Determining Pneumocystis jirovecii Colonisation from Infection Using PCR-Based Diagnostics in HIV-Negative Individuals
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. PCP PCR Assay
3. Analysis
4. Results
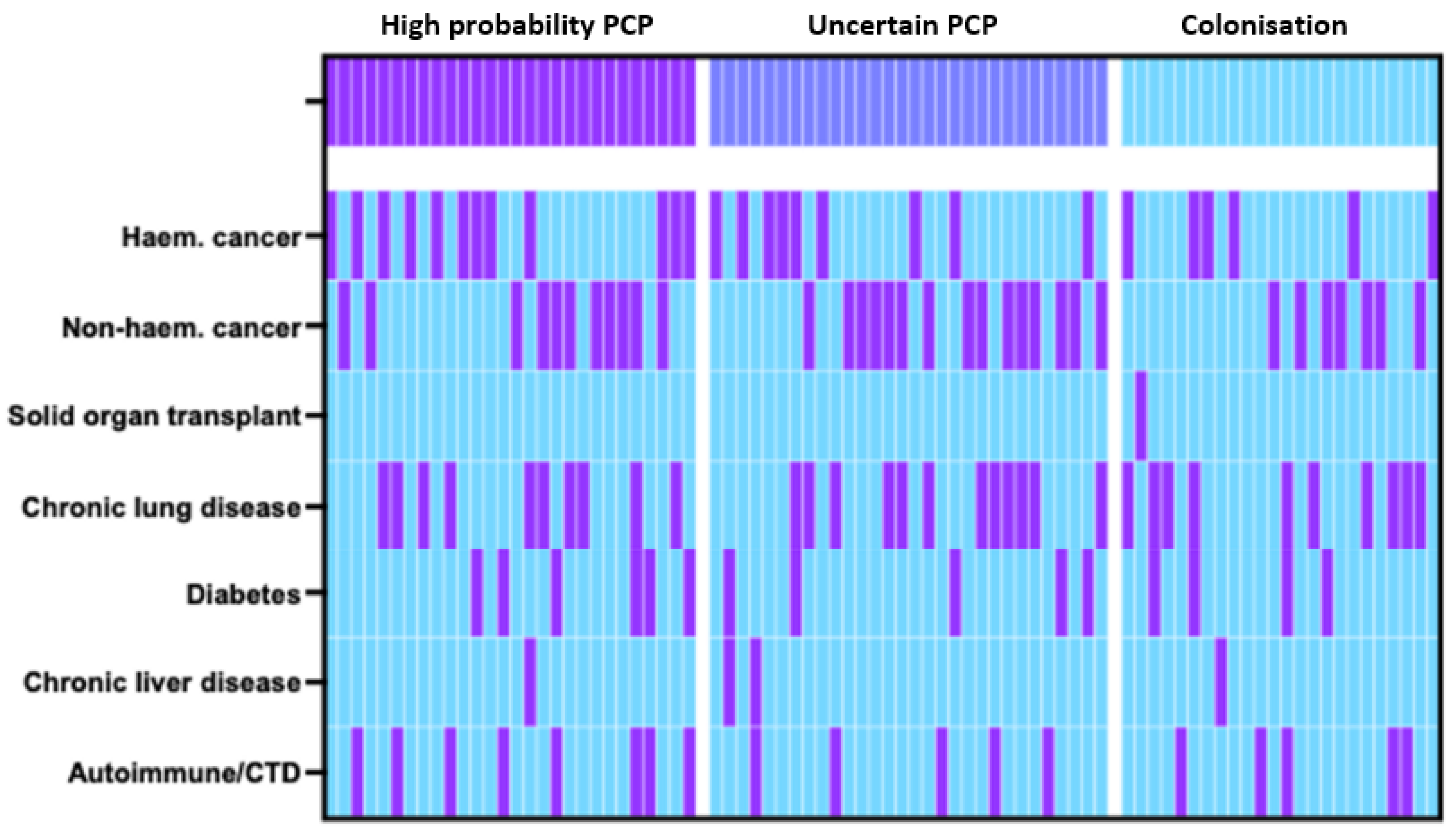
4.1. Study Population and Clinical Characteristics
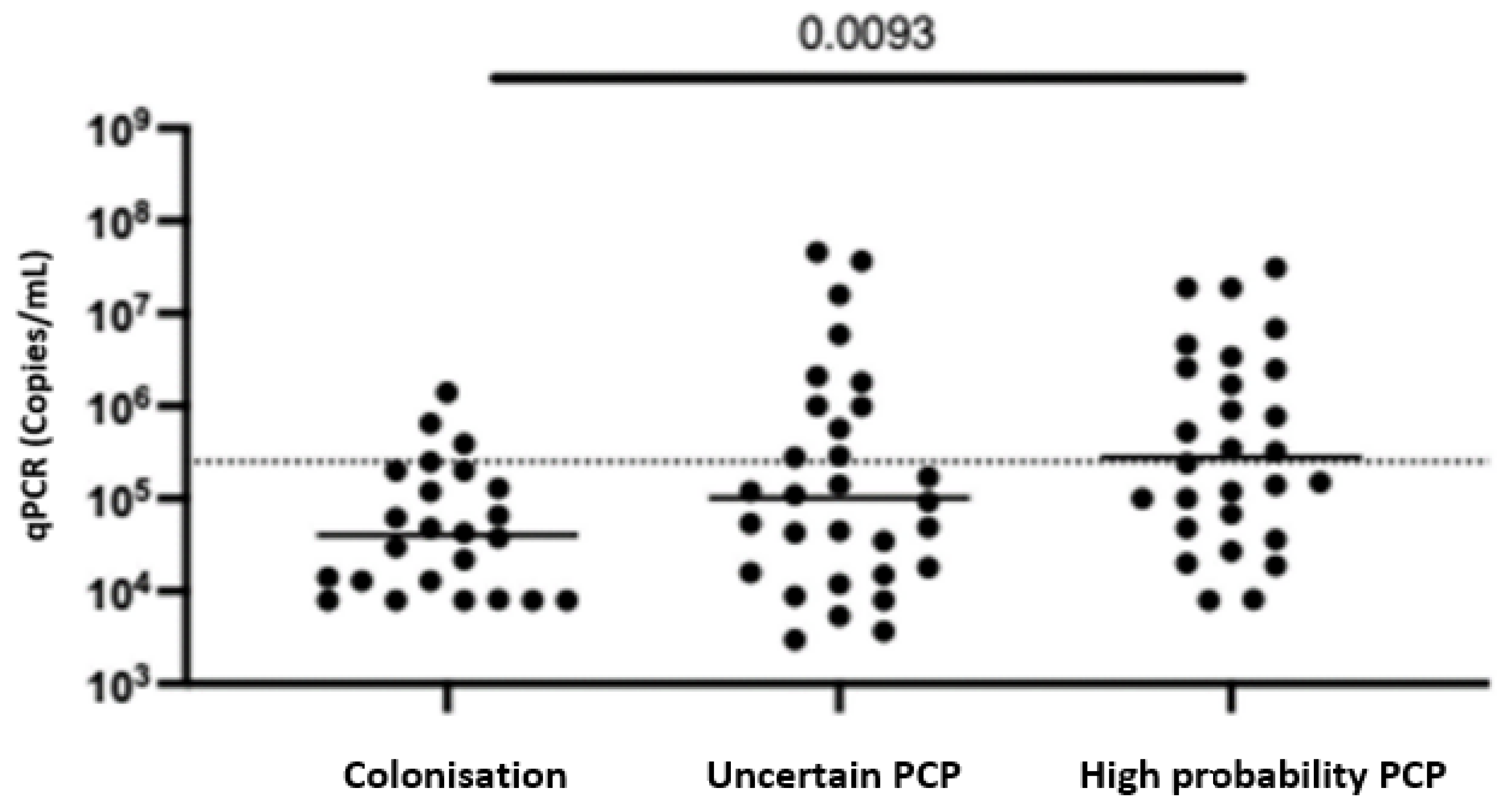
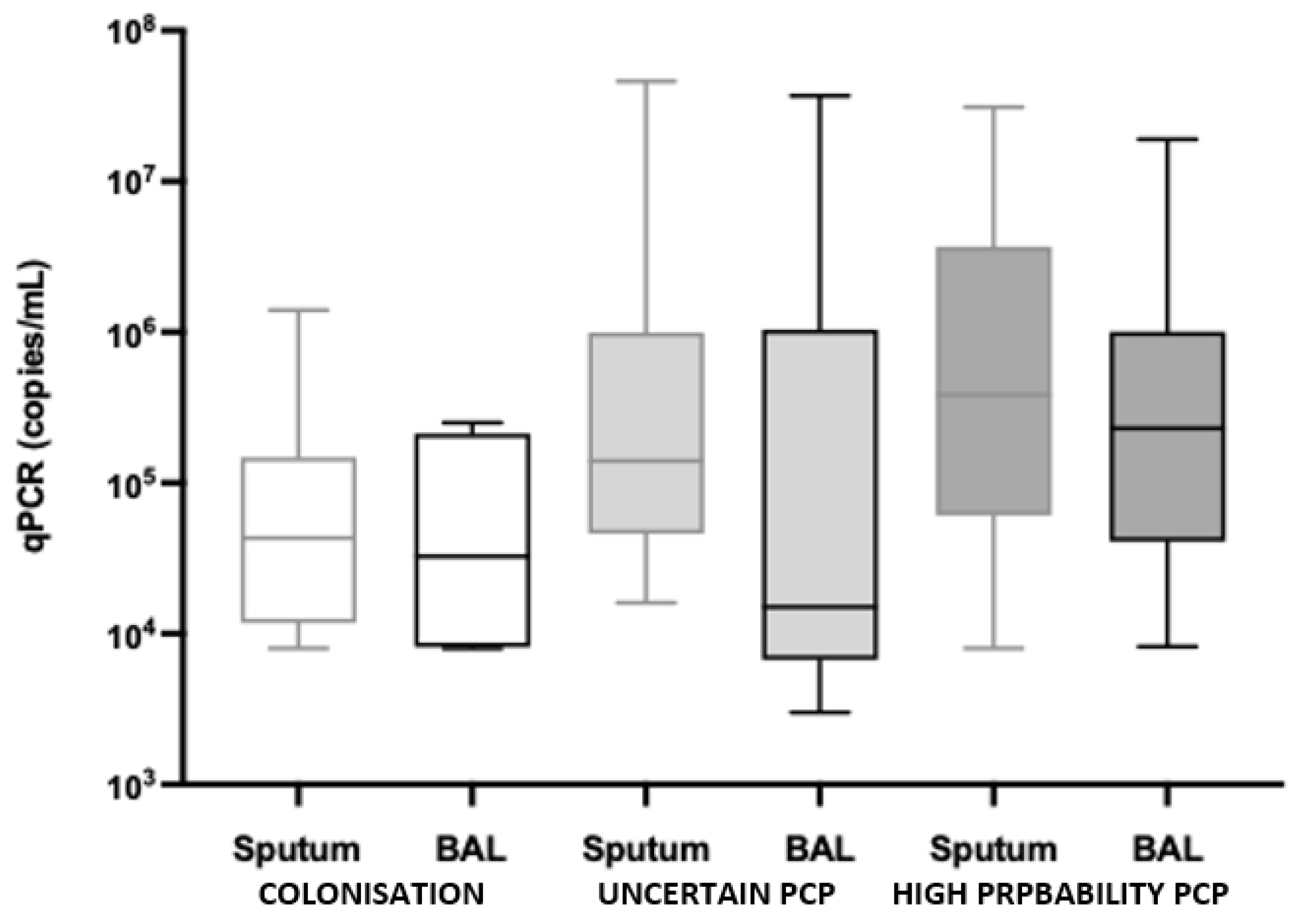
4.2. P. jirovecii Sampling and qPCR Results
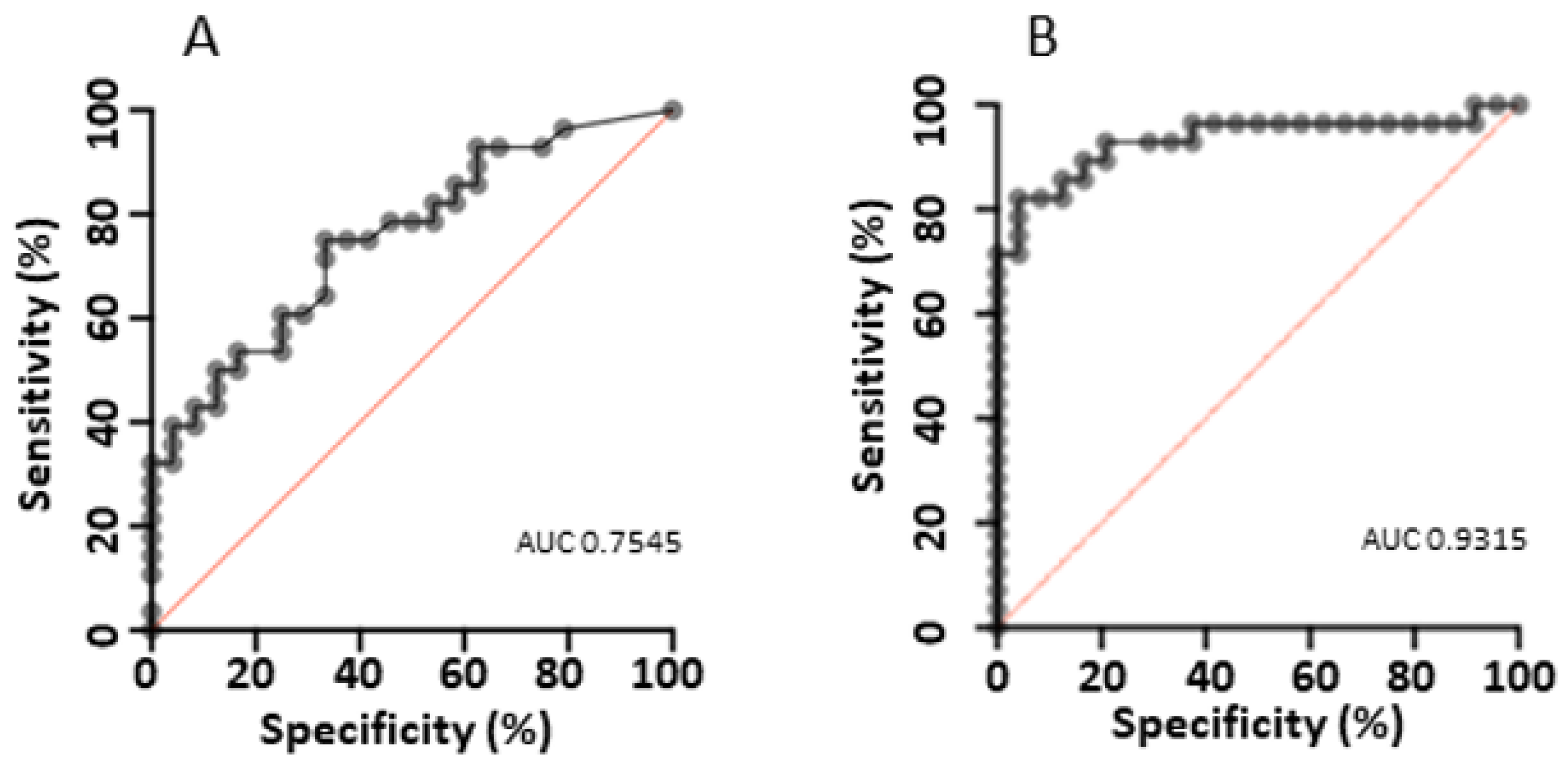
4.3. Assay Performance and Cutoff Optimisation
4.4. Exploratory Analysis
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kolbrink, B.; Scheikholeslami-Sabzewari, J.; Borzikowsky, C.; von Samson-Himmelstjerna, F.A.; Ullmann, A.J.; Kunzendorf, U.; Schulte, K. Evolving epidemiology of pneumocystis pneumonia: Findings from a longitudinal population-based study and a retrospective multi-center study in Germany. Lancet Reg. Health Eur. 2022, 18, 100400. [Google Scholar] [CrossRef] [PubMed]
- Lagrou, K.; Chen, S.; Masur, H.; Viscoli, C.; Decker, C.F.; Pagano, L.; Groll, A.H. Pneumocystis jirovecii Disease: Basis for the Revised EORTC/MSGERC Invasive Fungal Disease Definitions in Individuals without Human Immunodeficiency Virus. Clin. Infect. Dis. 2021, 72 (Suppl. S2), S114–S120. [Google Scholar] [CrossRef] [PubMed]
