Single Ventricle Reconstruction III: Brain Connectome and Neurodevelopmental Outcomes: Design, Recruitment, and Technical Challenges of a Multicenter, Observational Neuroimaging Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Funding
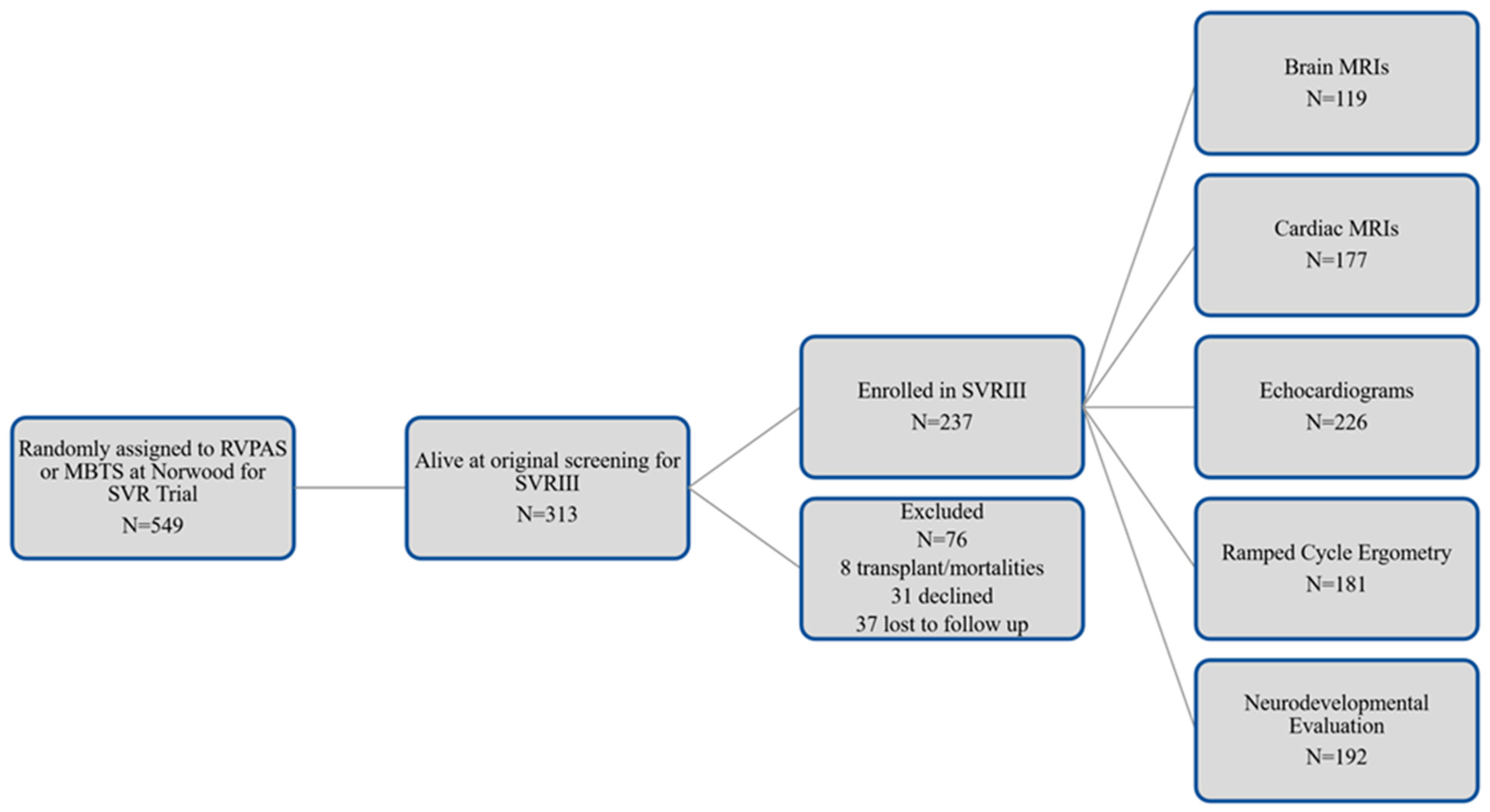
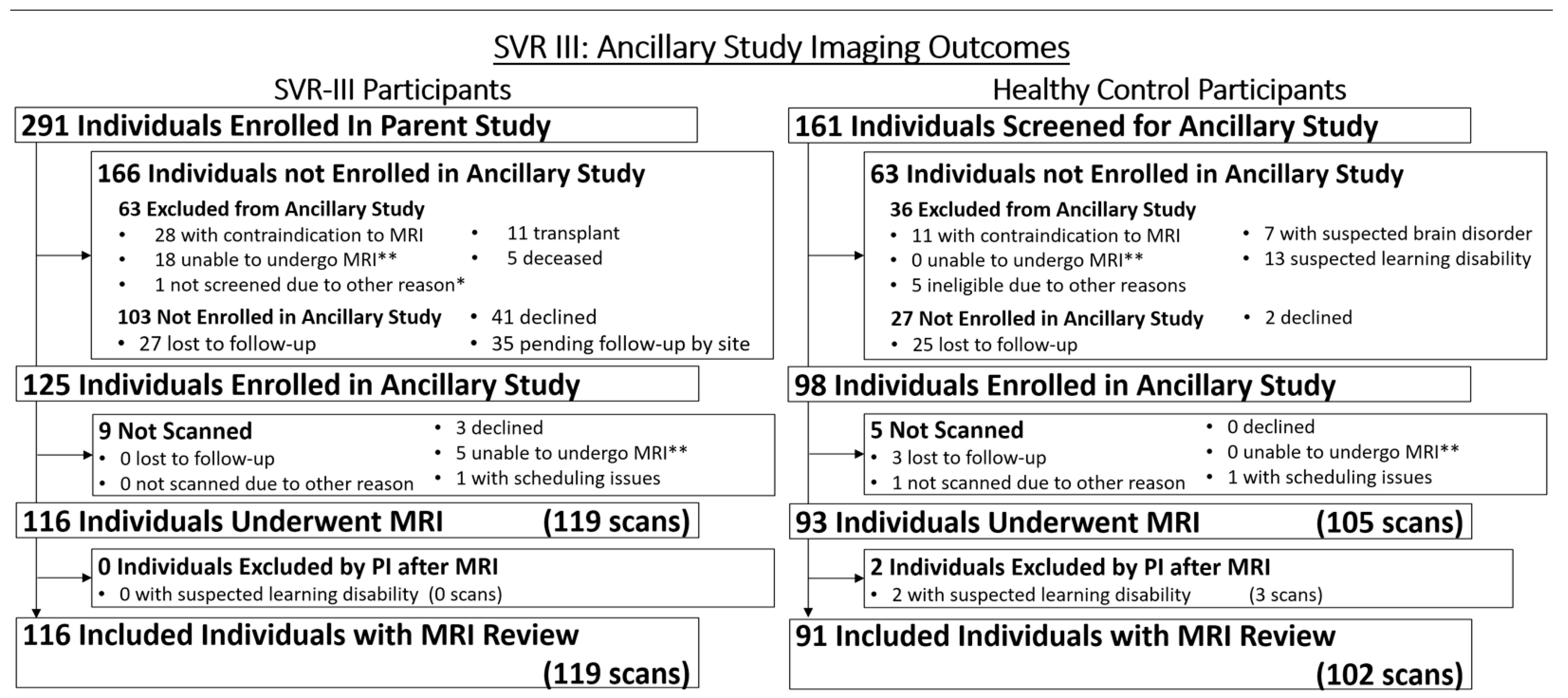
2.2. Screening, Consent, and Entry Criteria for Parent and Ancillary Study
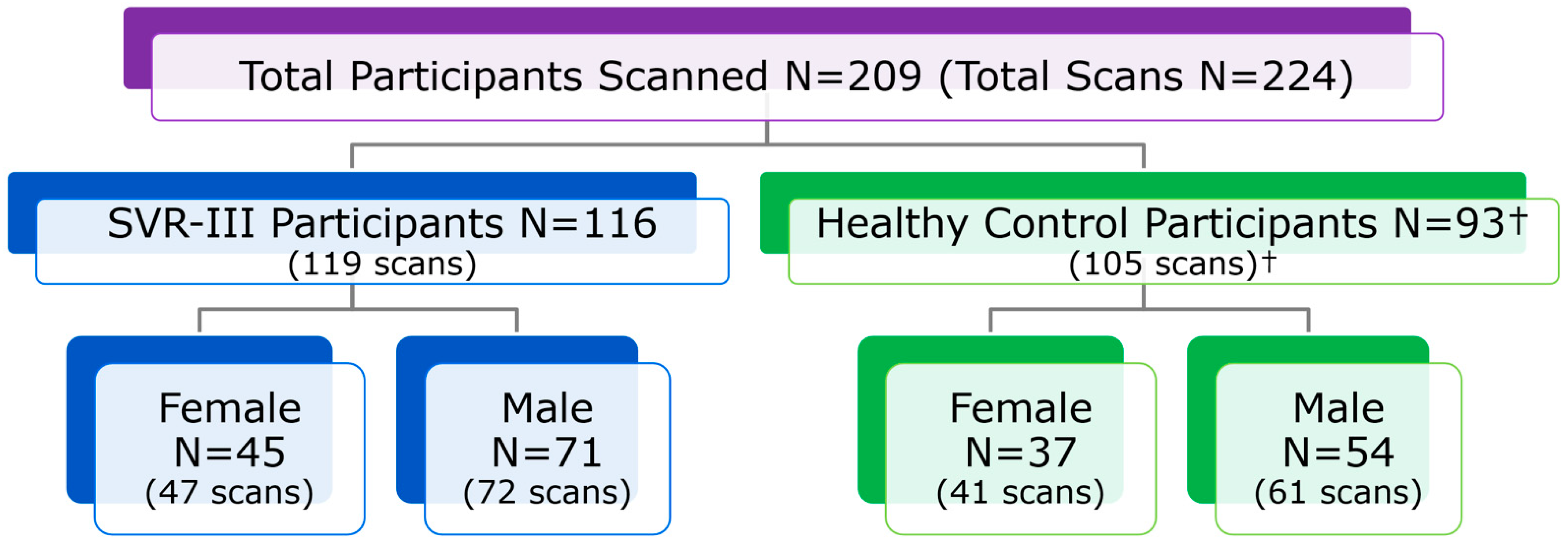
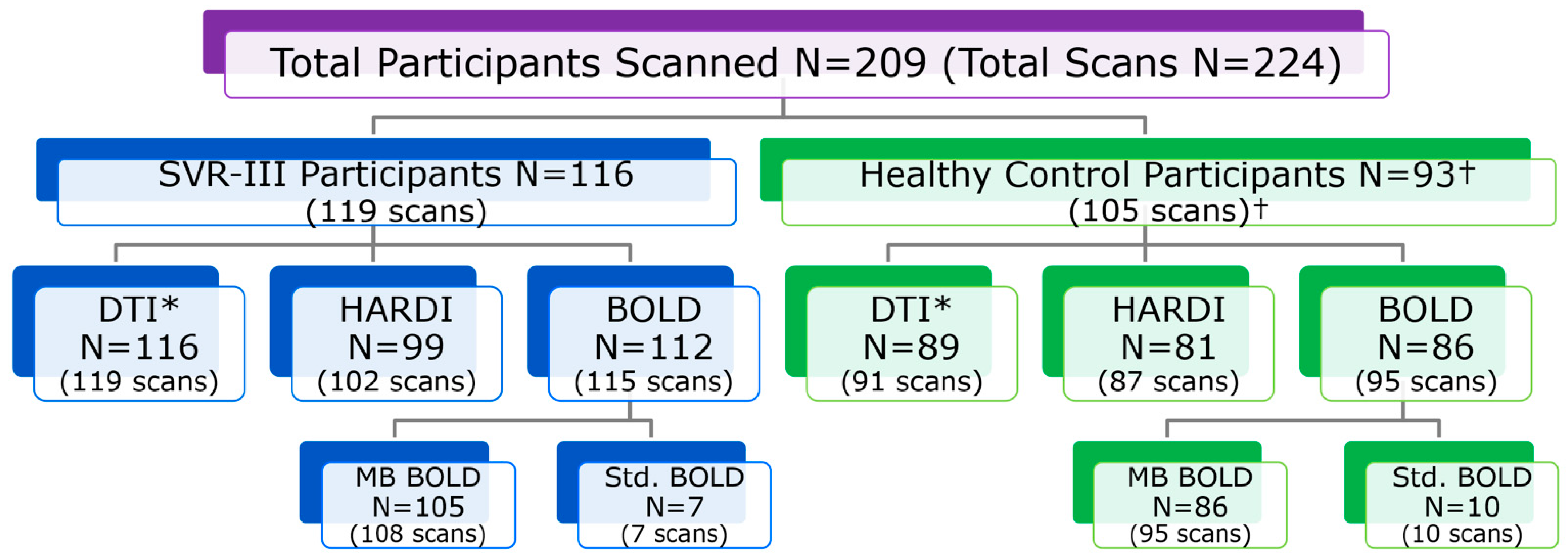
2.3. Sites, Participants and Imaging Acquisition Protocol
2.4. Multicenter MRI Quality Assurance and Quality Control (QA/QC)
2.5. Data Collection, Data Transfer and Participant Data Acceptance
2.6. Measures of Neurodevelopmental and Psychosocial Functioning
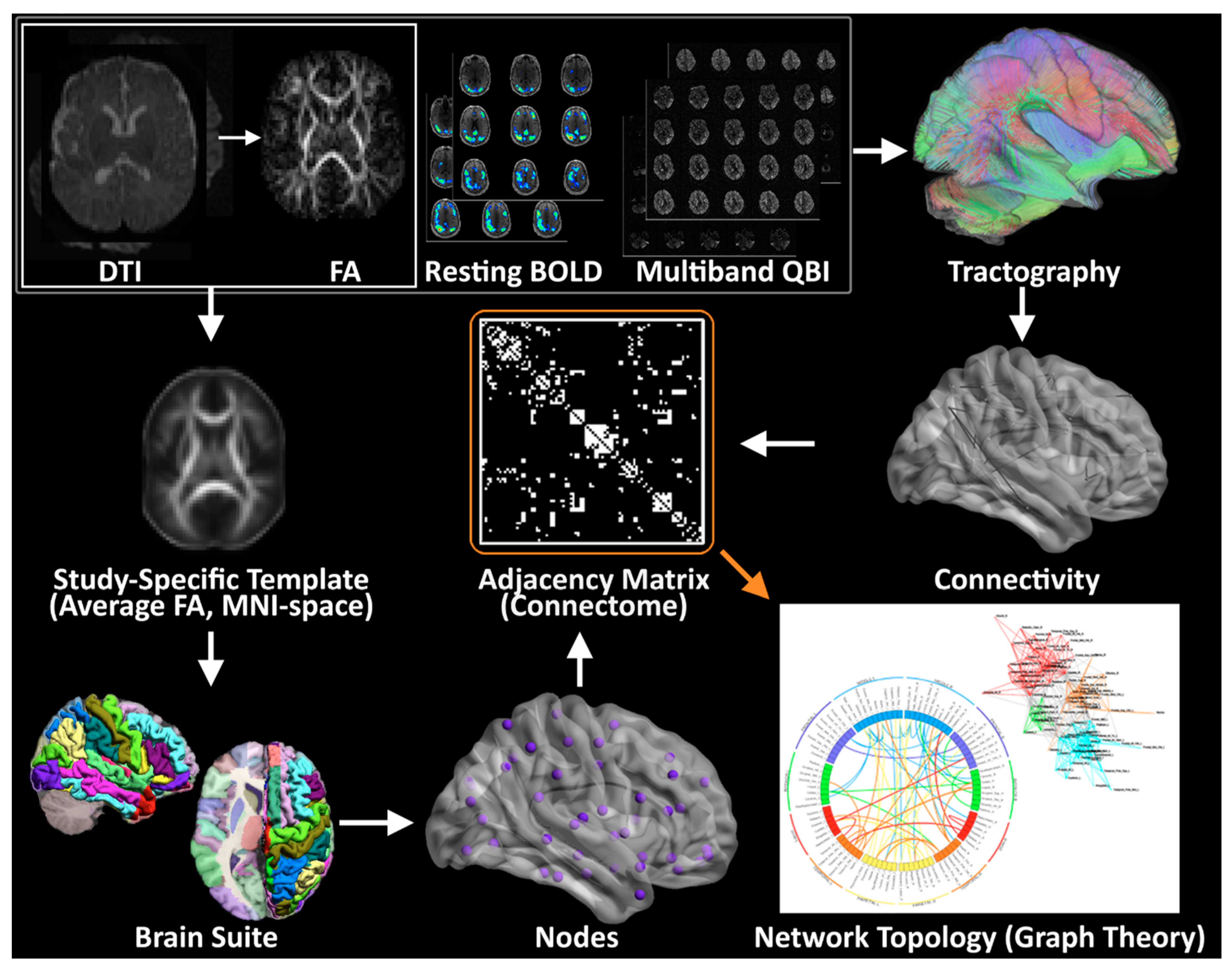
2.7. Planned Imaging Post-Processing
2.8. Planned Brain Connectome Outcome Measures
2.9. Planned Statistical Analysis
2.10. Initial Recruitment Challenges
2.11. Initial Technical Challenges
2.12. Timeline and Impact of COVID-19 Pandemic
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCrindle, B.W.; Williams, R.V.; Mitchell, P.D.; Hsu, D.T.; Paridon, S.M.; Atz, A.M.; Li, J.S.; Newburger, J.W. Relationship of patient and medical characteristics to health status in children and adolescents after the Fontan procedure. Circulation 2006, 113, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Mahle, W.T.; Clancy, R.R.; Moss, E.M.; Gerdes, M.; Jobes, D.R.; Wernovsky, G. Neurodevelopmental outcome and lifestyle assessment in school-aged and adolescent children with hypoplastic left heart syndrome. Pediatrics 2000, 105, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Marelli, A.J.; Ionescu-Ittu, R.; Mackie, A.S.; Guo, L.; Dendukuri, N.; Kaouache, M. Lifetime Prevalence of Congenital Heart Disease in the General Population from 2000 to 2010. Circulation 2014, 130, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, C.S.; Schwartz, E.M.; Brunberg, J.A.; Mosca, R.S.; Bove, E.L.; Schork, M.A.; Stetz, S.P.; Cheatham, J.P.; Kulik, T.J. Neurodevelopmental outcome of patients after the fontan operation: A comparison between children with hypoplastic left heart syndrome and other functional single ventricle lesions. J. Pediatr. 2000, 137, 646–652. [Google Scholar] [CrossRef]
- Tabbutt, S.; Nord, A.S.; Jarvik, G.P.; Bernbaum, J.; Wernovsky, G.; Gerdes, M.; Zackai, E.; Clancy, R.R.; Nicolson, S.C.; Spray, T.L.; et al. Neurodevelopmental outcomes after staged palliation for hypoplastic left heart syndrome. Pediatrics 2008, 121, 476–483. [Google Scholar] [CrossRef]
- Tabbutt, S.; Gaynor, J.W.; Newburger, J.W. Neurodevelopmental outcomes after congenital heart surgery and strategies for improvement. Curr. Opin. Cardiol. 2012, 27, 82–91. [Google Scholar] [CrossRef]
- Wernovsky, G.; Newburger, J. Neurologic and developmental morbidity in children with complex congenital heart disease. J. Pediatr. 2003, 142, 6–8. [Google Scholar] [CrossRef]
- Wernovsky, G.; Stiles, K.M.; Gauvreau, K.; Gentles, T.L.; du Plessis, A.J.; Bellinger, D.C.; Walsh, A.Z.; Burnett, J.; Jonas, R.A.; Mayer, J.E., Jr.; et al. Cognitive development after the Fontan operation. Circulation 2000, 102, 883–889. [Google Scholar] [CrossRef]
- Mellion, K.; Uzark, K.; Cassedy, A.; Drotar, D.; Wernovsky, G.; Newburger, J.W.; Mahony, L.; Mussatto, K.; Cohen, M.; Limbers, C.; et al. Health-related quality of life outcomes in children and adolescents with congenital heart disease. J. Pediatr. 2014, 164, 781–788.e1. [Google Scholar] [CrossRef]
- Shillingford, A.J.; Glanzman, M.M.; Ittenbach, R.F.; Clancy, R.R.; Gaynor, J.W.; Wernovsky, G. Inattention, hyperactivity, and school performance in a population of school-age children with complex congenital heart disease. Pediatrics 2008, 121, e759–e767. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Kathleen, M.; Daniel, L.; Gil, W. Neurodevelopment and quality of life for children with hypoplastic left heart syndrome: Current knowns and unknowns. Cardiol. Young 2011, 21 (Suppl. S2), 88–92. [Google Scholar] [CrossRef]
