Human Pathogenic Monkeypox Disease Recognition Using Q-Learning Approach
Abstract
1. Introduction
- The detailed survey relevant to the classification of monkeypox diseases was carried out. The authors’ contribution, limitations, and future scope are discussed;
- The proposed work is developed to recognize the Monkeypox Virus with respect to four classes;
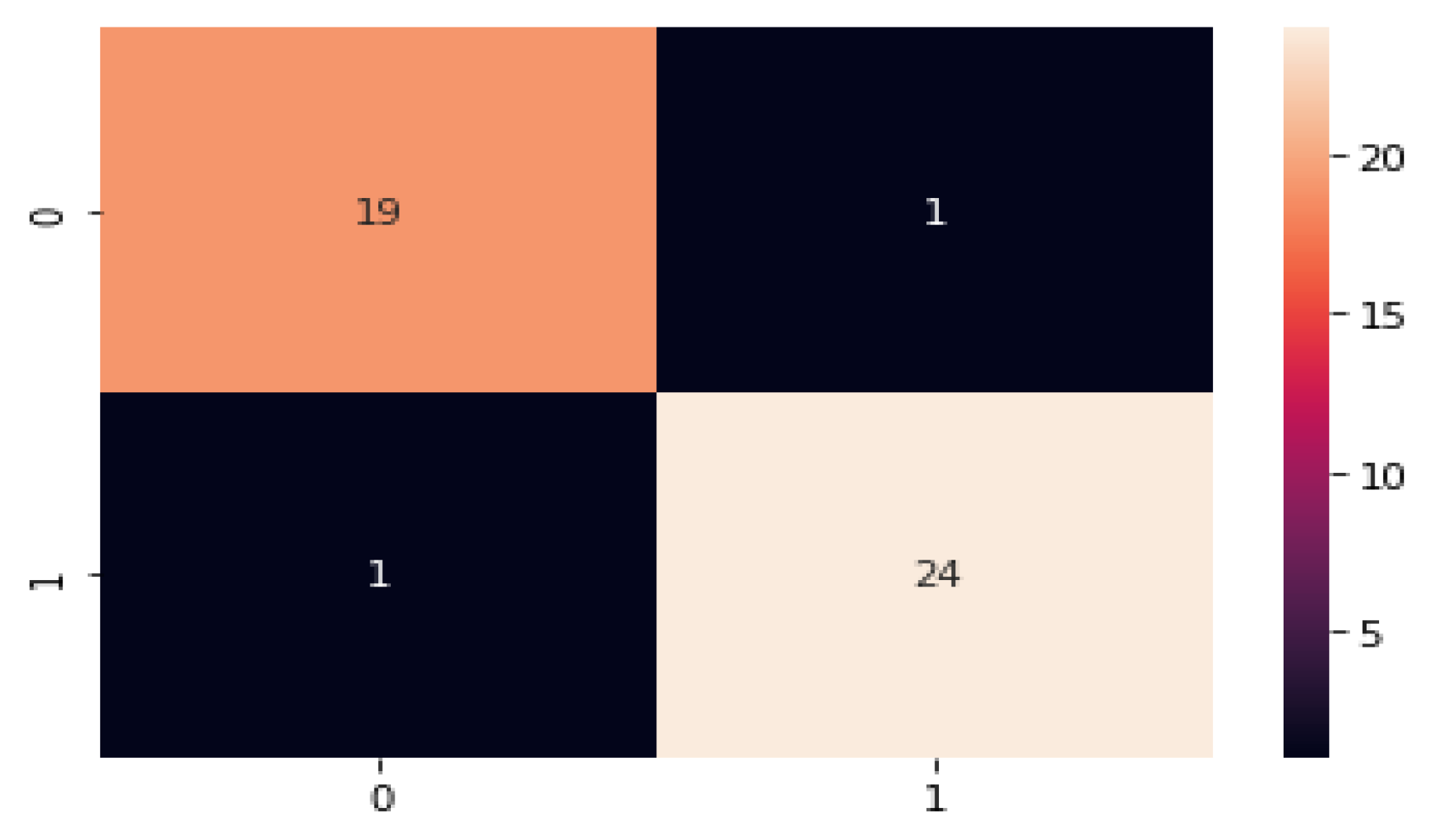
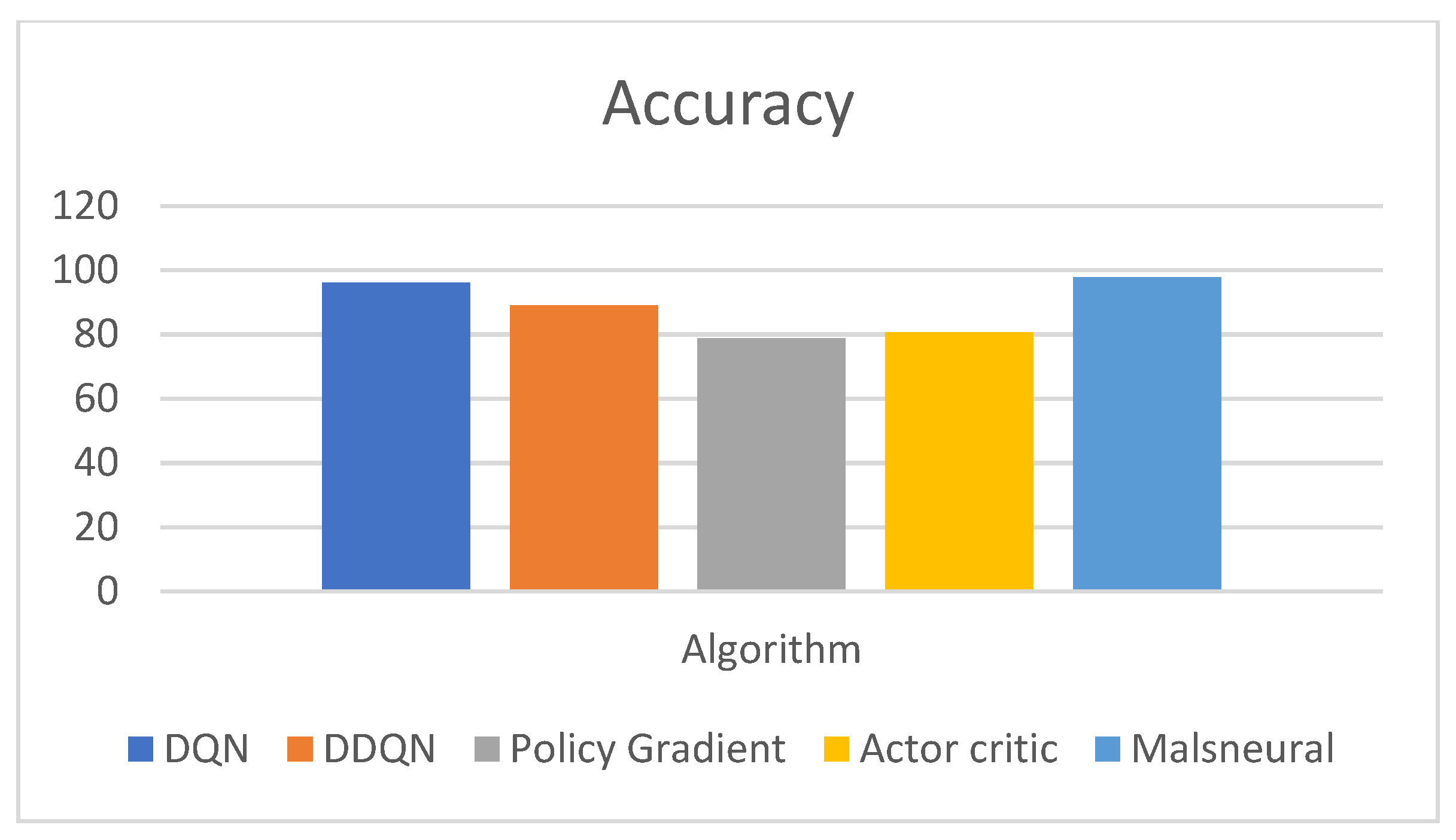
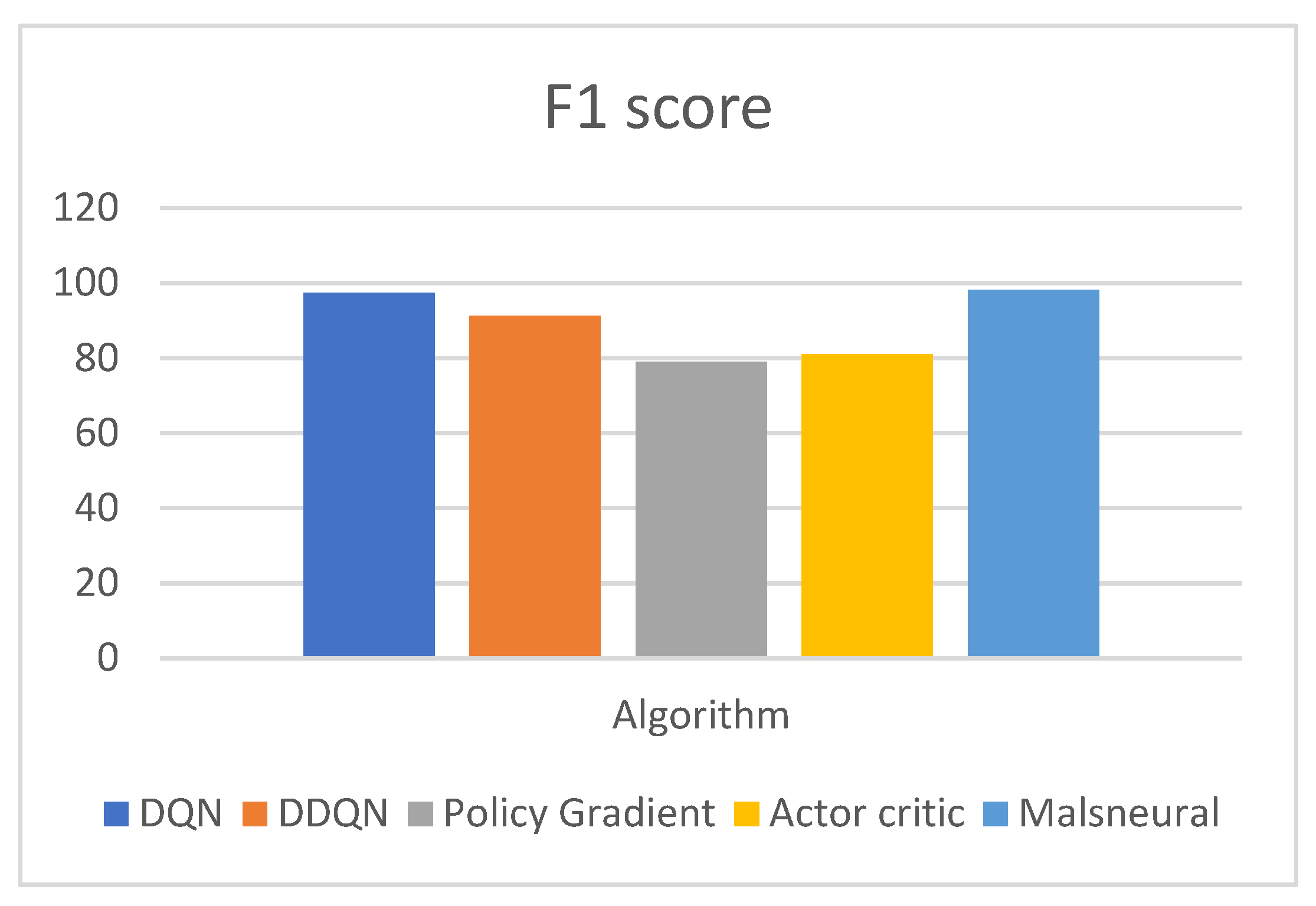