- Nevez, G.; Raccurt, C.; Vincent, P.; Jounieaux, V.; Dei-Cas, E. Pulmonary colonization with Pneumocystis carinii in human immunodeficiency virus-negative patients: Assessing risk with blood CD4+ T cell counts. Clin. Infect. Dis. 1999, 29, 1331–1332. [Google Scholar] [CrossRef] [PubMed]
- Helweg-Larsen, J.; Jensen, J.S.; Dohn, B.; Benfield, T.L.; Lundgren, B. Detection of Pneumocystis DNA in samples from patients suspected of bacterial pneumonia—A case-control study. BMC Infect. Dis. 2002, 2, 28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Limper, A.H.; Offord, K.P.; Smith, T.F.; Martin, W.J. Pneumocystis carinii pneumonia. Differences in lung parasite number and inflammation in patients with and without AIDS. Am. Rev. Respir. Dis. 1989, 140, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, M.; Marcati, P.; Malena, M.; Bosco, O.; Serpelloni, G.; Mengoli, C. Meta-analysis of diagnostic procedures for Pneumocystis carinii pneumonia in HIV-1-infected patients. Eur. Respir. J. 2002, 20, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.; Vogel, S.; Procop, G. Pneumocystis PCR: It Is Time to Make PCR the Test of Choice. Open Forum Infect. Dis. 2017, 4, ofx193. [Google Scholar] [CrossRef] [PubMed]
- Sing, A.; Trebesius, K.; Roggenkamp, A.; Rüssmann, H.; Tybus, K.; Pfaff, F.; Bogner, J.R.; Emminger, C.; Heesemann, J. Evaluation of diagnostic value and epidemiological implications of PCR for Pneumocystis carinii in different immunosuppressed and immunocompetent patient groups. J. Clin. Microbiol. 