- Donofrio, M.T.; Duplessis, A.J.; Limperopoulos, C. Impact of congenital heart disease on fetal brain development and injury. Curr. Opin. Pediatr. 2011, 23, 502–511. [Google Scholar] [CrossRef]
- Newburger, J.W.; Sleeper, L.A.; Frommelt, P.C.; Pearson, G.D.; Mahle, W.T.; Chen, S.; Dunbar-Masterson, C.; Mital, S.; Williams, I.A.; Ghanayem, N.S.; et al. Transplantation-free survival and interventions at 3 years in the single ventricle reconstruction trial. Circulation 2014, 129, 2013–2020. [Google Scholar] [CrossRef]
- Nathan, M.; Sleeper, L.A.; Ohye, R.G.; Frommelt, P.C.; Caldarone, C.A.; Tweddell, J.S.; Lu, M.; Pearson, G.D.; Gaynor, J.W.; Pizarro, C.; et al. Technical performance score is associated with outcomes after the Norwood procedure. J. Thorac. Cardiovasc. Surg. 2014, 148, 2208–2214. [Google Scholar] [CrossRef]
- Burch, P.T.; Gerstenberger, E.; Ravishankar, C.; Hehir, D.A.; Davies, R.R.; Colan, S.D.; Sleeper, L.A.; Newburger, J.W.; Clabby, M.L.; Williams, I.A.; et al. Longitudinal assessment of growth in hypoplastic left heart syndrome: Results from the single ventricle reconstruction trial. J. Am. Heart Assoc. 2014, 3, e000079. [Google Scholar] [CrossRef]
- Volpe, J.J. Encephalopathy of congenital heart disease- destructive and developmental effects intertwined. J. Pediatr. 2014, 164, 962–965. [Google Scholar] [CrossRef]
- Mahle, W.T.; Lu, M.; Ohye, R.G.; William Gaynor, J.; Goldberg, C.S.; Sleeper, L.A.; Pemberton, V.L.; Mussatto, K.A.; Williams, I.A.; Sood, E.; et al. A predictive model for neurodevelopmental outcome after the Norwood procedure. Pediatr. Cardiol. 2013, 34, 327–333. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Lu, M.; Sleeper, L.A.; Mahle, W.T.; Gaynor, J.W.; Williams, I.A.; Mussatto, K.A.; Ohye, R.G.; Graham, E.M.; Frank, D.U.; et al. Factors Associated with Neurodevelopment for Children with Single Ventricle Lesions. J. Pediatr. 2014, 165, 490–496. [Google Scholar] [CrossRef]
- Limperopoulos, C.; Tworetzky, W.; McElhinney, D.B.; Newburger, J.W.; Brown, D.W.; Robertson, R.L., Jr.; Guizard, N.; McGrath, E.; Geva, J.; Annese, D.; et al. Brain volume and metabolism in fetuses with congenital heart disease: Evaluation with quantitative magnetic resonance imaging and spectroscopy. Circulation 2010, 121, 26–33. [Google Scholar] [CrossRef]
- Clouchoux, C.; du Plessis, A.J.; Bouyssi-Kobar, M.; Tworetzky, W.; McElhinney, D.B.; Brown, D.W.; Gholipour, A.; Kudelski, D.; Warfield, S.K.; McCarter, R.J.; et al. Delayed cortical development in fetuses with complex congenital heart disease. Cereb. Cortex 2013, 23, 2932–2943. [Google Scholar] [CrossRef]
- Miller, S.P.; McQuillen, P.S.; Hamrick, S.; Xu, D.; Glidden, D.V.; Charlton, N.; Karl, T.; Azakie, A.; Ferriero, D.M.; Barkovich, A.J.; et al. Abnormal brain development in newborns with congenital heart disease. N. Engl. J. Med. 2007, 357, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Licht, D.J.; Shera, D.M.; Clancy, R.R.; Wernovsky, G.; Montenegro, L.M.; Nicolson, S.C.; Zimmerman, R.A.; Spray, T.L.; Gaynor, J.W.; Vossough, A. Brain maturation is delayed in infants with complex congenital heart defects. J. Thorac. Cardiovasc. Surg. 2009, 137, 529–536; Discussion 536–537. [Google Scholar] [CrossRef] [PubMed]
- Paquette, L.B.; Wisnowski, J.L.; Ceschin, R.; Pruetz, J.D.; Detterich, J.A.; Del Castillo, S.; Nagasunder, A.C.; Kim, R.; Painter, M.J.; Gilles, F.H.; et al. Abnormal cerebral microstructure in premature neonates with congenital heart disease. AJNR Am. J. Neuroradiol. 2013, 34, 2026–2033. [Google Scholar] [CrossRef] [PubMed]
