Content-Based Image Retrieval and Image Classification System for Early Prediction of Bladder Cancer
Abstract
1. Introduction
- In this study, which was conducted to detect bladder cancer, two systems were developed: classification and CBIR-based. In recent years, classification has become one of the most preferred methods in the literature. However, the most significant disadvantage is that the training of the models takes a long time in large datasets, and they cannot produce successful results in multi-class datasets. Therefore, it is crucial to use CBIR-based systems.
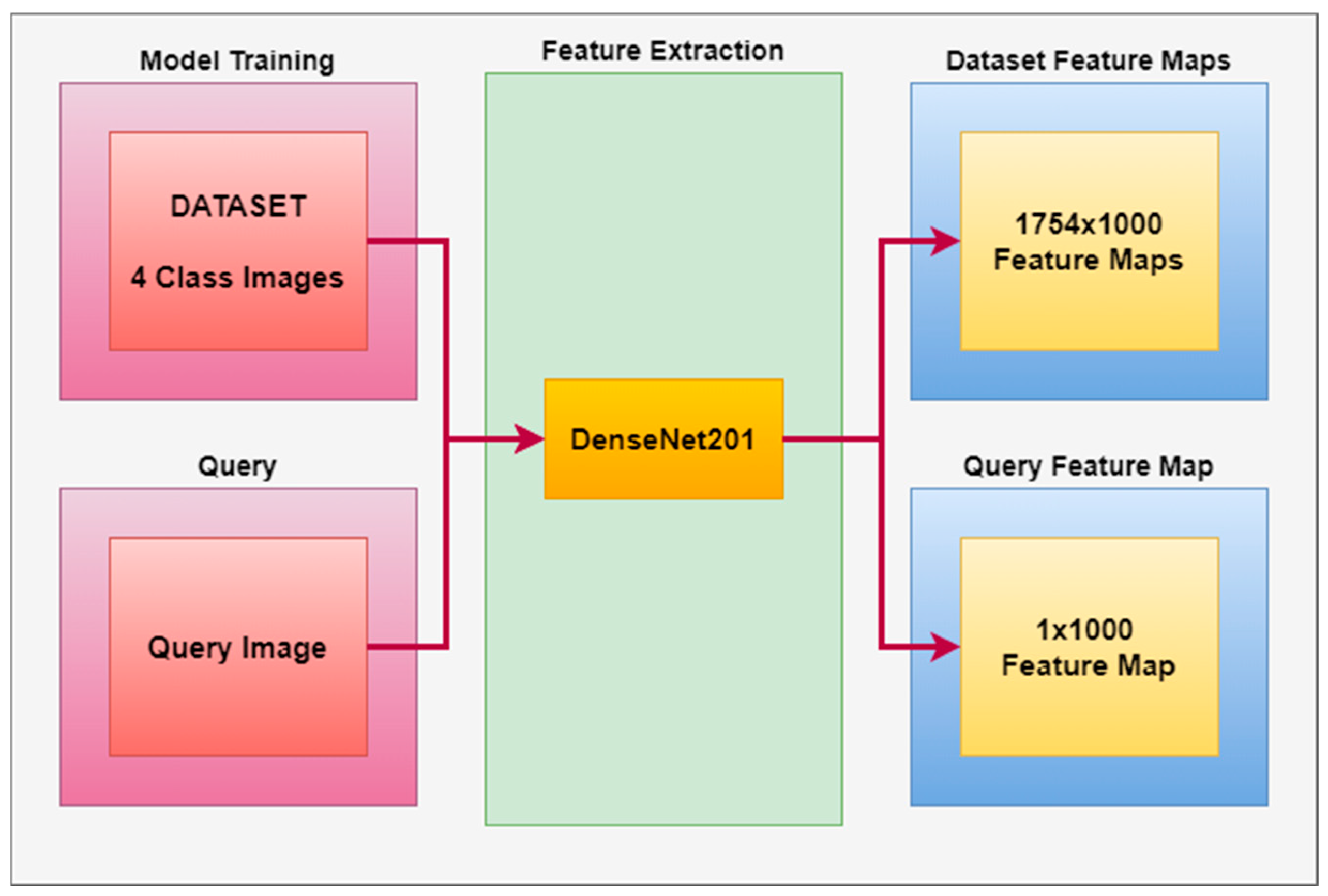
- For feature extraction in the CBIR-based system, LBP and HOG architectures were preferred for textural-based models, and Densenet201, GoogleNet, InceptionV3, GoogleNet-Places365, and NasnetLarge architectures were preferred for CNN architectures.
- Feature maps of the architectures were classified in machine learning classifiers, and the highest success was achieved in the Densenet201 + Subspace KNN duo with 99% accuracy.
