Image Quality Assessment and Reliability Analysis of Artificial Intelligence-Based Tumor Classification of Stimulated Raman Histology of Tumor Biobank Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Dataset
2.2. Biobank Dataset
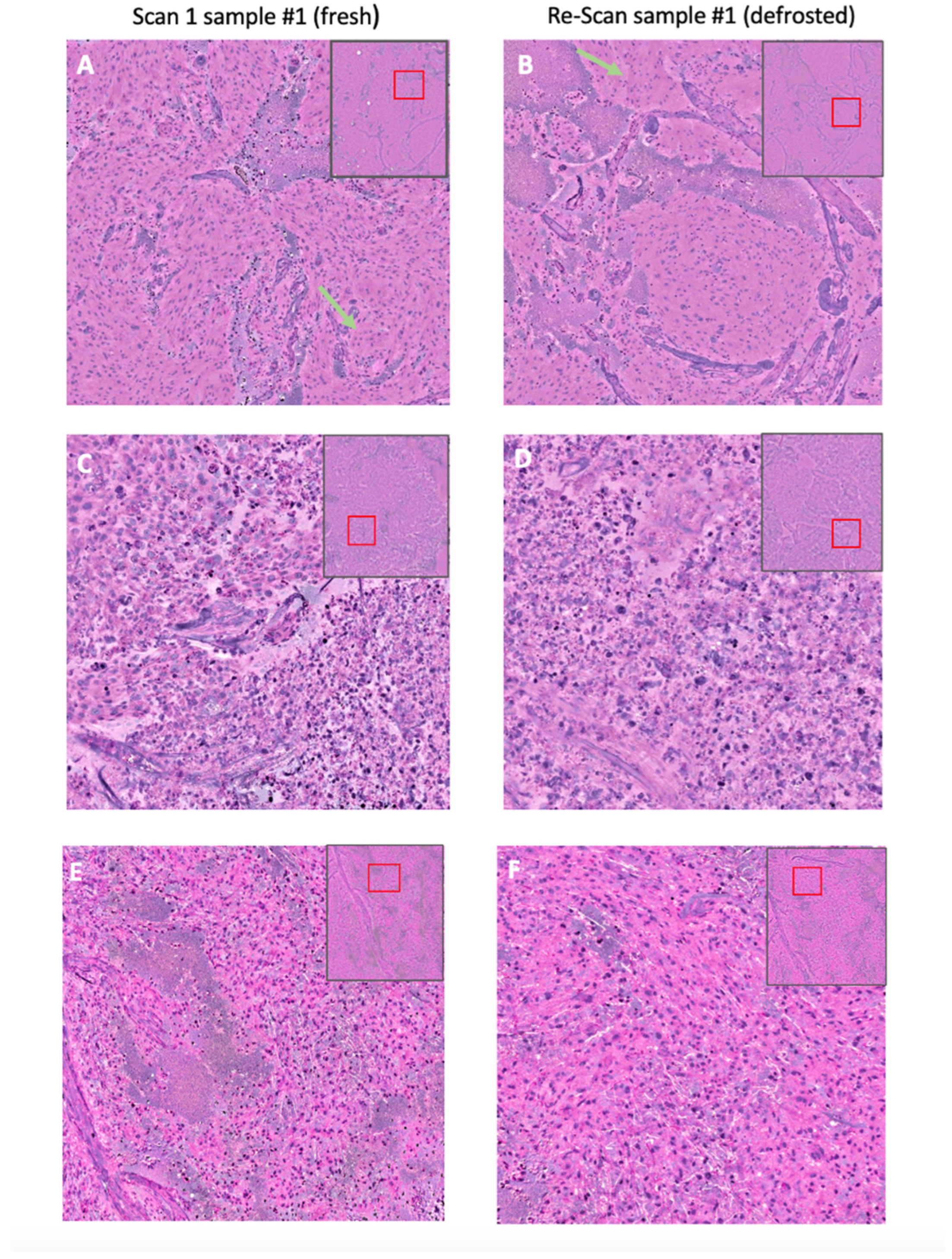
2.3. Specimen Collection and Stimulated Raman Histology
2.4. Image Quality Assessment
2.5. Image Analysis by Convolutional Neural Networks
Statistical Analysis
3. Results
3.1. Prospective Patient Dataset—Characteristics
3.2. Biobank Dataset—Characteristics
3.3. Image Quality Score
3.4. Prospective Patient Dataset—CNN Based Histological Tumor Class Prediction
3.5. Prospective Patient Dataset—CNN Based Molecular Subtype Classification (“Deep Glioma”)
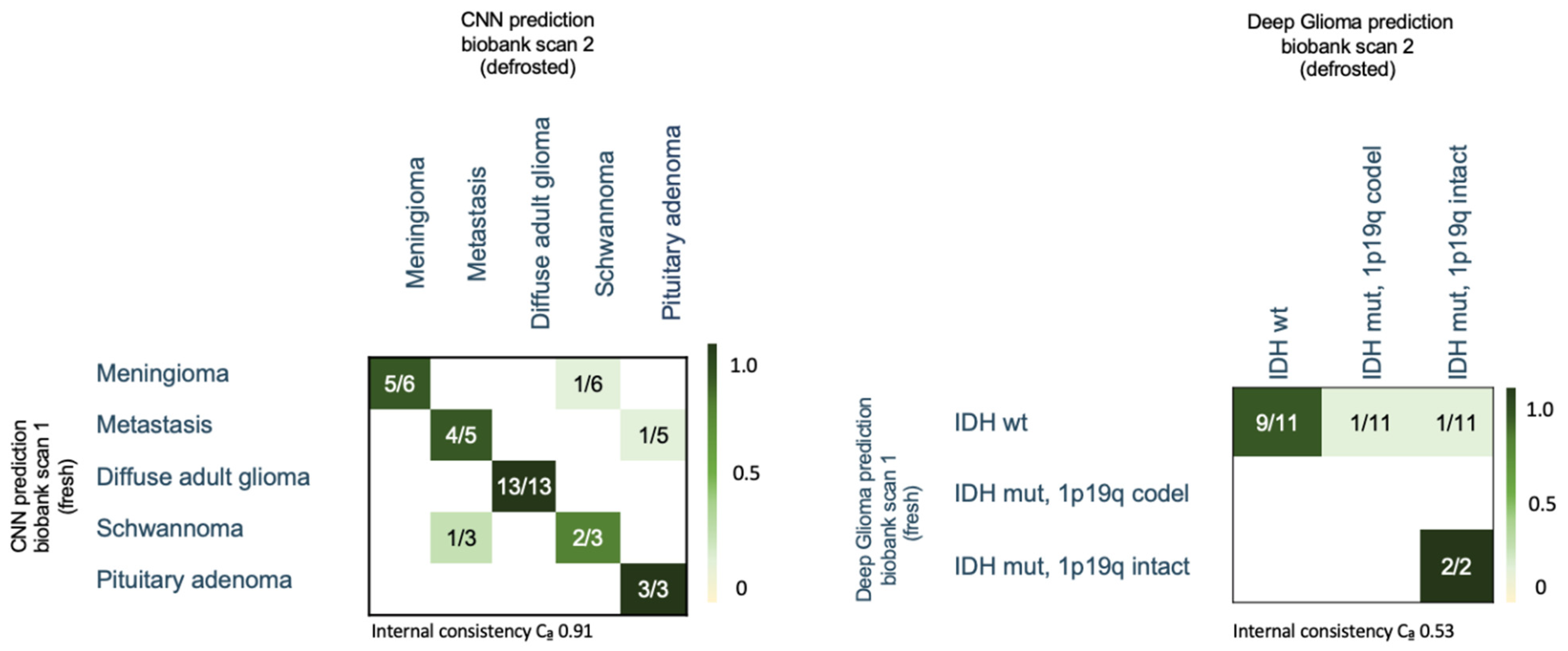
3.6. Biobank Dataset—CNN Based Histological Tumor Class Prediction
3.7. Biobank Dataset—CNN-Based Molecular Subtype Classification (“Deep Glioma”)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orringer, D.A.; Pandian, B.; Niknafs, Y.S.; Hollon, T.C.; Boyle, J.; Lewis, S.; Garrard, M.; Hervey-Jumper, S.L.; Garton, H.J.L.; Maher, C.O.; et al. Rapid Intraoperative Histology of Unprocessed Surgical Specimens via Fibre-Laser-Based Stimulated Raman Scattering Microscopy. Nat. Biomed. Eng. 2017, 1, 0027. [Google Scholar] [CrossRef] [PubMed]
- Freudiger, C.W.; Min, W.; Saar, B.G.; Lu, S.; Holtom, G.R.; He, C.; Tsai, J.C.; Kang, J.X.; Xie, X.S. Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science 2008, 322, 1857–1861. [Google Scholar] [CrossRef] [PubMed]
- Movahed-Ezazi, M.; Nasir-Moin, M.; Fang, C.; Pizzillo, I.; Galbraith, K.; Drexler, S.; Krasnozhen-Ratush, O.A.; Shroff, S.; Zagzag, D.; William, C.; et al. Clinical Validation of Stimulated Raman Histology for Rapid Intraoperative Diagnosis of Central Nervous System Tumors. Mod. Pathol. 2023, 36, 100219. [Google Scholar] [CrossRef] [PubMed]
- Eichberg, D.G.; Shah, A.H.; Di, L.; Semonche, A.M.; Jimsheleishvili, G.; Luther, E.M.; Sarkiss, C.A.; Levi, A.D.; Gultekin, S.H.; Komotar, R.J.; et al. Stimulated Raman Histology for Rapid and Accurate Intraoperative Diagnosis of CNS Tumors: Prospective Blinded Study. J. Neurosurg. 2019, 134, 137–143. [Google Scholar] [CrossRef]