- Von Rhein, M.; Buchmann, A.; Hagmann, C.; Huber, R.; Klaver, P.; Knirsch, W.; Latal, B. Brain volumes predict neurodevelopment in adolescents after surgery for congenital heart disease. Brain 2014, 137, 268–276. [Google Scholar] [CrossRef]
- Andropoulos, D.B.; Hunter, J.V.; Nelson, D.P.; Stayer, S.A.; Stark, A.R.; McKenzie, E.D.; Fraser, C.D., Jr. Brain immaturity is associated with brain injury before and after neonatal cardiac surgery with high-flow bypass and cerebral oxygenation monitoring. J. Thorac. Cardiovasc. Surg. 2010, 139, 543–556. [Google Scholar] [CrossRef]
- Dent, C.L.; Spaeth, J.P.; Jones, B.V.; Schwartz, S.M.; Glauser, T.A.; Hallinan, B.; Kurth, C.D. Brain magnetic resonance imaging abnormalities after the Norwood procedure using regional cerebral perfusion. J. Thorac. Cardiovasc. Surg. 2005, 130, 1523–1530. [Google Scholar] [CrossRef]
- Cordina, R.; Grieve, S.; Barnett, M.; Lagopoulos, J.; Malitz, N.; Celermajer, D.S. Brain volumetric, regional cortical thickness and radiographic findings in adults with cyanotic congenital heart disease. Neuroimage Clin. 2014, 4, 319–325. [Google Scholar] [CrossRef]
- McQuillen, P.S.; Miller, S.P. Congenital heart disease and brain development. Ann. New York Acad. Sci. 2010, 1184, 68–86. [Google Scholar] [CrossRef]
- Rollins, C.K.; Watson, C.G.; Asaro, L.A.; Wypij, D.; Vajapeyam, S.; Bellinger, D.C.; DeMaso, D.R.; Robertson, R.L., Jr.; Newburger, J.W.; Rivkin, M.J. White Matter Microstructure and Cognition in Adolescents with Congenital Heart Disease. J. Pediatr. 2014, 165, 936–944. [Google Scholar] [CrossRef]
- Bullmore, E.; Sporns, O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009, 10, 186–198. [Google Scholar] [CrossRef]
- Bullmore, E.; Sporns, O. The economy of brain network organization. Nat. Rev. Neurosci. 2012, 13, 336–349. [Google Scholar] [CrossRef]
- Griffa, A.; Baumann, P.S.; Thiran, J.-P.; Hagmann, P. Structural connectomics in brain diseases. Neuroimage 2013, 80, 515–526. [Google Scholar] [CrossRef]
- Sporns, O. Network attributes for segregation and integration in the human brain. Curr. Opin. Neurobiol. 2013, 23, 162–171. [Google Scholar] [CrossRef]
- Bullmore, E.T.; Bassett, D.S. Brain graphs: Graphical models of the human brain connectome. Annu. Rev. Clin. Psychol. 2011, 7, 113–140. [Google Scholar] [CrossRef]
- Fornito, A.; Zalesky, A.; Breakspear, M. Graph analysis of the human connectome: Promise, progress, and pitfalls. Neuroimage 2013, 80, 426–444. [Google Scholar] [CrossRef]
- Hagmann, P.; Kurant, M.; Gigandet, X.; Thiran, P.; Wedeen, V.J.; Meuli, R.; Thiran, J.P. Mapping human whole-brain structural networks with diffusion MRI. PLoS ONE 2007, 2, e597. [Google Scholar] [CrossRef]
- Bellinger, D.C.; Jonas, R.A.; Rappaport, L.A.; Wypij, D.; Wernovsky, G.; Kuban, K.C.; Barnes, P.D.; Holmes, G.L.; Hickey, P.R.; Strand, R.D.; et al. Developmental and neurologic status of children after heart surgery with hypothermic circulatory arrest or low-flow cardiopulmonary bypass. N. Engl. J. Med. 1995, 332, 549–555. [Google Scholar] [CrossRef]
- Bellinger, D.C.; Newburger, J.W.; Wypij, D.; Kuban, K.C.; du Plesssis, A.J.; Rappaport, L.A. Behaviour at eight years in children with surgically corrected transposition: The Boston Circulatory Arrest Trial. Cardiol. Young 2009, 19, 86–97. [Google Scholar] [CrossRef]
- Bellinger, D.C.; Wypij, D.; du Plessis, A.J.; Rappaport, L.A.; Jonas, R.A.; Wernovsky, G.; Newburger, J.W. Neurodevelopmental status at eight years in children with dextro-transposition of the great arteries: The Boston Circulatory Arrest Trial. J. Thorac. Cardiovasc. Surg. 2003, 126, 1385–1396. [Google Scholar] [CrossRef]
- Bellinger, D.C.; Wypij, D.; Rivkin, M.J.; DeMaso, D.R.; Robertson, R.L., Jr.; Dunbar-Masterson, C.; Rappaport, L.A.; Wernovsky, G.; Jonas, R.A.; Newburger, J.W. Adolescents with d-transposition of the great arteries corrected with the arterial switch procedure: Neuropsychological assessment and structural brain imaging. Circulation 2011, 124, 1361–1369. [Google Scholar] [CrossRef]