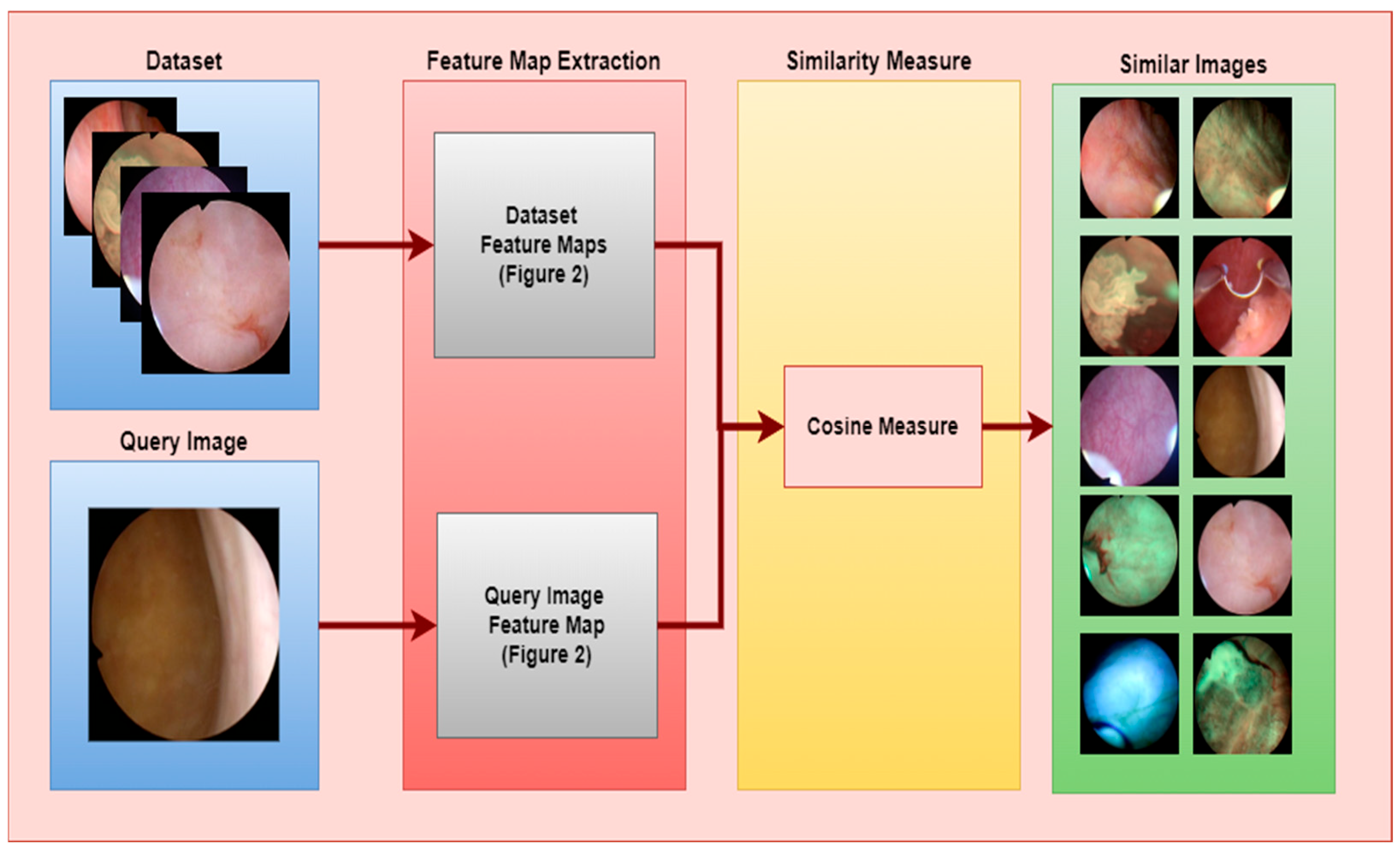
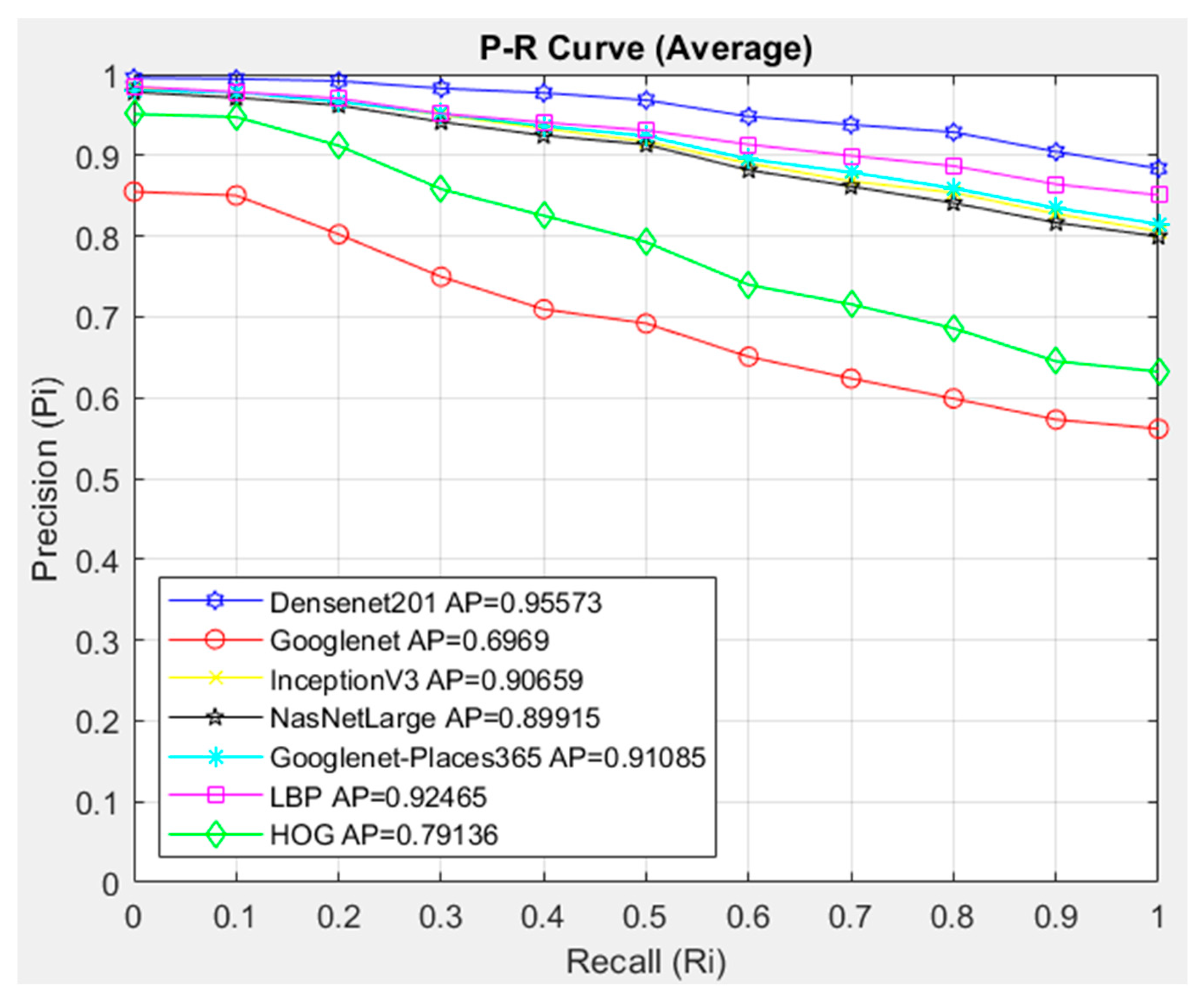
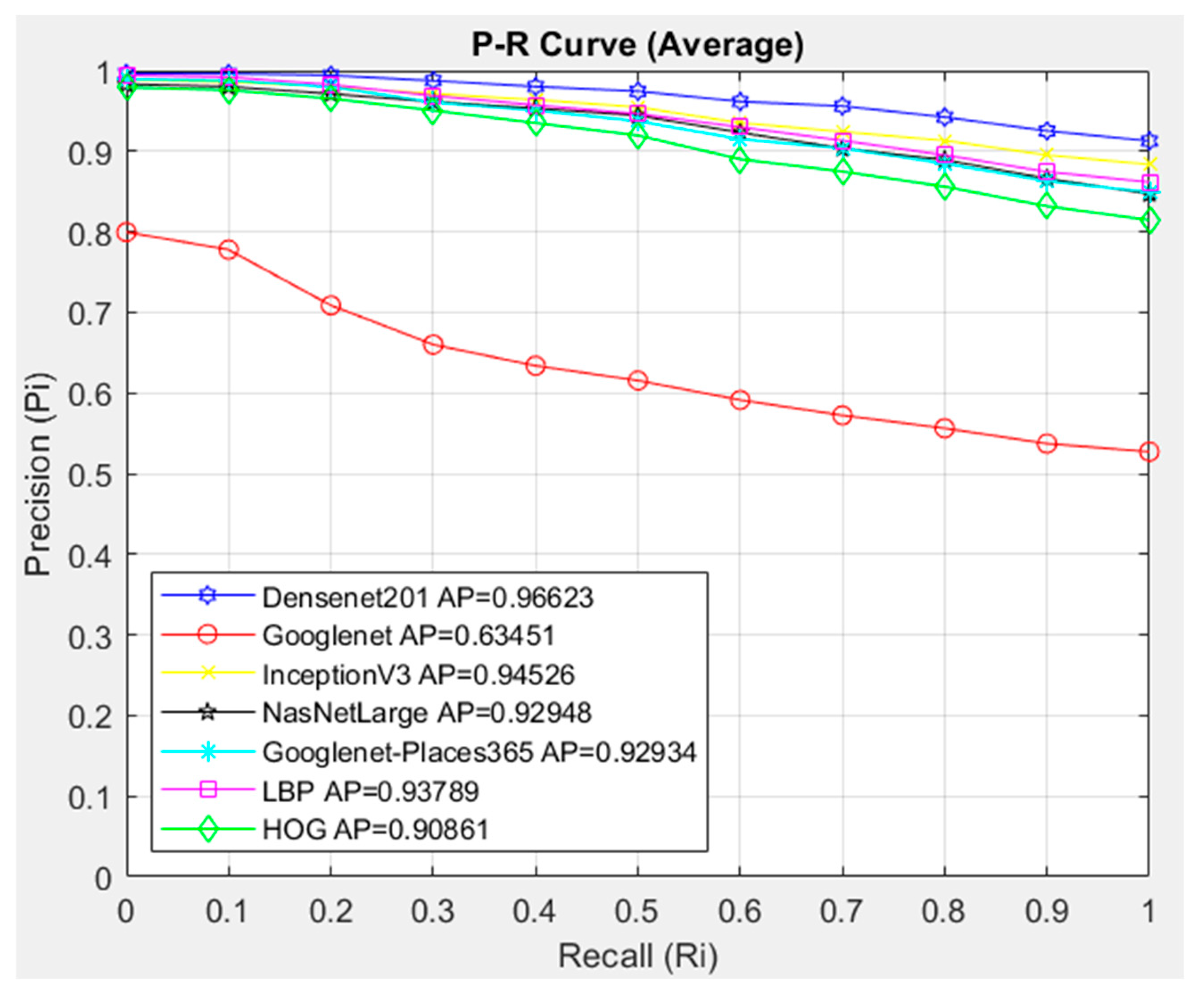
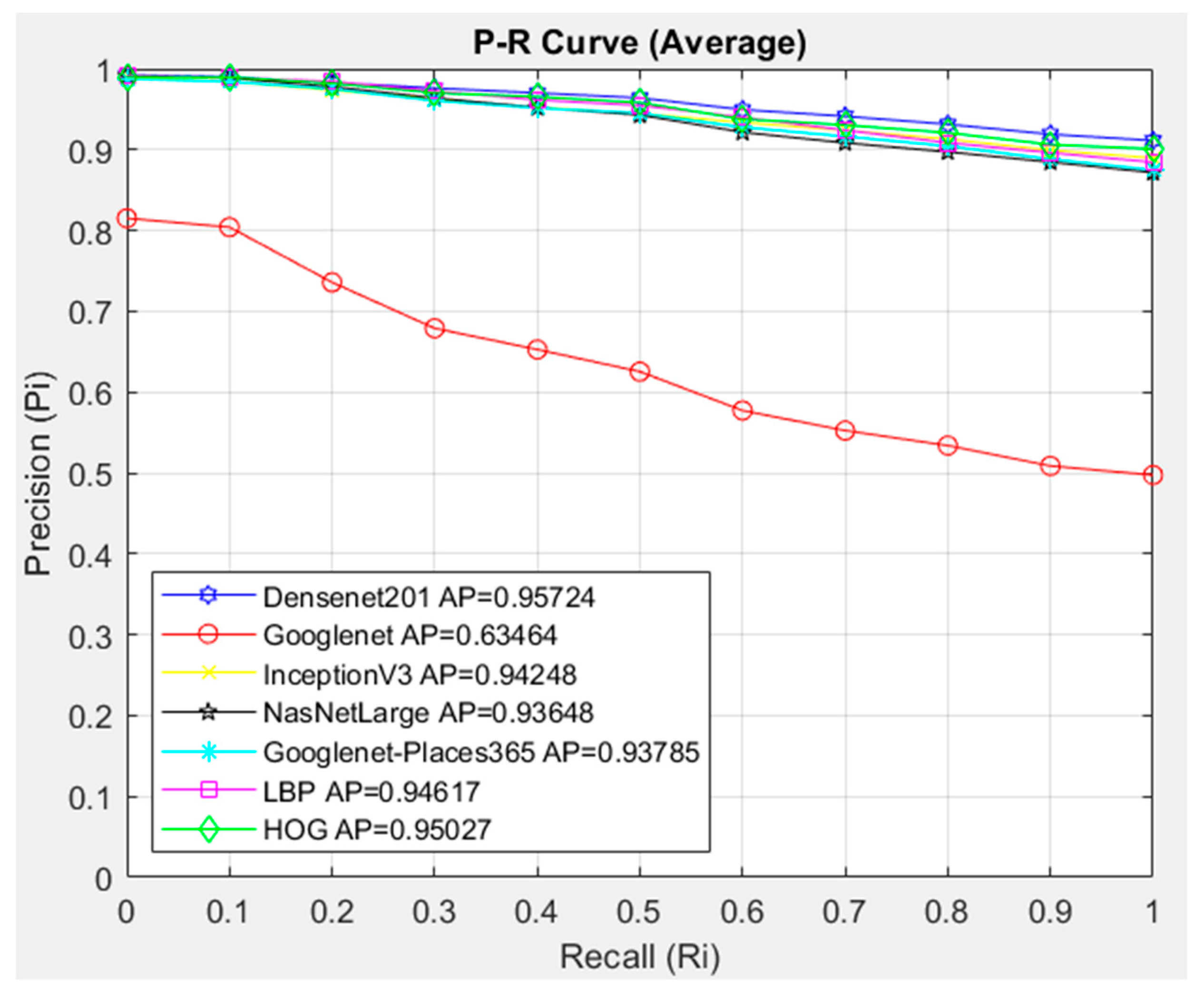
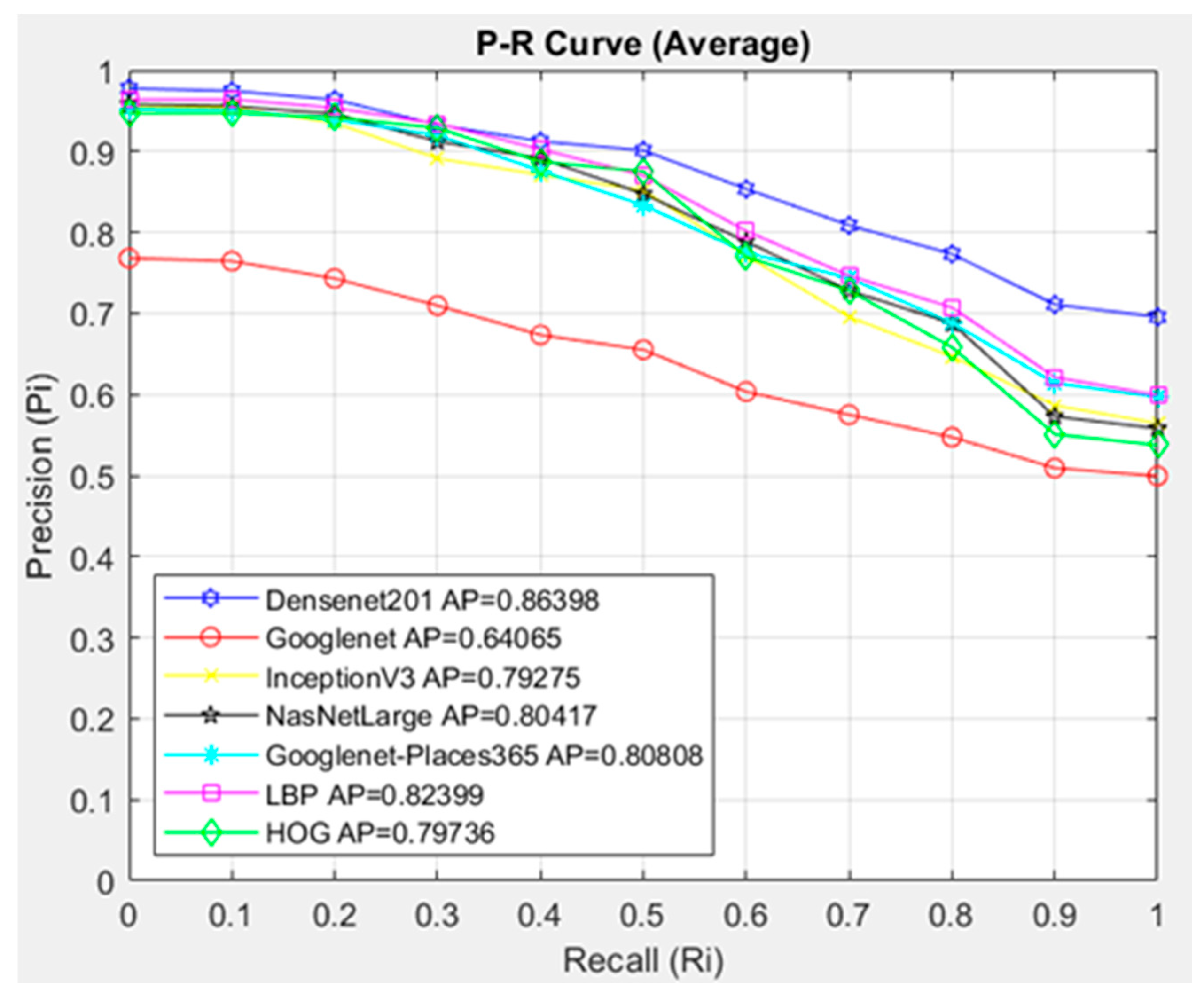
- The feature extraction model obtained with the Densenet201 was used in feature extraction in the proposed CBIR model, and the similarity and distance measurement methods in our CBIR system were compared.
- This paper examined the performance of seven different architectures and seven different similarity measurement metrics for the proposed CBIR system.
- An average AP value of 0.95302 was obtained by using the Densenet201 feature extraction and Cosine similarity measurement metric.