- Einstein, E.H.; Ablyazova, F.; Rosenberg, A.; Harshan, M.; Wahl, S.; Har-El, G.; Constantino, P.D.; Ellis, J.A.; Boockvar, J.A.; Langer, D.J.; et al. Stimulated Raman Histology Facilitates Accurate Diagnosis in Neurosurgical Patients: A One-to-One Noninferiority Study. J. Neurooncol. 2022, 159, 369–375. [Google Scholar] [CrossRef]
- Straehle, J.; Erny, D.; Neidert, N.; Henrik Heiland, D.; El Rahal, A.; Sacalean, V.; Steybe, D.; Schmelzeisen, R.; Vlachos, A.; Mizaikoff, B.; et al. Neuropathological Interpretation of Stimulated Raman Histology Images of Brain and Spine Tumors: Part B. Neurosurg. Rev. 2022, 45, 1721–1729. [Google Scholar] [CrossRef]
- Wadiura, L.I.; Kiesel, B.; Roetzer-Pejrimovsky, T.; Mischkulnig, M.; Vogel, C.C.; Hainfellner, J.A.; Matula, C.; Freudiger, C.W.; Orringer, D.A.; Wöhrer, A.; et al. Toward Digital Histopathological Assessment in Surgery for Central Nervous System Tumors Using Stimulated Raman Histology. Neurosurg. Focus 2022, 53, E12. [Google Scholar] [CrossRef]
- Hollon, T.C.; Lewis, S.; Pandian, B.; Niknafs, Y.S.; Garrard, M.R.; Garton, H.; Maher, C.O.; McFadden, K.; Snuderl, M.; Lieberman, A.P.; et al. Rapid Intraoperative Diagnosis of Pediatric Brain Tumors Using Stimulated Raman Histology. Cancer Res. 2018, 78, 278–289. [Google Scholar] [CrossRef]
- Di, L.; Eichberg, D.G.; Park, Y.J.; Shah, A.H.; Jamshidi, A.M.; Luther, E.M.; Lu, V.M.; Komotar, R.J.; Ivan, M.E.; Gultekin, S.H. Rapid Intraoperative Diagnosis of Meningiomas Using Stimulated Raman Histology. World Neurosurg. 2021, 150, e108–e116. [Google Scholar] [CrossRef]
- Pekmezci, M.; Morshed, R.A.; Chunduru, P.; Pandian, B.; Young, J.; Villanueva-Meyer, J.E.; Tihan, T.; Sloan, E.A.; Aghi, M.K.; Molinaro, A.M.; et al. Detection of Glioma Infiltration at the Tumor Margin Using Quantitative Stimulated Raman Scattering Histology. Sci. Rep. 2021, 11, 12162. [Google Scholar] [CrossRef]
- Jiang, C.; Bhattacharya, A.; Linzey, J.R.; Joshi, R.S.; Cha, S.J.; Srinivasan, S.; Alber, D.; Kondepudi, A.; Urias, E.; Pandian, B.; et al. Rapid Automated Analysis of Skull Base Tumor Specimens Using Intraoperative Optical Imaging and Artificial Intelligence. Neurosurgery 2022, 90, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, D.; Ruess, D.; Meissner, A.