Prediction of the Post-Pubertal Mandibular Length and Y Axis of Growth by Using Various Machine Learning Techniques: A Retrospective Longitudinal Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sample
2.2. Sample Size Justification
2.3. Data Collection
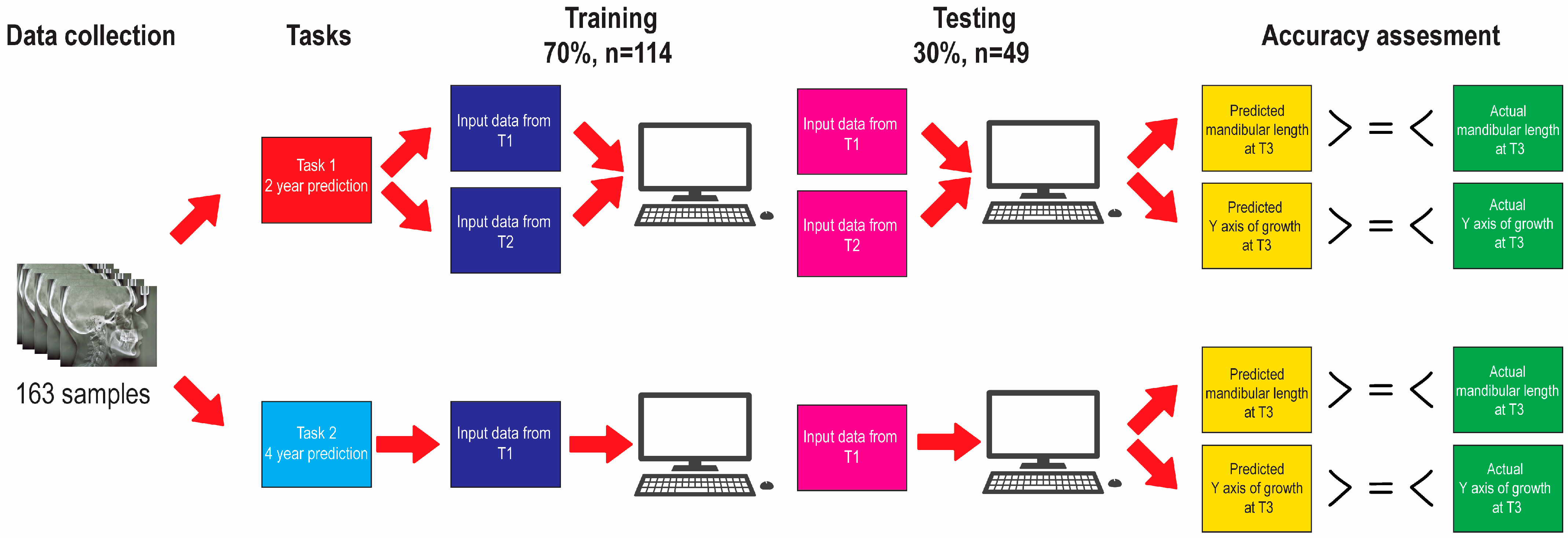
2.4. Algorithm Training and Testing
2.5. Statistical Analyses
3. Results
3.1. Reliability Analysis
3.2. Descriptive Statistics
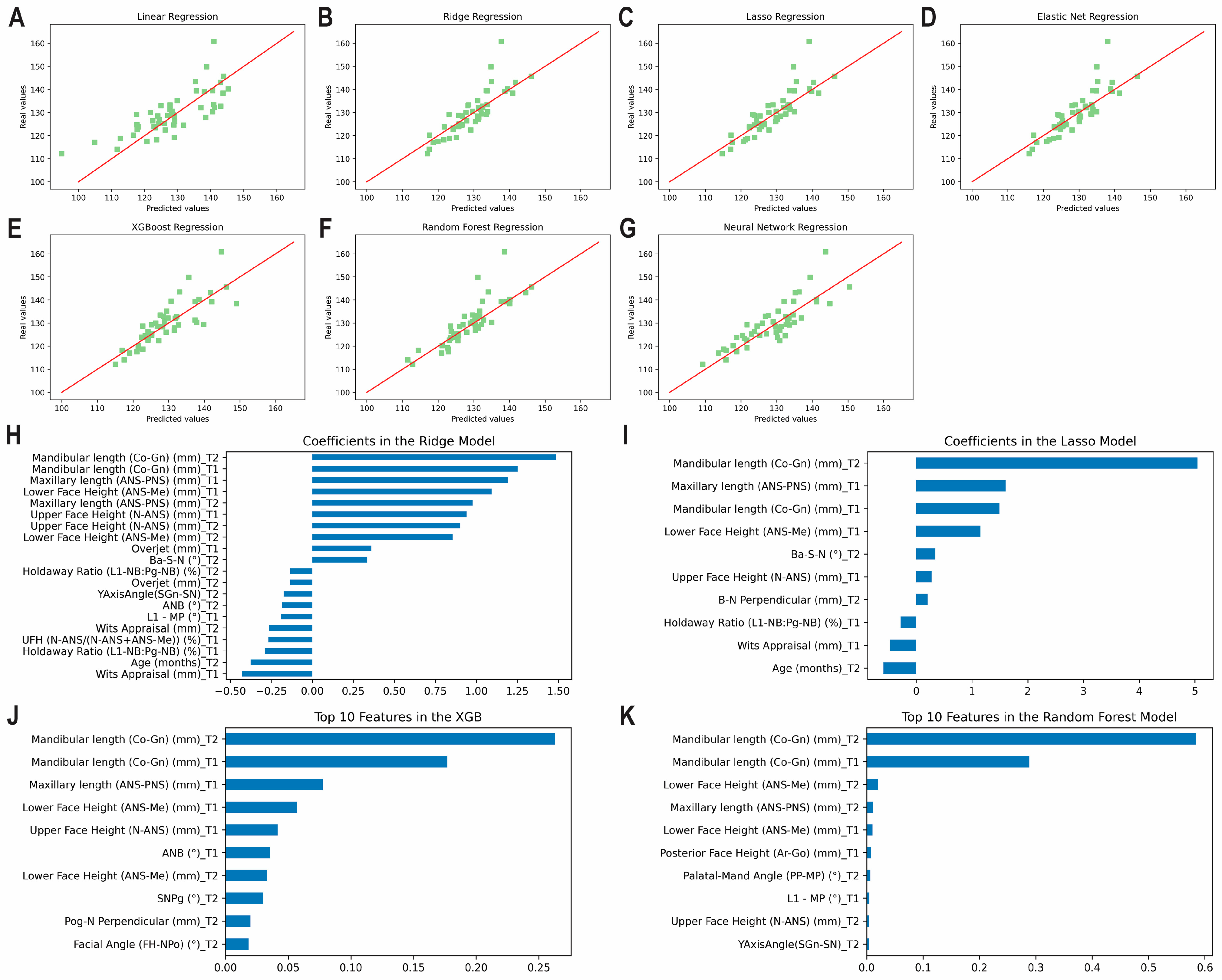
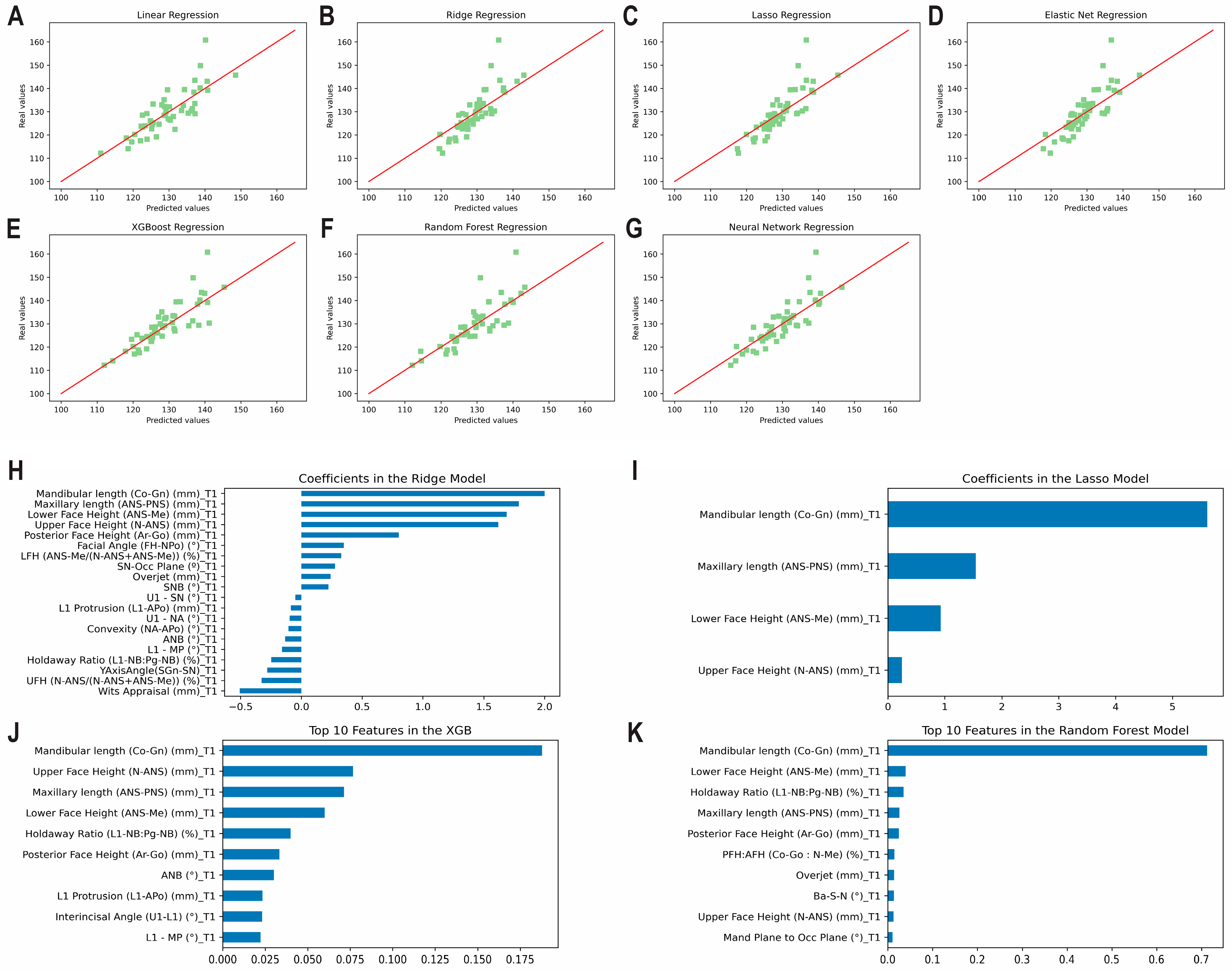
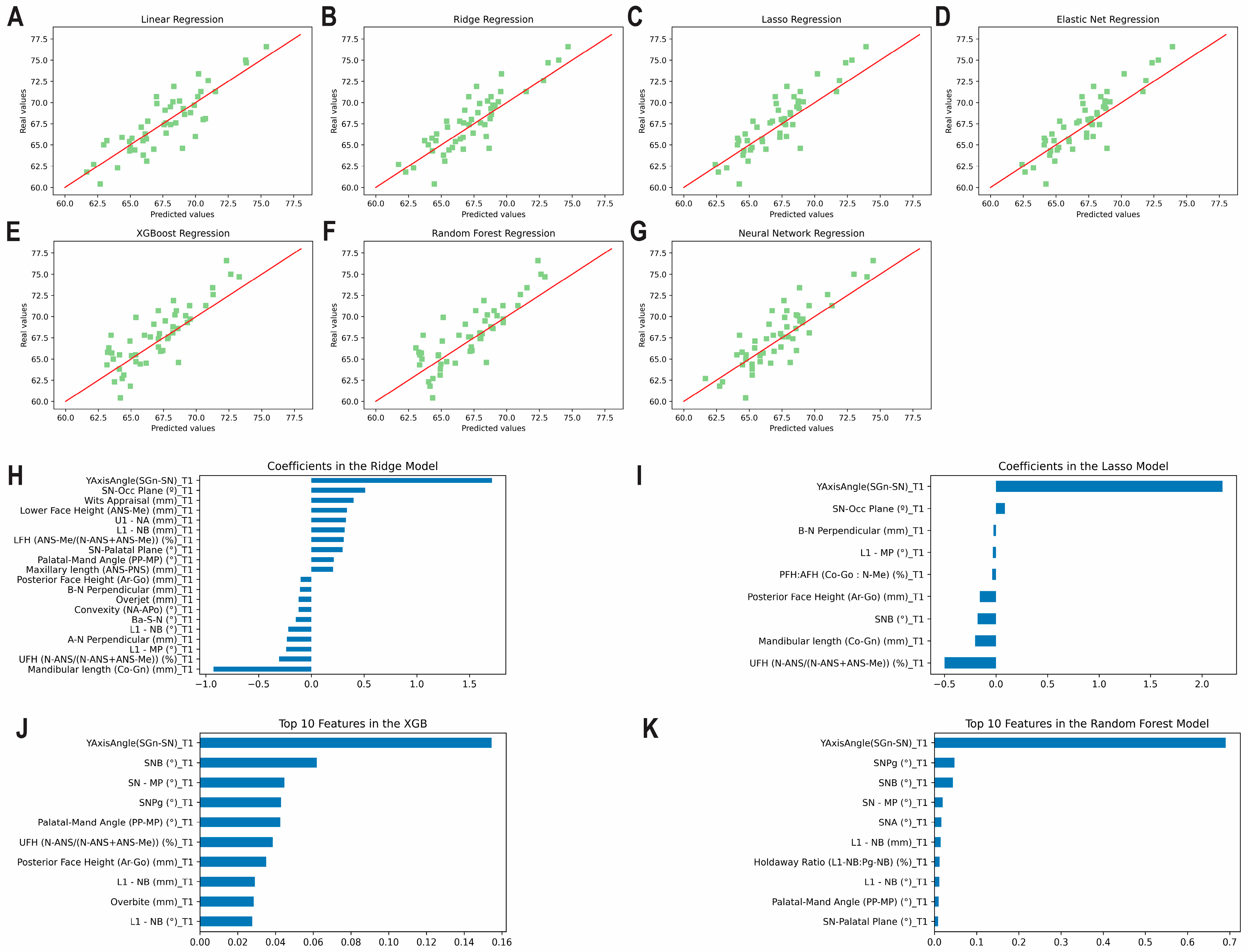
3.3. Prediction of the Post-Pubertal Mandibular Length
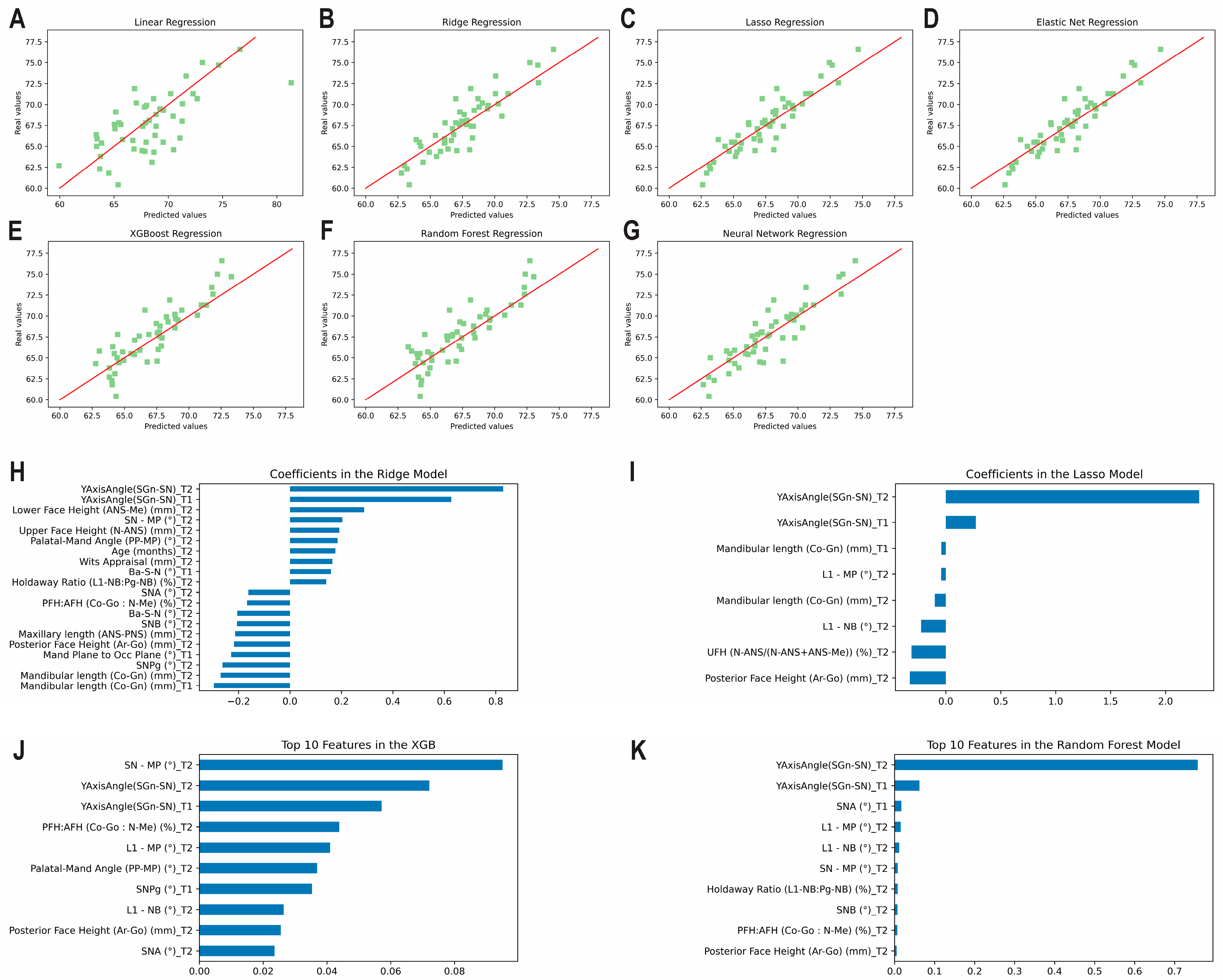
3.4. Prediction of the Post-Pubertal Y Axis of Growth
3.5. Overfitting
3.6. Method Comparison (ANOVA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manlove, A.E.; Romeo, G.; Venugopalan, S.R. Craniofacial Growth: Current Theories and Influence on Management. Oral Maxillofac. Surg. Clin. N. Am. 2020, 32, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Baumrind, S.; Korn, E.L.; West, E.E. Prediction of mandibular rotation: An empirical test of clinician performance. Am. J. Orthod. 1984, 86, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Bjork, A. Prediction of mandibular growth rotation. Am. J. Orthod. 1969, 55, 585–599. [Google Scholar] [CrossRef]
- Skieller, V.; Bjork, A.; Linde-Hansen, T. Prediction of mandibular growth rotation evaluated from a longitudinal implant sample. Am. J. Orthod. 1984, 86, 359–370. [Google Scholar] [CrossRef]