2. Materials and Methods
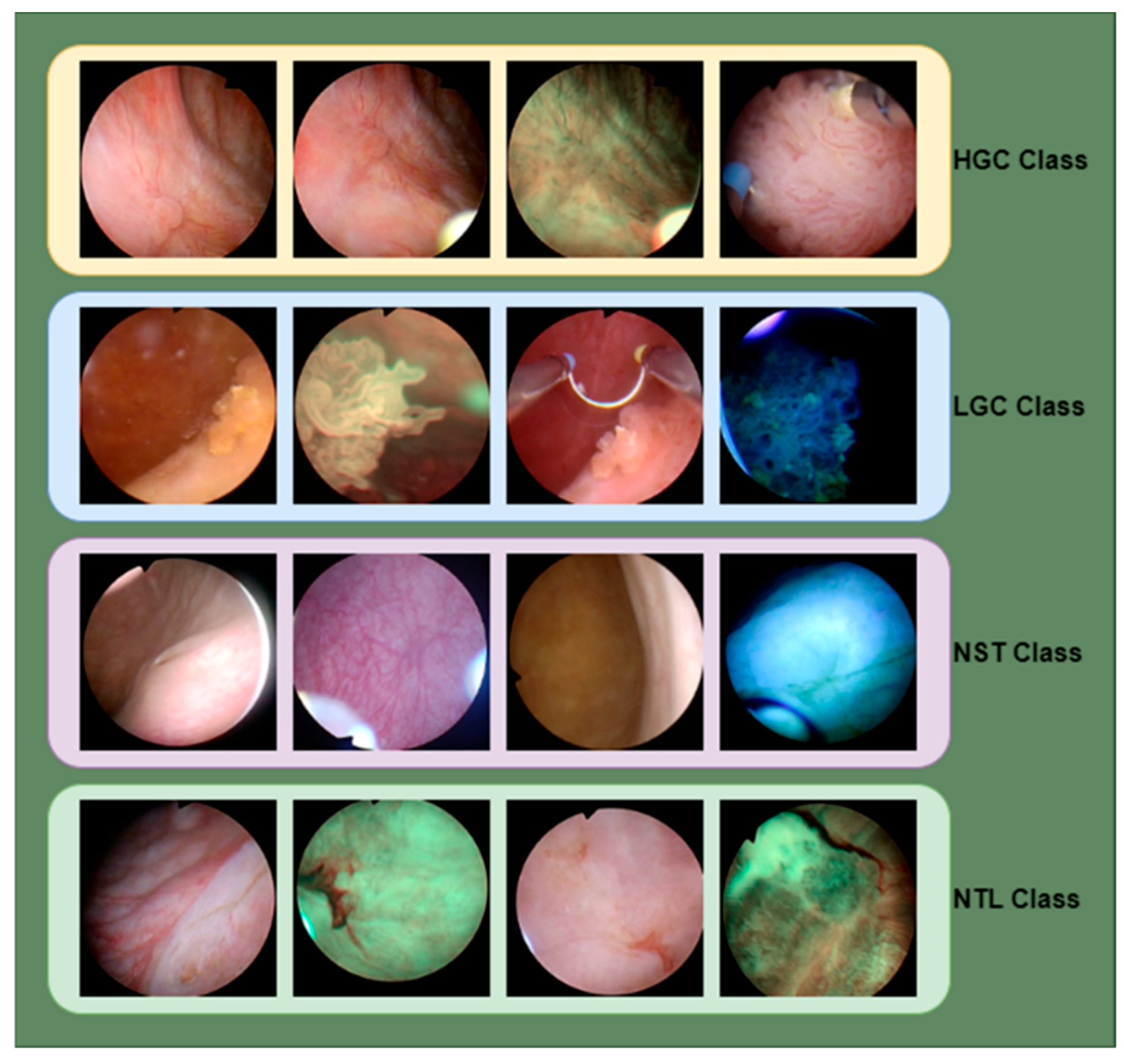
2.1. Bladder Dataset
2.2. Structures Used in Proposed Hybrid Systems
2.3. Developed CBIR System
3. Application Results
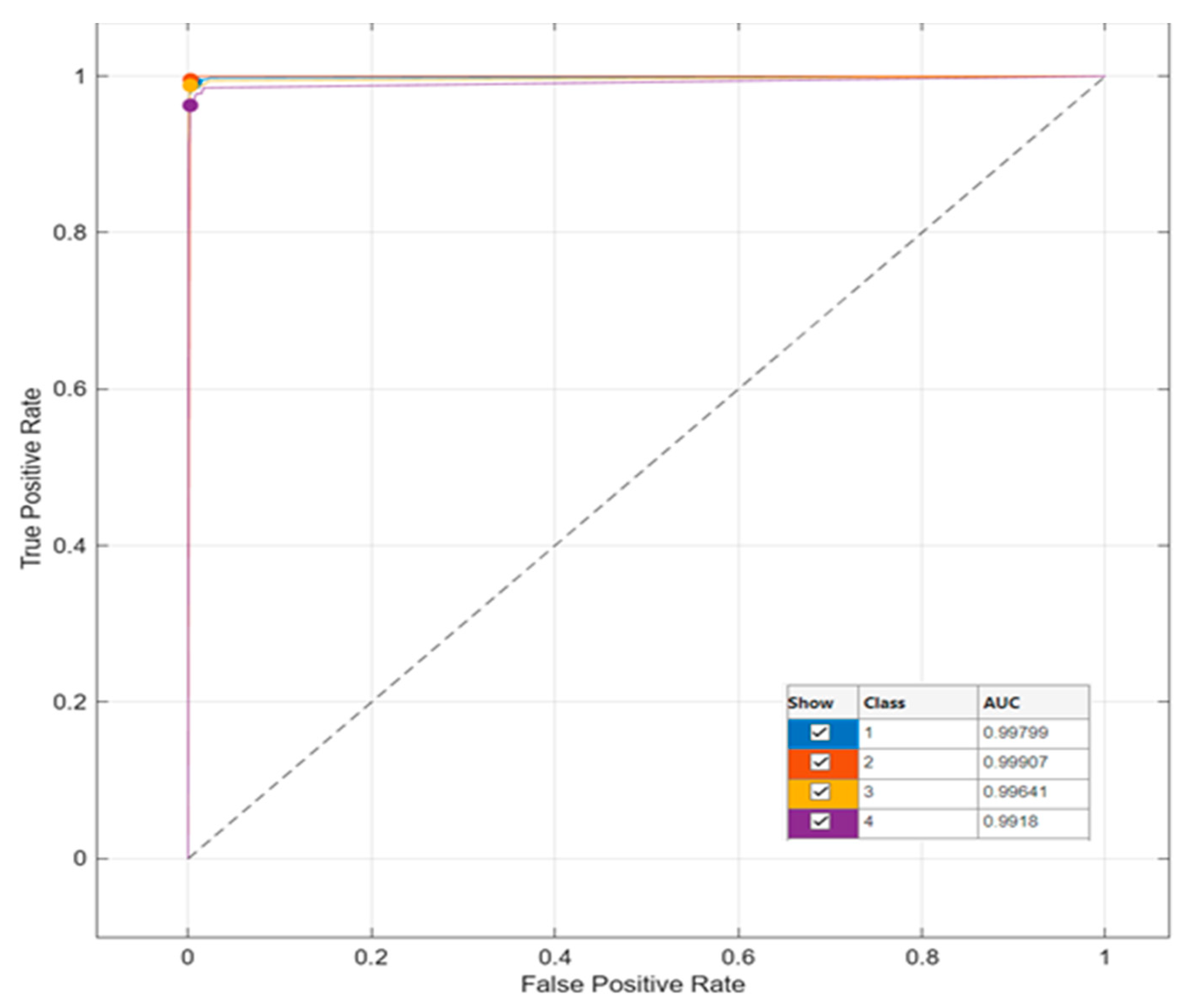
Deep Model and Classification Results
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khalaf, E.M.; Taherian, M.; Almalki, S.G.; Asban, P.; Kareem, A.K.; Alhachami, F.R.; Almulla, A.F.; Romero-Parra, R.M.; Jawhar, Z.H.; Kiani, F.; et al. Relationship between exposure to heavy metals on the increased health risk and carcinogenicity of urinary tract (kidney and bladder). Rev. Environ. Health 2024, 39, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, W.; Jiang, T.; Yang, B. Aquaporins in urinary system. In Aquaporins; Springer: Berlin/Heidelberg, Germany, 2023; pp. 155–177. [Google Scholar]
- Yunusa, B.; Abdullahi, M.; Mashi, S.A.; Aji, S.A.; Sani, A.; Giade, S.A.; Usman, S.A. Bladder cancer burden and challenges of management in Kano, North Western Nigeria. J. Cancer Sci. Clin. Ther. 2020, 4, 424–433. [Google Scholar] [CrossRef]
- Alderson, M.; Grivas, P.; Milowsky, M.I.; Wobker, S.E. Histologic variants of urothelial carcinoma: Morphology, molecular features and clinical implications. Bladder Cancer 2020, 6, 107–122. [Google Scholar] [CrossRef]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Padala, S.A.; Barsouk, A. Epidemiology of bladder cancer. Med. Sci. 2020, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Halaseh, S.A.; Halaseh, S.; Alali, Y.; Ashour, M.E.; Alharayzah, M.J. A review of the etiology and epidemiology of bladder cancer: All you need to know. Cureus 2022, 14, e27330. [Google Scholar] [CrossRef]
- Challacombe, B.; Bultitude, M.F. The Kidneys, Urinary Tract and Prostate, in Browse’s Introduction to the Symptoms & Signs of Surgical Disease; CRC Press: Boca Raton, FL, USA, 2021; pp. 573–587. [Google Scholar]
- Batirel, A.; Regmi, S.K.; Singh, P.; Mert, A.; Konety, B.R.; Kumar, R. Urological infections in the developing