-K.; Fürtjes, G.; von Spreckelsen, N.; Ion-Margineau, A.; Khalid, F.; Blau, T.; Stehle, T.; Al-Shughri, A.; et al. Streamlined Intraoperative Brain Tumor Classification and Molecular Subtyping in Stereotactic Biopsies Using Stimulated Raman Histology and Deep Learning. Clin. Cancer Res. 2024, 30, 3824–3836. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, D.; Spreckelsen, N.; Mawrin, C.; Ion-Margineanu, A.; Fürtjes, G.; Jünger, S.T.; Khalid, F.; Freudiger, C.W.; Timmer, M.; Ruge, M.I.; et al. Novel Rapid Intraoperative Qualitative Tumor Detection by a Residual Convolutional Neural Network Using Label-Free Stimulated Raman Scattering Microscopy. Acta Neuropathol. Commun. 2022, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Hollon, T.C.; Pandian, B.; Adapa, A.R.; Urias, E.; Save, A.V.; Khalsa, S.S.S.; Eichberg, D.G.; D’Amico, R.S.; Farooq, Z.U.; Lewis, S.; et al. Near Real-Time Intraoperative Brain Tumor Diagnosis Using Stimulated Raman Histology and Deep Neural Networks. Nat. Med. 2020, 26, 52–58. [Google Scholar] [CrossRef]
- Hollon, T.; Jiang, C.; Chowdury, A.; Nasir-Moin, M.; Kondepudi, A.; Aabedi, A.; Adapa, A.; Al-Holou, W.; Heth, J.; Sagher, O.; et al. Artificial-Intelligence-Based Molecular Classification of Diffuse Gliomas Using Rapid, Label-Free Optical Imaging. Nat. Med. 2023, 29, 828–832. [Google Scholar] [CrossRef]
- Hollon, T.C.; Pandian, B.; Urias, E.; Save, A.V.; Adapa, A.R.; Srinivasan, S.; Jairath, N.K.; Farooq, Z.; Marie, T.; Al-Holou, W.N.; et al. Rapid, Label-Free Detection of Diffuse Glioma Recurrence Using Intraoperative Stimulated Raman Histology and Deep Neural Networks. Neuro Oncol. 2021, 23, 144–155. [Google Scholar] [CrossRef]
- Fürtjes, G.; Reinecke, D.; Von Spreckelsen, N.; Meißner, A.-K.; Rueß, D.; Timmer, M.; Freudiger, C.; Ion-Margineanu, A.; Khalid, F.; Watrinet, K.; et al. Intraoperative Microscopic Autofluorescence Detection and Characterization in Brain Tumors Using Stimulated Raman Histology and Two-Photon Fluorescence. Front. Oncol. 2023, 13, 1146031. [Google Scholar] [CrossRef]
- Neidert, N.; Straehle, J.; Erny, D.; Sacalean, V.; El Rahal, A.; Steybe, D.; Schmelzeisen, R.; Vlachos, A.; Reinacher, P.C.; Coenen, V.A.; et al. Stimulated Raman Histology in the Neurosurgical Workflow of a Major European Neurosurgical Center—Part A. Neurosurg. Rev. 2022, 45, 1731–1739. [Google Scholar] [CrossRef]