- Leslie, L.R.; Southard, T.E.; Southard, K.A.; Casko, J.S.; Jakobsen, J.R.; Tolley, E.A.; Hillis, S.L.; Carolan, C.; Logue, M. Prediction of mandibular growth rotation: Assessment of the Skieller, Björk, and Linde-Hansen method. Am. J. Orthod. Dentofac. Orthop. 1998, 114, 659–667. [Google Scholar] [CrossRef]
- Buschang, P.H.; Tanguay, R.; LaPalme, L.; Demirjian, A. Mandibular growth prediction: Mean growth increments versus mathematical models. Eur. J. Orthod. 1990, 12, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Oueis, H.; Ono, Y.; Takagi, Y. Prediction of mandibular growth in Japanese children age 4 to 9 years. Pediatr. Dent. 2002, 24, 264–268. [Google Scholar]
- Kunz, F.; Stellzig-Eisenhauer, A.; Zeman, F.; Boldt, J. Artificial intelligence in orthodontics: Evaluation of a fully automated cephalometric analysis using a customized convolutional neural network. J. Orofac. Orthop. 2020, 81, 52–68. [Google Scholar] [CrossRef]
- Kim, H.; Shim, E.; Park, J.; Kim, Y.J.; Lee, U.; Kim, Y. Web-based fully automated cephalometric analysis by deep learning. Comput. Methods Programs Biomed. 2020, 194, 105513. [Google Scholar] [CrossRef]
- Rousseau, M.; Retrouvey, J.-M. Machine learning in orthodontics: Automated facial analysis of vertical dimension for increased precision and efficiency. Am. J. Orthod. Dentofac. Orthop. 2022, 161, 445–450. [Google Scholar] [CrossRef]