world: An increasing problem in developed countries. World J. Urol. 2020, 38, 2681–2691. [Google Scholar] [CrossRef]
- Lavallee, E.; Sfakianos, J.P.; Mulholland, D.J. Tumor heterogeneity and consequences for bladder cancer treatment. Cancers 2021, 13, 5297. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Cimadamore, A.; Blanca, A.; Massari, F.; Vau, N.; Scarpelli, M.; Cheng, L.; Montironi, R. Immune checkpoint inhibitors for the treatment of bladder cancer. Cancers 2021, 13, 131. [Google Scholar] [CrossRef]
- Dyrskjøt, L.; Hansel, D.E.; Efstathiou, J.A.; Knowles, M.A.; Galsky, M.D.; Teoh, J.; Theodorescu, D. Bladder cancer. Nat. Rev. Dis. Primers 2023, 9, 58. [Google Scholar] [CrossRef]
- Eroglu, O.; Eroglu, Y.; Yildirim, M.; Karlidag, T.; Cinar, A.; Akyigit, A.; Kaygusuz, I.; Yildirim, H.; Keles, E.; Yalcin, S. Comparison of computed tomography-based artificial intelligence modeling and magnetic resonance imaging in diagnosis of cholesteatoma. J. Int. Adv. Otol. 2023, 19, 342. [Google Scholar]
- Tuncer, S.A.; Yildirim, M.; Tuncer, T.; Mülayim, M.K. YOLOv8-Based System for Nail Capillary Detection on a Single-Board Computer. Diagnostics 2024, 14, 1843. [Google Scholar] [CrossRef] [PubMed]
- Endoscopic Bladder Tissue Classification Dataset. Available online: https://zenodo.org/records/7741476 (accessed on 20 September 2024).