2000, 38, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Kitada, K.; Oka, S.; Kimura, S.; Shimada, K.; Serikawa, T.; Yamada, J.; Tsunoo, H.; Egawa, K.; Nakamura, Y. Detection of Pneumocystis carinii sequences by polymerase chain reaction: Animal models and clinical application to noninvasive specimens. J. Clin. Microbiol. 1991, 29, 1985–1990. [Google Scholar] [CrossRef] [PubMed]
- Roblot, F.; Godet, C.; Le Moal, G.; Garo, B.; Faouzi Souala, M.; Dary, M.; De Gentile, L.; Gandji, J.; Guimard, Y.; Lacroix, C.; et al. Analysis of underlying diseases and prognosis factors associated with Pneumocystis carinii pneumonia in immunocompromised HIV-negative patients. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 523–531. [Google Scholar] [PubMed]
- Alanio, A.; Desoubeaux, G.; Sarfati, C.; Hamane, S.; Bergeron, A.; Azoulay, E.; Molina, J.M.; Derouin, F.; Menotti, J. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin. Microbiol. Infect. 2011, 17, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Ito, Y.; Iinuma, Y.; Yasuma, K.; Yamamoto, M.; Matsushima, A.; Nagao, M.; Takakura, S.; Ichiyama, S. Quantitative real-time PCR and the (1 → 3)-β-d-glucan assay for differentiation between Pneumocystis jirovecii pneumonia and colonization. Clin. Microbiol. Infect. 