- Yu, X.; Liu, B.; Pei, Y.; Xu, T. Evaluation of facial attractiveness for patients with malocclusion: A machine-learning technique employing Procrustes. Angle Orthod. 2014, 84, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Nino-Sandoval, T.C.; Guevara Perez, S.V.; Gonzalez, F.A.; Jaque, R.A.; Infante-Contreras, C. An automatic method for skeletal patterns classification using craniomaxillary variables on a Colombian population. Forensic Sci. Int. 2016, 261, 159.e1–159.e6. [Google Scholar] [CrossRef] [PubMed]
- Nino-Sandoval, T.C.; Guevara Perez, S.V.; Gonzalez, F.A.; Jaque, R.A.; Infante-Contreras, C. Use of automated learning techniques for predicting mandibular morphology in skeletal class I, II and III. Forensic Sci. Int. 2017, 281, 187.e1–187.e7. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ahmad, S.; Frazier, M.; Dundar, M.M.; Turkkahraman, H. A novel machine learning model for class III surgery decision. J. Orofac. Orthop. 2022. [Google Scholar] [CrossRef]
- Choi, H.I.; Jung, S.K.; Baek, S.H.; Lim, W.H.; Ahn, S.J.; Yang, I.H.; Kim, T.W. Artificial Intelligent Model With Neural Network Machine Learning for the Diagnosis of Orthognathic Surgery. J. Craniofac. Surg. 2019, 30, 1986–1989. [Google Scholar] [CrossRef] [PubMed]
- El Bsat, A.R.; Shammas, E.; Asmar, D.; Sakr, G.E.; Zeno, K.G.; Macari, A.T.; Ghafari, J.G. Semantic Segmentation of Maxillary Teeth and Palatal Rugae in Two-Dimensional Images. Diagnostics 2022, 12, 2176. [Google Scholar] [CrossRef]