- Bhroin, M.N.; Seada, S.A.; Bonthrone, A.F.; Kelly, C.J.; Christiaens, D.; Schuh, A.; Pietsch, M.; Hutter, J.; Tournier, J.-D.; Cordero-Grande, L. Reduced structural connectivity in cortico-striatal-thalamic network in neonates with congenital heart disease. NeuroImage Clin. 2020, 28, 102423. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, M.; Guo, T.; Miller, S.P.; Knirsch, W.; Kottke, R.; Hagmann, C.; Latal, B.; Jakab, A. Delayed maturation of the structural brain connectome in neonates with congenital heart disease. Brain Commun. 2020, 2, fcaa209. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Ferdman, D.; Copel, J.; Scheinost, D.; Shabanova, V.; Brueckner, M.; Khokha, M.K.; Ment, L.R. De novo damaging variants associated with congenital heart diseases contribute to the connectome. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.; Peyvandi, S.; Cox, S.; Gano, D.; Xu, D.; Tymofiyeva, O.; McQuillen, P.S. Neonatal brain injury influences structural connectivity and childhood functional outcomes. PLoS ONE 2022, 17, e0262310. [Google Scholar] [CrossRef]
- Schmithorst, V.J.; Votava-Smith, J.K.; Tran, N.; Kim, R.; Lee, V.; Ceschin, R.; Lai, H.; Johnson, J.A.; De Toledo, J.S.; Blüml, S. Structural network topology correlates of microstructural brain dysmaturation in term infants with congenital heart disease. Hum. Brain Mapp. 2018, 39, 4593–4610. [Google Scholar] [CrossRef]
- Panigrahy, A.; Schmithorst, V.J.; Wisnowski, J.L.; Watson, C.G.; Bellinger, D.C.; Newburger, J.W.; Rivkin, M.J. Relationship of white matter network topology and cognitive outcome in adolescents with d-transposition of the great arteries. Neuroimage Clin. 2015, 7, 438–448. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Gaynor, J.W.; Mahle, W.T.; Ravishankar, C.; Frommelt, P.; Ilardi, D.; Bellinger, D.; Paridon, S.; Taylor, M.; Hill, K.D. The pediatric heart network’s study on long-term outcomes of children with HLHS and the impact of Norwood Shunt type in the single ventricle reconstruction trial cohort (SVRIII): Design and adaptations. Am. Heart J. 2022, 254, 216–227. [Google Scholar] [CrossRef]
- Hagmann, P.; Cammoun, L.; Gigandet, X.; Gerhard, S.; Ellen Grant, P.; Wedeen, V.; Meuli, R.; Thiran, J.-P.; Honey, C.J.; Sporns, O. MR connectomics: Principles and challenges. J. Neurosci. Methods 2010, 194, 34–45. [Google Scholar] [CrossRef]
- Hagmann, P.; Cammoun, L.; Gigandet, X.; Meuli, R.; Honey, C.J.; Wedeen, V.J.; Sporns, O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008, 6, e159. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, Y.; Sheng, J.; Kronenberger, W.G.; Mathews, V.P.; Hummer, T.A.; Saykin, A.J. Characteristics and variability of structural networks derived from diffusion tensor imaging. Neuroimage 2012, 61, 1153–1164. [Google Scholar] [CrossRef]
- Achard, S.; Salvador, R.; Whitcher, B.; Suckling, J.; Bullmore, E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 2006, 26, 63–72. [Google Scholar] [CrossRef]
- Biswal, B.; Zerrin Yetkin, F.; Haughton, V.M.; Hyde, J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995, 34, 537–541. [Google Scholar] [CrossRef]
- Cordes, D.; Haughton, V.M.; Arfanakis, K.; Carew, J.D.; Turski, P.A.; Moritz, C.H.; Quigley, M.A.; Meyerand, M.E. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. Am. J. Neuroradiol. 2001, 22, 1326–1333. [Google Scholar]
- Cordes, D.; Haughton, V.M.; Arfanakis, K.; Wendt, G.J.; Turski, P.A.; Moritz, C.H.; Quigley, M.A.; Meyerand, M.E. Mapping functionally related regions of brain with functional connectivity MR imaging. Am. J. Neuroradiol. 2000, 21, 1636–1644. [Google Scholar]
