | 0.83825649 | 0.05 | Not Rejected |
| Recall (RFC is the control classifier) | |||||
| i | Algorithm | p-value | ÷ i | Hypothesis | |
| 7 | GNB | 2.85773803 | 0.00426672 | 0.00714286 | Rejected |
| 6 | DT | 2.44948974 | 0.01430588 | 0.00833333 | Rejected |
| 5 | SVM | 1.83711731 | 0.06619258 | 0.01 | Not Rejected |
| 4 | ANN | 1.83711731 | 0.06619258 | 0.0125 | Not Rejected |
| 3 | LR | 1.22474487 | 0.22067136 | 0.01666667 | Not Rejected |
| 2 | GBC | 0.81649658 | 0.41421618 | 0.025 | Not Rejected |
| 1 | KNC | 0.40824829 | 0.68309140 | 0.05 | Not Rejected |
| F1 score (RFC is the control classifier) | |||||
| i | Algorithm | p-value | ÷ i | Hypothesis | |
| 7 | GNB | 2.85773803 | 0.004266725 | 0.00714286 | Rejected |
| 6 | DT | 2.44948974 | 0.014305878 | 0.00833333 | Rejected |
| 5 | ANN | 2.04124145 | 0.041226833 | 0.01 | Not Rejected |
| 4 | SVM | 1.63299316 | 0.102470435 | 0.0125 | Not Rejected |
| 3 | LR | 1.22474487 | 0.220671362 | 0.01666667 | Not Rejected |
| 2 | GBC | 0.61237244 | 0.540291375 | 0.025 | Not Rejected |
| 1 | KNC | 0.61237244 | 0.540291375 | 0.05 | Not Rejected |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elashmawi, W.H.; Djellal, A.; Sheta, A.; Surani, S.; Aljahdali, S. Machine Learning for Enhanced COPD Diagnosis: A Comparative Analysis of Classification Algorithms. Diagnostics 2024, 14, 2822. https://doi.org/10.3390/diagnostics14242822
Elashmawi WH, Djellal A, Sheta A, Surani S, Aljahdali S. Machine Learning for Enhanced COPD Diagnosis: A Comparative Analysis of Classification Algorithms. Diagnostics. 2024; 14(24):2822. https://doi.org/10.3390/diagnostics14242822
Chicago/Turabian StyleElashmawi, Walaa H., Adel Djellal, Alaa Sheta, Salim Surani, and Sultan Aljahdali. 2024. "Machine Learning for Enhanced COPD Diagnosis: A Comparative Analysis of Classification Algorithms" Diagnostics 14, no. 24: 2822. https://doi.org/10.3390/diagnostics14242822
APA StyleElashmawi, W. H., Djellal, A., Sheta, A., Surani, S., & Aljahdali, S. (2024). Machine Learning for Enhanced COPD Diagnosis: A Comparative Analysis of Classification Algorithms. Diagnostics, 14(24), 2822. https://doi.org/10.3390/diagnostics14242822