- Chen, S.; Wang, L.; Li, G.; Wu, T.-H.; Diachina, S.; Tejera, B.; Jungeun Kwon, J.; Lin, F.-C.; Lee, Y.-T.; Xu, T.; et al. Machine learning in orthodontics: Introducing a 3D auto-segmentation and auto-landmark finder of CBCT images to assess maxillary constriction in unilateral impacted canine patients. Angle Orthod. 2020, 90, 77–84. [Google Scholar] [CrossRef]
- Atici, S.F.; Ansari, R.; Allareddy, V.; Suhaym, O.; Cetin, A.E.; Elnagar, M.H. Fully automated determination of the cervical vertebrae maturation stages using deep learning with directional filters. PLoS ONE 2022, 17, e0269198. [Google Scholar] [CrossRef]
- Dallora, A.L.; Anderberg, P.; Kvist, O.; Mendes, E.; Diaz Ruiz, S.; Sanmartin Berglund, J. Bone age assessment with various machine learning techniques: A systematic literature review and meta-analysis. PLoS ONE 2019, 14, e0220242. [Google Scholar] [CrossRef]
- Wu, T.J.; Tsai, C.L.; Gao, Q.Z.; Chen, Y.P.; Kuo, C.F.; Huang, Y.H. The Application of Artificial-Intelligence-Assisted Dental Age Assessment in Children with Growth Delay. J. Pers. Med. 2022, 12, 1158. [Google Scholar] [CrossRef]
- Li, P.; Kong, D.; Tang, T.; Su, D.; Yang, P.; Wang, H.; Zhao, Z.; Liu, Y. Orthodontic treatment planning based on artificial neural networks. Sci. Rep. 2019, 9, 2037. [Google Scholar] [CrossRef]