- Hollon, T.C.; Orringer, D.A. An Automated Tissue-to-Diagnosis Pipeline Using Intraoperative Stimulated Raman Histology and Deep Learning. Mol. Cell Oncol. 2020, 7, 1736742. [Google Scholar] [CrossRef]
- Hollon, T.; Orringer, D.A. Label-Free Brain Tumor Imaging Using Raman-Based Methods. J. Neurooncol. 2021, 151, 393–402. [Google Scholar] [CrossRef]
- Shrestha, P.; Kneepkens, R.; van Elswijk, G.; Vrijnsen, J.; Ion, R.; Verhagen, D.; Abels, E.; Vossen, D.; Hulsken, B. Objective and Subjective Assessment of Digital Pathology Image Quality. AIMS Med. Sci. 2015, 2, 65–78. [Google Scholar] [CrossRef]
- Hashimoto, N.; Bautista, P.A.; Yamaguchi, M.; Ohyama, N.; Yagi, Y. Referenceless Image Quality Evaluation for Whole Slide Imaging. J. Pathol. Inform. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Forero, C.G. Cronbach’s Alpha. In Encyclopedia of Quality of Life and Well-Being Research; Springer: Dordrecht, The Netherlands, 2014; pp. 1357–1359. [Google Scholar]
- Khonglah, Y.; Lyngdoh, B.S.; Kakati, A.; Mishra, J.; Al Aman, M.M.; Phukan, P. Intraoperative Diagnosis of Central Nervous System Tumors: Challenges, Errors, Lessons Learned, and the Surgeon’s Perspective. Cureus 2021, 13, e17823. [Google Scholar] [CrossRef] [PubMed]
- Meißner, A.-K.; Goldbrunner, R.; Neuschmelting, V. Intraoperative Stimulated Raman Histology for Personalized Brain Tumor Surgery. Chirurgie 2024, 95, 274–279. [Google Scholar] [CrossRef]
- Klamminger, G.G.; Gérardy, J.-J.; Jelke, F.; Mirizzi, G.; Slimani, R.; Klein, K.; Husch, A.; Hertel, F.; Mittelbronn, M.; Kleine-Borgmann, F.B. Application of Raman Spectroscopy for Detection of Histologically Distinct Areas in Formalin-Fixed Paraffin-Embedded Glioblastoma. Neurooncol. Adv. 2021, 3, vdab077. [Google Scholar] [CrossRef]
- Kalkanis, S.N.; Kast, R.E.; Rosenblum, M.L.; Mikkelsen, T.; Yurgelevic, S.M.; Nelson, K.M.; Raghunathan, A.; Poisson, L.M.; Auner, G.W. Raman Spectroscopy to Distinguish Grey Matter, Necrosis, and Glioblastoma Multiforme in Frozen Tissue Sections. J. Neurooncol. 2014, 116, 477–485. [Google Scholar] [CrossRef]
- Kast, R.E.; Auner, G.W.; Rosenblum, M.L.; Mikkelsen, T.; Yurgelevic, S.M.; Raghunathan, A.; Poisson, L.M.; Kalkanis, S.N. Raman Molecular Imaging of Brain Frozen Tissue Sections. J. Neurooncol. 2014, 120, 55–62. [Google Scholar] [CrossRef]