2012, 18, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Teh, B.W.; Azzato, F.A.; Lingaratnam, S.M.; Thursky, K.A.; Slavin, M.A.; Worth, L.J. Molecular diagnosis of Pneumocystis jirovecii in patients with malignancy: Clinical significance of quantitative polymerase chain reaction. Med. Mycol. 2014, 52, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Mühlethaler, K.; Bögli-Stuber, K.; Wasmer, S.; von Garnier, C.; Dumont, P.; Rauch, A.; Mühlemann, K.; Garzoni, C. Quantitative PCR to diagnose Pneumocystis pneumonia in immunocompromised non-HIV patients. Eur. Respir. J. 2012, 39, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Hauser, P.; Lagrou, L.; Melchers, W.J.; Helweg-Larsen, J.; Matos, O.; Cesaro, S.; Maschmeyer, G.; Einsele, H.; Donnelly, J.P.; et al. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J. Antimicrob. Chemother. 2016, 71, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Gits-Muselli, M.; White, P.; Mengoli, C.; Chen, S.; Crowley, B.; Dingemans, G.; Fréalle, E.; Gorton, R.L.; Guiver, M.; Hagen, F.; et al. The Fungal PCR Initiative’s evaluation of in-house and commercial Pneumocystis jirovecii qPCR assays: Toward a standard for a diagnostics assay. Med. Mycol. 2020, 58, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Onishi, A.; Sugiyama, D.; Kogata, Y.; Saegusa, J.; Sugimoto, T.; Kawano, S.; Morinobu, A.; Nishimura, K.; Kumagai, S. Diagnostic accuracy of serum 1,3-β-D-glucan for pneumocystis jiroveci pneumonia, invasive candidiasis, and invasive aspergillosis: Systematic review and meta-analysis. J. Clin. Microbiol. 