- Suhail, Y.; Upadhyay, M.; Chhibber, A.; Kshitiz. Machine Learning for the Diagnosis of Orthodontic Extractions: A Computational Analysis Using Ensemble Learning. Bioengineering 2020, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.K.; Kim, T.W. New approach for the diagnosis of extractions with neural network machine learning. Am. J. Orthod. Dentofac. Orthop. 2016, 149, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Real, A.D.; Real, O.D.; Sardina, S.; Oyonarte, R. Use of automated artificial intelligence to predict the need for orthodontic extractions. Korean J. Orthod. 2022, 52, 102–111. [Google Scholar] [CrossRef]
- Xie, X.; Wang, L.; Wang, A. Artificial neural network modeling for deciding if extractions are necessary prior to orthodontic treatment. Angle Orthod. 2010, 80, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, L.; Volovic, J.; Steinhauer, L.; Mason, T.; Eckert, G.; Dean, J.A.; Dundar, M.M.; Turkkahraman, H. Can we predict orthodontic extraction patterns by using machine learning? Orthod Craniofac. Res. 2023. [Google Scholar] [CrossRef] [PubMed]
- Jiwa, S. Applicability of Deep Learning for Mandibular Growth Prediction. M.S.D. Thesis, Boston University, Ann Arbor, MI, USA, 2020. [Google Scholar]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Stahl, F.; Baccetti, T.; Franchi, L.; McNamara, J.A., Jr. Longitudinal growth changes in untreated subjects with Class II Division 1 malocclusion. Am. J. Orthod. Dentofac. Orthop. 2008, 134, 125–137. [Google Scholar] [CrossRef]