- Lowe, M.; Mock, B.; Sorenson, J. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. Neuroimage 1998, 7, 119–132. [Google Scholar] [CrossRef]
- Casey, B.J.; Cannonier, T.; Conley, M.I.; Cohen, A.O.; Barch, D.M.; Heitzeg, M.M.; Soules, M.E.; Teslovich, T.; Dellarco, D.V.; Garavan, H. The adolescent brain cognitive development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018, 32, 43–54. [Google Scholar] [CrossRef]
- Owen, J.P.; Ziv, E.; Bukshpun, P.; Pojman, N.; Wakahiro, M.; Berman, J.I.; Roberts, T.P.; Friedman, E.J.; Sherr, E.H.; Mukherjee, P. Test–Retest Reliability of Computational Network Measurements Derived from the Structural Connectome of the Human Brain. Brain Connect. 2013, 3, 160–176. [Google Scholar] [CrossRef]
- Walker, L.; Curry, M.; Nayak, A.; Lange, N.; Pierpaoli, C. A framework for the analysis of phantom data in multicenter diffusion tensor imaging studies. Hum. Brain Mapp. 2013, 34, 2439–2454. [Google Scholar] [CrossRef]
- Mulkern, R.V.; Forbes, P.; Dewey, K.; Osganian, S.; Clark, M.; Wong, S.; Ramamurthy, U.; Kun, L.; Poussaint, T.Y. Establishment and Results of a Magnetic Resonance Quality Assurance Program for the Pediatric Brain Tumor Consortium. Acad. Radiol. 2008, 15, 1099–1110. [Google Scholar] [CrossRef]
- Poussaint, T.Y.; Phillips, P.C.; Vajapeyam, S.; Fahey, F.H.; Robertson, R.L.; Osganian, S.; Ramamurthy, U.; Mulkern, R.V.; Treves, S.T.; Boyett, J.M.; et al. The Neuroimaging Center of the Pediatric Brain Tumor Consortium-collaborative neuroimaging in pediatric brain tumor research: A work in progress. AJNR Am. J. Neuroradiol. 2007, 28, 603–607. [Google Scholar]
- Marcus, D.S.; Harms, M.P.; Snyder, A.Z.; Jenkinson, M.; Wilson, J.A.; Glasser, M.F.; Barch, D.M.; Archie, K.A.; Burgess, G.C.; Ramaratnam, M.; et al. Human Connectome Project informatics: Quality control, database services, and data visualization. Neuroimage 2013, 80, 202–219. [Google Scholar] [CrossRef] [PubMed]
- Elam, J.S.; Van Essen, D. Human Connectome Project, Encyclopedia of Computational Neuroscience; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–4. [Google Scholar]
- Sotiropoulos, S.N.; Jbabdi, S.; Xu, J.; Andersson, J.L.; Moeller, S.; Auerbach, E.J.; Glasser, M.F.; Hernandez, M.; Sapiro, G.; Jenkinson, M. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage 2013, 80, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Van Essen, D.C.; Smith, S.M.; Barch, D.M.; Behrens, T.E.; Yacoub, E.; Ugurbil, K. The WU-Minn human connectome project: An overview. Neuroimage 2013, 80, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.-N.; Xing, X.-X. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: A systems neuroscience perspective. Neurosci. Biobehav. Rev. 2014, 45, 100–118. [Google Scholar] [CrossRef]
- Dams-O’Connor, K.; Spielman, L.; Singh, A.; Gordon, W.A.; Lingsma, H.F.; Maas, A.I.; Manley, G.T.; Mukherjee, P.; Okonkwo, D.O.; Puccio, A.M. The Impact of Previous Traumatic Brain Injury on Health and Functioning: A TRACK-TBI Study. J. Neurotrauma 2013, 30, 2014–2020. [Google Scholar] [CrossRef]
- Yue, J.K.; Vassar, M.J.; Lingsma, H.F.; Cooper, S.R.; Okonkwo, D.O.; Valadka, A.B.; Gordon, W.A.; Maas, A.I.; Mukherjee, P.; Yuh, E.L. Transforming research and clinical knowledge in traumatic brain injury pilot: Multicenter implementation of the common data elements for traumatic brain injury. J. Neurotrauma 2013, 30, 1831–1844. [Google Scholar] [CrossRef]
- Yuh, E.L.; Cooper, S.R.; Mukherjee, P.; Yue, J.K.; Lingsma, H.; Gordon, W.; Valadka, A.; Okonkwo, D.O.; Schnyer, D.M.; Vassar, M.J. Diffusion Tensor Imaging for Outcome Prediction in Mild Traumatic Brain Injury: A TRACK-TBI Study. J. Neurotrauma 2014, 31, 1457–1477. [Google Scholar] [CrossRef]
- Keenan, K. Quantitative Magnetic Resonance Imaging and Phantom Development. Bull. Am. Phys. Soc. 2014, 59, Y38-001. [Google Scholar]