2012, 50, 7–15. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Colonisation (n = 24) | Uncertain PCP (n = 30) | High-Probability PCP (n = 28) | 3-Way Comparison | 2-Way Comparison (High-Probability PCP vs. Colonised) |
|---|---|---|---|---|---|
| Sex (male) | 70.8% (17/24) | 66.6% (20/30) | 57.1% (16/28) | 0.5641 | 0.3911 |
| Age (median, IQR) | 57 (42–66) | 68 (62–73) | 63 (53–74) | 0.0192 | 0.1137 |
| Sample type (BAL) | 25% (6/24) | 43.3% (13/30) | 35.7% (10/28) | 0.3748 | 0.5487 |
| qPCR (median, IQR) | 40,500 (11,800–147,500) | 101,500 (16,500–885,000) | 280,000 (63,750–2,525,000) | 0.0093 | 0.0013 |
| BAL qPCR | 32,500 (11,650–160,750) | 15,000 (8000–280,000) | 230,000 (66,000–665,000) | 0.1913 | 0.0978 |
| Sputum qPCR | 43,000 (13,000–127,500) | 140,000 (49,000–990,000) | 385,000 (76,750–3,200,000) | 0.0113 | 0.0056 |
| Co-morbidities Haem. cancer | 25% (6/24) | 30% (9/30) | 42.9% (12/28) | 0.3589 | 0.2452 |
| Non-haem. cancer | 29.2% (7/24) | 50% (15/30) | 39.3% (11/28) | 0.2943 | 0.4615 |
| Solid organ transplant | 4.2% (1/24) | 0% (0/30) | 0% (0/28) | 0.2978 | 0.5622 |
| Chronic lung disease | 41.7% (10/24) | 40% (12/30) | 35.7% (10/28) | 0.8997 | 0.7772 |
| Diabetes | 16.7% (4/24) | 16.7% (5/30) | 21.4% (6/28) | - | >0.9999 |
| Diabetes and concurrent steroid use | 25% (1/4) | 20% (1/5) | 66.7% (4/6) | ||