- Baccetti, T.; Franchi, L.; McNamara, J.A. Growth in the Untreated Class III Subject. Semin. Orthod. 2007, 13, 130–142. [Google Scholar] [CrossRef]
- Davidovitch, M.; Eleftheriadi, I.; Kostaki, A.; Shpack, N. The use of Björk’s indications of growth for evaluation of extremes of skeletal morphology. Eur. J. Orthod. 2016, 38, 555–562. [Google Scholar] [CrossRef]
| Using Variables from T1 and T2 | Using Variables from T1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | ME (SD) | MAE | RMSE | ICC | Accuracy % | ME (SD) | MAE | RMSE | ICC | Accuracy % |
| Least Squares | −1.32 (0.99) | 5.47 | 6.95 | 0.76 | 95.80 | −0.22 (0.77) | 3.90 | 5.35 | 0.80 | 97.00 |
| Ridge | −0.37 (0.74) | 3.45 | 5.17 | 0.80 | 97.35 | −0.16 (0.82) | 3.99 | 5.69 | 0.72 | 96.94 |
| Lasso | −0.66 (0.70) | 3.31 | 4.91 | 0.82 | 97.46 | −0.34 (0.79) | 3.67 | 5.49 | 0.75 | 97.18 |
| Elastic Net | −0.57 (0.72) | 3.35 | 5.05 | 0.81 | 97.43 | −0.36 (0.81) | 3.96 | 5.61 | 0.74 | 96.96 |
| XGBoost | −0.48 (0.75) | 3.78 | 5.20 | 0.82 | 97.10 | −0.72 (0.73) | 3.52 | 5.11 | 0.82 | 97.30 |
| Random Forest | −1.03 (0.72) | 3.07 | 5.11 | 0.81 | 97.64 | −0.35 (0.77) | 3.51 | 5.32 | 0.79 | 97.30 |
| Neural Net | −0.80 (0.72) | 4.10 | 5.08 | 0.84 | 96.85 | −0.20 (0.72) | 3.47 | 4.99 | 0.82 | 97.33 |