- Draux, F.; Gobinet, C.; Sulé-Suso, J.; Trussardi, A.; Manfait, M.; Jeannesson, P.; Sockalingum, G.D. Raman Spectral Imaging of Single Cancer Cells: Probing the Impact of Sample Fixation Methods. Anal. Bioanal. Chem. 2010, 397, 2727–2737. [Google Scholar] [CrossRef]
- Huang, Z.; McWilliams, A.; Lam, S.; English, J.; McLean, D.; Lui, H.; Zeng, H. Effect of Formalin Fixation on the Near-Infrared Raman Spectroscopy of Normal and Cancerous Human Bronchial Tissues. Int. J. Oncol. 2003, 23, 649–655. [Google Scholar] [CrossRef]
- Desciak, E.B.; Maloney, M.E. Artifacts in Frozen Section Preparation. Dermatol. Surg. 2000, 26, 500–504. [Google Scholar] [CrossRef]
- Nackenhorst, M.C.; Kasiri, M.; Gollackner, B.; Regele, H. Ex Vivo Fluorescence Confocal Microscopy: Chances and Changes in the Analysis of Breast Tissue. Diagn. Pathol. 2022, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Shabihkhani, M.; Lucey, G.M.; Wei, B.; Mareninov, S.; Lou, J.J.; Vinters, H.V.; Singer, E.J.; Cloughesy, T.F.; Yong, W.H. The Procurement, Storage, and Quality Assurance of Frozen Blood and Tissue Biospecimens in Pathology, Biorepository, and Biobank Settings. Clin. Biochem. 2014, 47, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, C.; Franzén, B.; Nistér, M. Frozen Tissue Biobanks. Tissue Handling, Cryopreservation, Extraction, and Use for Proteomic Analysis. Acta Oncol. 2006, 45, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Estrada, L.I.; Robinson, A.A.; Amaral, A.C.; Giannaris, E.L.; Heyworth, N.C.; Mortazavi, F.; Ngwenya, L.B.; Roberts, D.E.; Cabral, H.J.; Killiany, R.J.; et al. Evaluation of Long-Term Cryostorage of Brain Tissue Sections for Quantitative Histochemistry. J. Histochem. Cytochem. 2017, 65, 153–171. [Google Scholar] [CrossRef]
- Harms, J.W.A.; Streckert, E.M.S.; Kiolbassa, N.M.; Thomas, C.; Grauer, O.; Oertel, M.; Eich, H.T.; Stummer, W.; Paulus, W.; Brokinkel, B. Confounders of Intraoperative Frozen Section Pathology during Glioma Surgery. Neurosurg. Rev. 2023, 46, 286. [Google Scholar] [CrossRef]