| Chronic liver disease | 4.2% (1/24) | 6.7% (2/30) | 3.6% (1/28) | 0.8453 | >0.9999 |
| Autoimmune/CTD | 20.8% (5/24) | 16.7% (5/30) | 28.6% (8/28) | 0.5426 | 0.749 |
| Immunosuppression medications | 58.3% (14/24) | 80% (24/30) | 67.9% (19/28) | 0.2222 | 0.5685 |
| Steroids ≥ 2 wks > 15 mg/day | 25% (6/24) | 36.7% (11/30) | 28.6% (8/28) | 0.6281 | >0.9999 |
| Prophylaxis | 8.3% (2/24) | 13.3% (4/30) | 17.9% (5/28) | 0.6037 | 0.43 |
| Clinical presentation Fever | 20.8% (5/24) | 46.7% (14/30) | 64.3% (18/28) | 0.0071 | 0.0022 |
| Dyspnoea | 75% (18/24) | 96.7% (29/30) | 85.7% (24/28) | 0.0666 | 0.4829 |
| Cough | 66.7% (16/24) | 73.3% (22/30) | 64.3% (18/28) | 0.7449 | >0.9999 |
| Hypoxia (<95%) | 41.7% (10/24) | 80% (24/30) | 67.9% (19/28) | 0.0125 | 0.0926 |
| Bacterial | 12.5% (3/24) | 40% (12/30) | 0% (0/28) | 0.0003 | 0.0916 |
| Viral | 20.8% (5/24) | 30% (9/30) | 0% (0/28) | 0.0085 | 0.0164 |
| Other infective | 16.7% (4/24) | 10% (3/30) | 0% (0/28) | 0.0941 | 0.0393 |
| Non-infective | 75% (18/24) | 53.3% (16/30) | 0% (0/28) | <0.0001 | <0.0001 |
| Radiological findings | |||||
| Ground glass | 20.8% (5/24) | 60% (18/30) | 67.9% (19/28) | 0.0016 | 0.0009 |
| Reticular opacities | 0% (0/24) | 3.3% (1/30) | 3.6% (1/28) | 0.6532 | >0.9999 |
| Nodules | 25% (6/24) | 20% (6/30) | 21.4% (6/28) | 0.9042 | >0.9999 |