| Using Variables from T1 and T2 | Using Variables from T1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | ME (SD) | MAE | RMSE | ICC | Accuracy % | ME (SD) | MAE | RMSE | ICC | Accuracy % |
| Least Squares | 0.52 (0.41) | 2.29 | 2.91 | 0.68 | 96.60 | −0.09 (0.26) | 1.45 | 1.82 | 0.85 | 97.85 |
| Ridge | −0.15 (0.23) | 1.29 | 1.61 | 0.87 | 98.09 | −0.38 (0.25) | 1.42 | 1.77 | 0.85 | 97.89 |
| Lasso | −0.17 (0.21) | 1.12 | 1.43 | 0.90 | 98.34 | −0.51 (0.25) | 1.43 | 1.79 | 0.84 | 97.88 |
| Elastic Net | −0.17 (0.21) | 1.12 | 1.43 | 0.90 | 98.34 | −0.51 (0.25) | 1.43 | 1.79 | 0.84 | 97.88 |
| XGBoost | −0.46 (0.24) | 1.35 | 1.74 | 0.85 | 98.00 | −0.70 (0.28) | 1.67 | 2.08 | 0.78 | 97.52 |
| Random Forest | −0.41 (0.25) | 1.38 | 1.75 | 0.85 | 97.95 | −0.62 (0.28) | 1.64 | 2.03 | 0.80 | 97.57 |
| Neural Net | −0.05 (0.23) | 1.24 | 1.59 | 0.88 | 98.16 | −0.45 (0.27) | 1.49 | 1.90 | 0.82 | 97.79 |
| Mandibular Length | Y Axis of Growth | |||||||
|---|---|---|---|---|---|---|---|---|
| Method | ME | MAE | RMSE | Accuracy % | ME | MAE | RMSE | Accuracy % |
| Least Squares | 0.69 | 3.67 | 5.39 | 97.18 | −0.11 | 1.01 | 1.32 | 98.50 |
| Ridge | 1.11 | 3.77 | 5.37 | 97.10 | −0.11 | 1.01 | 1.32 | 98.50 |
| Lasso | 1.18 | 3.88 | 5.91 | 97.02 | −0.23 | 1.00 | 1.34 | 98.52 |
| Elastic Net | 1.22 | 3.78 | 5.53 | 97.10 | −0.20 | 0.99 | 1.32 | 98.53 |
| XGBoost | 2.79 | 4.69 | 7.31 | 96.40 | 0.05 | 1.24 | 1.58 | 98.16 |
| Random Forest | 1.86 | 4.22 | 6.74 | 96.76 | −0.39 | 1.22 | 1.58 | 98.19 |