- Selwyn, R. Phantoms for Magnetic Resonance Imaging, The Phantoms of Medical and Health Physics; Springer: Berlin/Heidelberg, Germany, 2014; pp. 181–199. [Google Scholar]
- Vannier, M.W. Traumatic Brain Injury Diffusion Magnetic Resonance Imaging Research Roadmap Development Project; DTIC Document; Chicago University: Chicago, IL, USA, 2010. [Google Scholar]
- Glover, G.H.; Mueller, B.A.; Turner, J.A.; van Erp, T.G.; Liu, T.T.; Greve, D.N.; Voyvodic, J.T.; Rasmussen, J.; Brown, G.G.; Keator, D.B. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. J. Magn. Reson. Imaging 2012, 36, 39–54. [Google Scholar] [CrossRef]
- Friedman, L.; Glover, G.H. Report on a multicenter fMRI quality assurance protocol. J. Magn. Reson. Imaging 2006, 23, 827–839. [Google Scholar] [CrossRef]
- Friedman, L.; Glover, G.H.; Consortium, F. Reducing interscanner variability of activation in a multicenter fMRI study: Controlling for signal-to-fluctuation-noise-ratio (SFNR) differences. Neuroimage 2006, 33, 471–481. [Google Scholar] [CrossRef]
- Friedman, L.; Glover, G.H.; Krenz, D.; Magnotta, V. Reducing inter-scanner variability of activation in a multicenter fMRI study: Role of smoothness equalization. Neuroimage 2006, 32, 1656–1668. [Google Scholar] [CrossRef]
- Friedman, L.; Stern, H.; Brown, G.G.; Mathalon, D.H.; Turner, J.; Glover, G.H.; Gollub, R.L.; Lauriello, J.; Lim, K.O.; Cannon, T. Test–retest and between-site reliability in a multicenter fMRI study. Hum. Brain Mapp. 2008, 29, 958–972. [Google Scholar] [CrossRef]
- Keator, D.B.; Grethe, J.S.; Marcus, D.; Ozyurt, B.; Gadde, S.; Murphy, S.; Pieper, S.; Greve, D.; Notestine, R.; Bockholt, H.J. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). Inf. Technol. Biomed. IEEE Trans. 2008, 12, 162–172. [Google Scholar] [CrossRef]
- Marcus, D.S.; Harwell, J.; Olsen, T.; Hodge, M.; Glasser, M.F.; Prior, F.; Jenkinson, M.; Laumann, T.; Curtiss, S.W.; Van Essen, D.C. Informatics and data mining tools and strategies for the human connectome project. Front. Neuroinform. 2011, 5, 4. [Google Scholar] [CrossRef]
- Marcus, D.S.; Olsen, T.R.; Ramaratnam, M.; Buckner, R.L. The extensible neuroimaging archive toolkit. Neuroinformatics 2007, 5, 11–33. [Google Scholar] [CrossRef]
- Haldar, J.P.; Leahy, R.M. Linear transforms for Fourier data on the sphere: Application to high angular resolution diffusion MRI of the brain. Neuroimage 2013, 71, 233–247. [Google Scholar] [CrossRef]
- Shattuck, D.W.; Prasad, G.; Mirza, M.; Narr, K.L.; Toga, A.W. Online resource for validation of brain segmentation methods. Neuroimage 2009, 45, 431–439. [Google Scholar] [CrossRef]
- Shattuck, D.W.; Leahy, R.M. BrainSuite: An automated cortical surface identification tool. Med. Image Anal. 2002, 6, 129–142. [Google Scholar] [CrossRef]
- Joshi, A.A.; Pantazis, D.; Li, Q.; Damasio, H.; Shattuck, D.W.; Toga, A.W.; Leahy, R.M. Sulcal set optimization for cortical surface registration. Neuroimage 2010, 50, 950–959. [Google Scholar] [CrossRef]
- Fennema-Notestine, C.; Ozyurt, I.B.; Clark, C.P.; Morris, S.; Bischoff-Grethe, A.; Bondi, M.W.; Jernigan, T.L.; Fischl, B.; Segonne, F.; Shattuck, D.W.; et al. Quantitative evaluation of automated skull-stripping methods applied to contemporary and legacy images: Effects of diagnosis, bias correction, and slice location. Hum. Brain Mapp. 2006, 27, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.A.; Shattuck, D.W.; Thompson, P.M.; Leahy, R.M. Surface-constrained volumetric brain registration using harmonic mappings. Med. Imaging IEEE Trans. 2007, 26, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.A.; Shattuck, D.W.; Thompson, P.M.; Leahy, R.M. A parameterization-based numerical method for isotropic and anisotropic diffusion smoothing on non-flat surfaces. Image Process. IEEE Trans. 2009, 18, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Pantazis, D.; Joshi, A.; Jiang, J.; Shattuck, D.W.; Bernstein, L.E.; Damasio, H.; Leahy, R.M. Comparison of landmark-based and automatic methods for cortical surface registration. Neuroimage 2010, 49, 2479–2493. [Google Scholar] [CrossRef]