- Lazo, J.F.; Rosa, B.; Catellani, M.; Fontana, M.; Mistretta, F.A.; Musi, G.; de Cobelli, O.; de Mathelin, M.; De Momi, E. Semi-supervised Bladder Tissue Classification in Multi-Domain Endoscopic Images. IEEE Trans. Biomed. Eng. 2023, 70, 2822–2833. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Punn, N.S.; Agarwal, S. Automated diagnosis of COVID-19 with limited posteroanterior chest X-ray images using fine-tuned deep neural networks. Appl. Intell. 2021, 51, 2689–2702. [Google Scholar] [CrossRef]
- Popescu, D.; El-Khatib, M.; Ichim, L. Skin lesion classification using collective intelligence of multiple neural networks. Sensors 2022, 22, 4399. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikäinen, M.; Harwood, D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–26 June 2005; IEEE: New York, NY, USA, 2005. [Google Scholar]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep feature extraction and classification of hyperspectral images based on convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef]
- Yildirim, M. Image Visualization and Classification Using Hydatid Cyst Images with an Explainable Hybrid Model. Appl. Sci. 2023, 13, 9926. [Google Scholar] [CrossRef]
- Hong, X.; Zhao, G.; Pietikainen, M.; Chen, X. Combining LBP difference and feature correlation for texture description. IEEE Trans. Image Process. 2014, 23, 2557–2568. [Google Scholar] [CrossRef]
- Ghahremannezhad, H.; Shi, H.; Liu, C. Object detection in traffic videos: A survey. IEEE Trans. Intell. Transp. Syst. 2023, 24, 6780–6799. [Google Scholar] [CrossRef]
- Bagwari, N.; Kumar, S.; Verma, V.S. Verma, A comprehensive review on segmentation techniques for satellite images. Arch. Comput. Methods Eng. 2023, 30, 4325–4358. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Cookson, M.S.; Guercio, B.J.; Cheng, L. Advances in diagnosis and treatment of bladder cancer. BMJ 2024, 384, e076743. [Google Scholar] [CrossRef] [PubMed]
- Cochetti, G.; de Vermandois, J.A.R.; Maulà, V.; Cari, L.; Cagnani, R.; Suvieri, C.; Balducci, P.M.; Paladini, A.; Del Zingaro, M.; Nocentini, G.; et al. Diagnostic performance of the Bladder EpiCheck methylation test and photodynamic diagnosis-guided cystoscopy in the surveillance of high-risk non-muscle invasive bladder cancer: A single centre, prospective, blinded clinical trial. In Urologic Oncology: Seminars and Original Investigations; Elsevier: Amsterdam, The Netherlands, 2022; Volume 40, pp. 105.e11–105.e18. [Google Scholar]
- Shamsipour, G.; Fekri-Ershad, S.; Sharifi, M.; Alaei, A. Improve the efficiency of handcrafted features in image retrieval by adding selected feature generating layers of deep convolutional neural networks. Signal Image Video Process. 2024, 18, 2607–2620. [Google Scholar] [CrossRef]
- Fekri-Ershad, S.; Alsaffar, M.F. Developing a Tuned Three-Layer Perceptron Fed with Trained Deep Convolutional Neural Networks for Cervical Cancer Diagnosis. Diagnostics 2023, 13, 686. [Google Scholar] [CrossRef] [PubMed]
- Eroğlu, O.; Yildirim, M. Automatic detection of eardrum otoendoscopic images in patients with otitis media using hybrid-based deep models. Int. J. Imaging Syst. Technol. 2022, 32, 717–727. [Google Scholar] [CrossRef]



| SUBSPACE KNN | QUADRATİC SVM | CUBİC SVM | MEDIUM GAUSSIAN SVM | FINE KNN | SUBSPACE DISCRIMINANT | |
|---|---|---|---|---|---|---|
| DENSENET201 | 99.0 | 97.8 | 98.1 | 97.7 | 98.2 | 97.2 |
| INCEPTIONV3 | 95.8 | 95.3 | 96.5 | 94.9 | 95.9 | 95.0 |
| GOOGLENET | 97.0 | 96.0 | 96.1 | 95.6 | 97.0 | 95.7 |
| NASNETLARGE | 95.4 | 96.1 | 96.2 | 95.2 | 95.7 | 96.0 |
| GOOGLENET-PLACES365 | 96.2 | 95.7 | 96.3 | 94.4 | 96.5 | 93.9 |
| LBP | 97.7 | 95.3 | 96.3 | 95.3 | 97.9 | 82.6 |
| HOG | 94.5 | 92.7 | 94.2 | 90.9 | 94.0 | 89.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yildirim, M. Content-Based Image Retrieval and Image Classification System for Early Prediction of Bladder Cancer. Diagnostics 2024, 14, 2637. https://doi.org/10.3390/diagnostics14232637
Yildirim M. Content-Based Image Retrieval and Image Classification System for Early Prediction of Bladder Cancer. Diagnostics. 2024; 14(23):2637. https://doi.org/10.3390/diagnostics14232637
Chicago/Turabian StyleYildirim, Muhammed. 2024. "Content-Based Image Retrieval and Image Classification System for Early Prediction of Bladder Cancer" Diagnostics 14, no. 23: 2637. https://doi.org/10.3390/diagnostics14232637
APA StyleYildirim, M. (2024). Content-Based Image Retrieval and Image Classification System for Early Prediction of Bladder Cancer. Diagnostics, 14(23), 2637. https://doi.org/10.3390/diagnostics14232637