| Neural Net | 1.50 | 4.25 | 6.64 | 96.74 | −0.15 | 1.04 | 1.37 | 98.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wood, T.; Anigbo, J.O.; Eckert, G.; Stewart, K.T.; Dundar, M.M.; Turkkahraman, H. Prediction of the Post-Pubertal Mandibular Length and Y Axis of Growth by Using Various Machine Learning Techniques: A Retrospective Longitudinal Study. Diagnostics 2023, 13, 1553. https://doi.org/10.3390/diagnostics13091553
Wood T, Anigbo JO, Eckert G, Stewart KT, Dundar MM, Turkkahraman H. Prediction of the Post-Pubertal Mandibular Length and Y Axis of Growth by Using Various Machine Learning Techniques: A Retrospective Longitudinal Study. Diagnostics. 2023; 13(9):1553. https://doi.org/10.3390/diagnostics13091553
Chicago/Turabian StyleWood, Tyler, Justina O. Anigbo, George Eckert, Kelton T. Stewart, Mehmet Murat Dundar, and Hakan Turkkahraman. 2023. "Prediction of the Post-Pubertal Mandibular Length and Y Axis of Growth by Using Various Machine Learning Techniques: A Retrospective Longitudinal Study" Diagnostics 13, no. 9: 1553. https://doi.org/10.3390/diagnostics13091553
APA StyleWood, T., Anigbo, J. O., Eckert, G., Stewart, K. T., Dundar, M. M., & Turkkahraman, H. (2023). Prediction of the Post-Pubertal Mandibular Length and Y Axis of Growth by Using Various Machine Learning Techniques: A Retrospective Longitudinal Study. Diagnostics, 13(9), 1553. https://doi.org/10.3390/diagnostics13091553