- Malykhin, N.; Concha, L.; Seres, P.; Beaulieu, C.; Coupland, N.J. Diffusion tensor imaging tractography and reliability analysis for limbic and paralimbic white matter tracts. Psychiatry Res. Neuroimaging 2008, 164, 132–142. [Google Scholar] [CrossRef]
- Jiang, H.; van Zijl, P.; Kim, J.; Pearlson, G.D.; Mori, S. DtiStudio: Resource program for diffusion tensor computation and fiber bundle tracking. Comput. Methods Programs Biomed. 2006, 81, 106–116. [Google Scholar] [CrossRef]
- Yeh, F.-C.; Wedeen, V.J.; Tseng, W.-Y.I. Estimation of fiber orientation and spin density distribution by diffusion deconvolution. Neuroimage 2011, 55, 1054–1062. [Google Scholar] [CrossRef]
- Power, J.D.; Mitra, A.; Laumann, T.O.; Snyder, A.Z.; Schlaggar, B.L.; Petersen, S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 2014, 84, 320–341. [Google Scholar] [CrossRef]
- Yan, C.-G.; Craddock, R.C.; He, Y.; Milham, M.P. Addressing head motion dependencies for small-world topologies in functional connectomics. Front. Hum. Neurosci. 2013, 7, 910. [Google Scholar] [CrossRef]
- Achard, S.; Bullmore, E. Efficiency and cost of economical brain functional networks. PLoS Comput. Biol. 2007, 3, e17. [Google Scholar] [CrossRef]
- Xia, M.; Wang, J.; He, Y. BrainNet Viewer: A network visualization tool for human brain connectomics. PLoS ONE 2013, 8, e68910. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Ginestet, C.E.; Nichols, T.E.; Bullmore, E.T.; Simmons, A. Brain network analysis: Separating cost from topology using cost-integration. PLoS ONE 2011, 6, e21570. [Google Scholar] [CrossRef]
- Fruchterman, T.M.; Reingold, E.M. Graph drawing by force-directed placement. Softw. Pract. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Schmithorst, V.J.; Badaly, D.; Beers, S.R.; Lee, V.K.; Weinberg, J.; Lo, C.W.; Panigrahy, A. Relationships between Regional Cerebral Blood Flow and Neurocognitive Outcomes in Children and Adolescents with Congenital Heart Disease, Seminars in Thoracic and Cardiovascular Surgery; Elsevier: Amsterdam, The Netherlands, 2021; Volume 34, pp. 1285–1295. [Google Scholar]
- DiCiccio, T.J.; Efron, B. Bootstrap Confidence Intervals. Stat. Sci. 1996, 11, 189–228. [Google Scholar] [CrossRef]
- Hayes, A.F.; Scharkow, M. The relative trustworthiness of inferential tests of the indirect effect in statistical mediation analysis: Does method really matter? Psychol. Sci. 2013, 24, 1918–1927. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. On the Adaptive Control of the False Discovery Rate in Multiple Testing with Independent Statistics. J. Educ. Behav. Stat. 2000, 25, 60–83. [Google Scholar] [CrossRef]
- Meyers, B.D.; Lee, V.K.; Dennis, L.G.; Wallace, J.; Schmithorst, V.; Votava-Smith, J.K.; Rajagopalan, V.; Herrup, E.; Baust, T.; Tran, N.N. Harmonization of Multi-Center Diffusion Tensor Tractography in Neonates with Congenital Heart Disease: Optimizing Post-Processing and Application of ComBat. Neuroimage Rep. 2022, 2, 100114. [Google Scholar] [CrossRef]


| 8AM | 9AM | 10AM | 11AM | 12PM | 1PM | 2PM | 3PM | 4PM | |
|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Ramped Cycle Exercise Test | Cardiac MRI | Lunch | Echo | |||||
| Day 2 | Neurodevelopmental Testing | Lunch | ND Testing (Cont’d) | Brain MRI | |||||
| Domain | Instruments and Subtests | Completion Time- Child | Completion Time- Respondent |
|---|---|---|---|
| Intelligence | Wechsler Intelligence Scale for Children-V (WISC-V; original test kit) Block design Similarities Matrix reasoning Digit span forward Backward Sequencing Coding Vocabulary Figure weights Visual puzzles Picture span Symbol search | 65–80 min | |