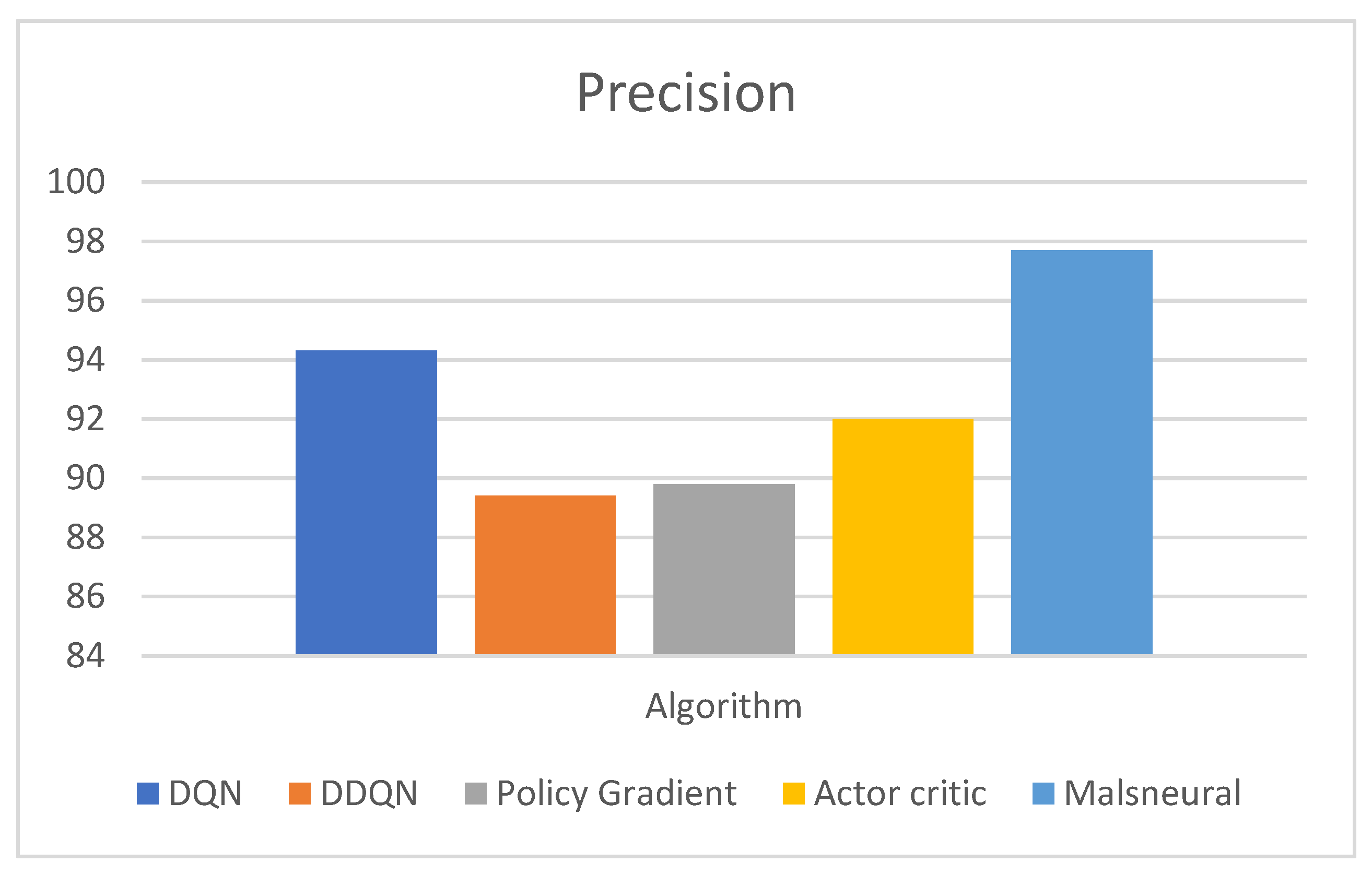
- The performance of the model can be measured with the help of evaluation metrics, namely, AUC, CA, F1, precision, and Recall. The DQN approach achieves a classification accuracy (C.A.) of 0.975;
- The comparison of the proposed work with the benchmark mark algorithms, namely, DQN, DDQN, Policy Gradient, and Actor–Critic. Compared with other state-of-the-art methods, the proposed DQN outperforms others with higher accuracy and AUC.