- Yadav, M.; Sharma, P.; Singh, V.; Tewari, R.; Mishra, P.S.; Roy, K. An Audit of Diagnostic Disparity between Intraoperative Frozen Section Diagnosis and Final Histopathological Diagnosis of Central Nervous System Lesions at a Tertiary Care Center. J. Lab. Physicians 2022, 14, 384–393. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Wright, A.I.; Dunn, C.M.; Hale, M.; Hutchins, G.G.A.; Treanor, D.E. The Effect of Quality Control on Accuracy of Digital Pathology Image Analysis. IEEE J. Biomed. Health Inform. 2021, 25, 307–314. [Google Scholar] [CrossRef]
- Hosseini, M.S.; Brawley-Hayes, J.A.Z.; Zhang, Y.; Chan, L.; Plataniotis, K.; Damaskinos, S. Focus Quality Assessment of High-Throughput Whole Slide Imaging in Digital Pathology. IEEE Trans. Med. Imaging 2020, 39, 62–74. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meißner, A.-K.; Blau, T.; Reinecke, D.; Fürtjes, G.; Leyer, L.; Müller, N.; von Spreckelsen, N.; Stehle, T.; Al Shugri, A.; Büttner, R.; et al. Image Quality Assessment and Reliability Analysis of Artificial Intelligence-Based Tumor Classification of Stimulated Raman Histology of Tumor Biobank Samples. Diagnostics 2024, 14, 2701. https://doi.org/10.3390/diagnostics14232701
Meißner A-K, Blau T, Reinecke D, Fürtjes G, Leyer L, Müller N, von Spreckelsen N, Stehle T, Al Shugri A, Büttner R, et al. Image Quality Assessment and Reliability Analysis of Artificial Intelligence-Based Tumor Classification of Stimulated Raman Histology of Tumor Biobank Samples. Diagnostics. 2024; 14(23):2701. https://doi.org/10.3390/diagnostics14232701
Chicago/Turabian StyleMeißner, Anna-Katharina, Tobias Blau, David Reinecke, Gina Fürtjes, Lili Leyer, Nina Müller, Niklas von Spreckelsen, Thomas Stehle, Abdulkader Al Shugri, Reinhard Büttner, and et al. 2024. "Image Quality Assessment and Reliability Analysis of Artificial Intelligence-Based Tumor Classification of Stimulated Raman Histology of Tumor Biobank Samples" Diagnostics 14, no. 23: 2701. https://doi.org/10.3390/diagnostics14232701
APA StyleMeißner, A.-K., Blau, T., Reinecke, D., Fürtjes, G., Leyer, L., Müller, N., von Spreckelsen, N., Stehle, T., Al Shugri, A., Büttner, R., Goldbrunner, R., Timmer, M., & Neuschmelting, V. (2024). Image Quality Assessment and Reliability Analysis of Artificial Intelligence-Based Tumor Classification of Stimulated Raman Histology of Tumor Biobank Samples. Diagnostics, 14(23), 2701. https://doi.org/10.3390/diagnostics14232701