| Math | Wechsler Individual Achievement Tests III (WIAT-III) Math problem solving Numerical operations | 30 min | |
| Reading | Wechsler Individual Achievement Tests III (WIAT-III) Word reading Reading comprehension Pseudoword decoding | 32 min | |
| Language | NEPSY-II Comprehension of instructions Oromotor sequence | 13 min | |
| Executive Function and Attention | Delis-Kaplan Executive Function System (DKEFS) Tower Trail making (all 5 Trials) Verbal fluency (Letter, Category, Switching) Behavior Rating Inventory of Executive Functioning (BRIEF) Parent and teacher report Conners’ III ADHD Index Parent and teacher report | 35 min | 15 min 8 min |
| Visual and Perceptual Skills | Beery Developmental Test of Visual Visual- Motor Integration (VMI-6) Beery VMI Visual perception | 8 min | |
| Fine Motor | Lafayette Grooved Pegboard (use administration instructions from manual record raw time in seconds) | 5 min | |
| Memory | Wide Range Assessment of Memory and Learning (WRAML-2) Story memory, Story memory recall, Story recognition; Design memory, Design recognition; Verbal learning, Verbal Learning recall, Verbal learning recognition; Picture memory, Picture memory recognition; Finger windows; Number letter; Verbal working memory; Symbolic working memory; Sentence memory | 11 min | 7 min |
| Social Skills | NEPSY-II Theory of Mind and Affect Recognition Autism Spectrum Rating Scale (ASRS) Parent report | ||
| Behavior | Behavioral Assessment System for Children – Second Edition (BASC-2) Parent and teacher report | 20 min | |
| Quality of Life | PedsQL Generic and Cardiac Modules Parent and Child reports | 15 min | |
| Adaptive Function | Adaptive Behavior Assessment System—Third Edition (ABAS-3) Parent report | 20 min | |
| Total Time Required | 199–214 min | 85 min |
| Pre-Stage I and Demographics | Stage I, II, III Hospitalization | Medication Use | Procedural History Data |
|---|---|---|---|
|
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmithorst, V.; Ceschin, R.; Lee, V.; Wallace, J.; Sahel, A.; Chenevert, T.L.; Parmar, H.; Berman, J.I.; Vossough, A.; Qiu, D.; et al. Single Ventricle Reconstruction III: Brain Connectome and Neurodevelopmental Outcomes: Design, Recruitment, and Technical Challenges of a Multicenter, Observational Neuroimaging Study. Diagnostics 2023, 13, 1604. https://doi.org/10.3390/diagnostics13091604
Schmithorst V, Ceschin R, Lee V, Wallace J, Sahel A, Chenevert TL, Parmar H, Berman JI, Vossough A, Qiu D, et al. Single Ventricle Reconstruction III: Brain Connectome and Neurodevelopmental Outcomes: Design, Recruitment, and Technical Challenges of a Multicenter, Observational Neuroimaging Study. Diagnostics. 2023; 13(9):1604. https://doi.org/10.3390/diagnostics13091604
Chicago/Turabian StyleSchmithorst, Vanessa, Rafael Ceschin, Vincent Lee, Julia Wallace, Aurelia Sahel, Thomas L. Chenevert, Hemant Parmar, Jeffrey I. Berman, Arastoo Vossough, Deqiang Qiu, and et al. 2023. "Single Ventricle Reconstruction III: Brain Connectome and Neurodevelopmental Outcomes: Design, Recruitment, and Technical Challenges of a Multicenter, Observational Neuroimaging Study" Diagnostics 13, no. 9: 1604. https://doi.org/10.3390/diagnostics13091604
APA StyleSchmithorst, V., Ceschin, R., Lee, V., Wallace, J., Sahel, A., Chenevert, T. L., Parmar, H., Berman, J. I., Vossough, A., Qiu, D., Kadom, N., Grant, P. E., Gagoski, B., LaViolette, P. S., Maheshwari, M., Sleeper, L. A., Bellinger, D. C., Ilardi, D., O’Neil, S., ... Panigrahy, A. (2023). Single Ventricle Reconstruction III: Brain Connectome and Neurodevelopmental Outcomes: Design, Recruitment, and Technical Challenges of a Multicenter, Observational Neuroimaging Study. Diagnostics, 13(9), 1604. https://doi.org/10.3390/diagnostics13091604