| Consolidation | 45.8% (11/24) | 40% (12/30) | 57.1% (16/28) | 0.4175 | 0.5783 |
| Deceased D+30 | 12.5% (3/24) | 26.7% (8/30) | 17.9% (5/28) | 0.4111 | 0.7109 |
| PCP treatment | 0% (0/24) | 93.3% (28/30) | 100% (28/28) | <0.0001 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watson, A.L.; Woodford, J.; Britton, S.; Gupta, R.; Whiley, D.; McCarthy, K. Determining Pneumocystis jirovecii Colonisation from Infection Using PCR-Based Diagnostics in HIV-Negative Individuals. Diagnostics 2024, 14, 114. https://doi.org/10.3390/diagnostics14010114
Watson AL, Woodford J, Britton S, Gupta R, Whiley D, McCarthy K. Determining Pneumocystis jirovecii Colonisation from Infection Using PCR-Based Diagnostics in HIV-Negative Individuals. Diagnostics. 2024; 14(1):114. https://doi.org/10.3390/diagnostics14010114
Chicago/Turabian StyleWatson, Anna Louise, John Woodford, Sumudu Britton, Rita Gupta, David Whiley, and Kate McCarthy. 2024. "Determining Pneumocystis jirovecii Colonisation from Infection Using PCR-Based Diagnostics in HIV-Negative Individuals" Diagnostics 14, no. 1: 114. https://doi.org/10.3390/diagnostics14010114
APA StyleWatson, A. L., Woodford, J., Britton, S., Gupta, R., Whiley, D., & McCarthy, K. (2024). Determining Pneumocystis jirovecii Colonisation from Infection Using PCR-Based Diagnostics in HIV-Negative Individuals. Diagnostics, 14(1), 114. https://doi.org/10.3390/diagnostics14010114