2. Related Works
3. Materials and Methods
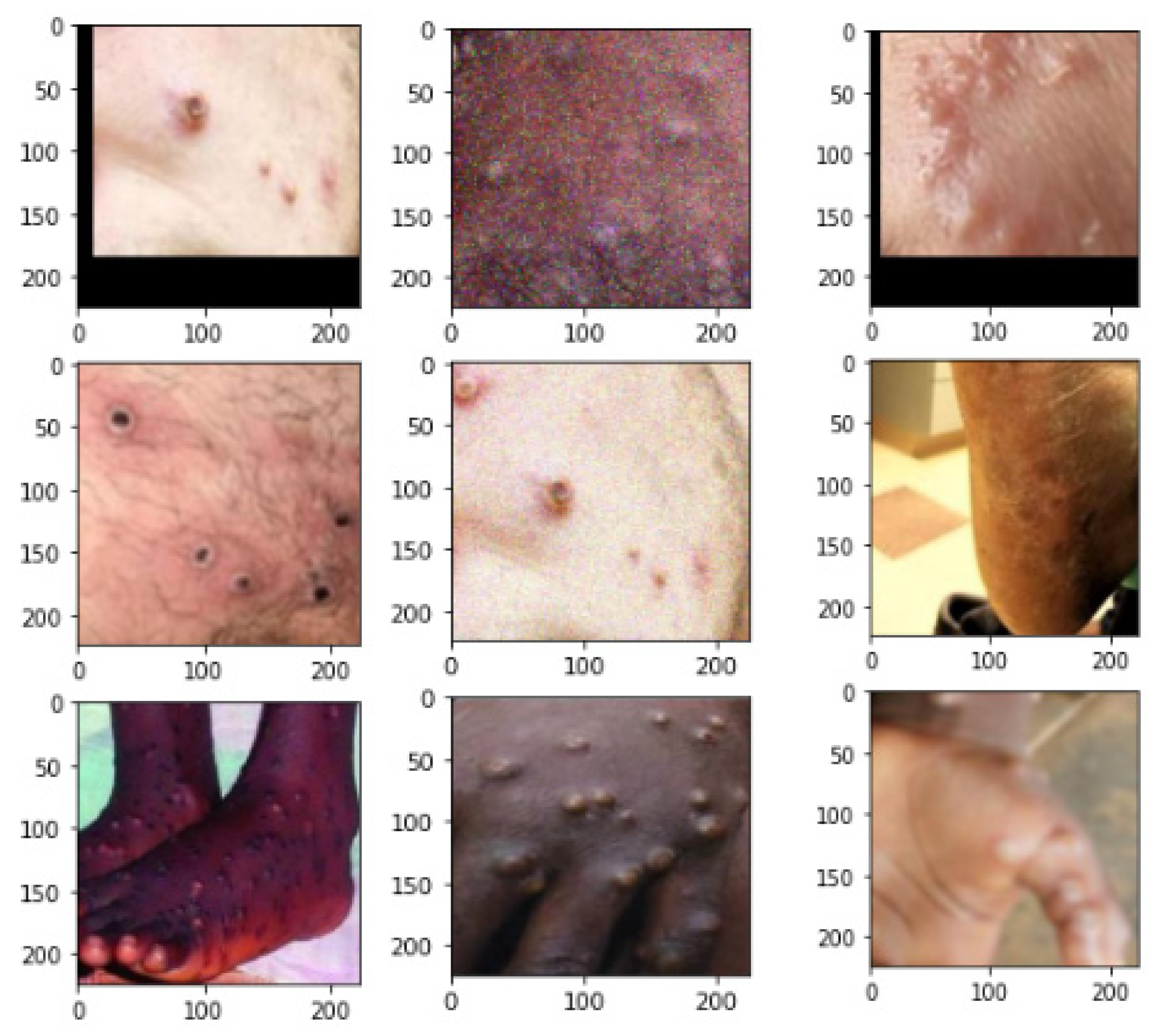
3.1. Dataset
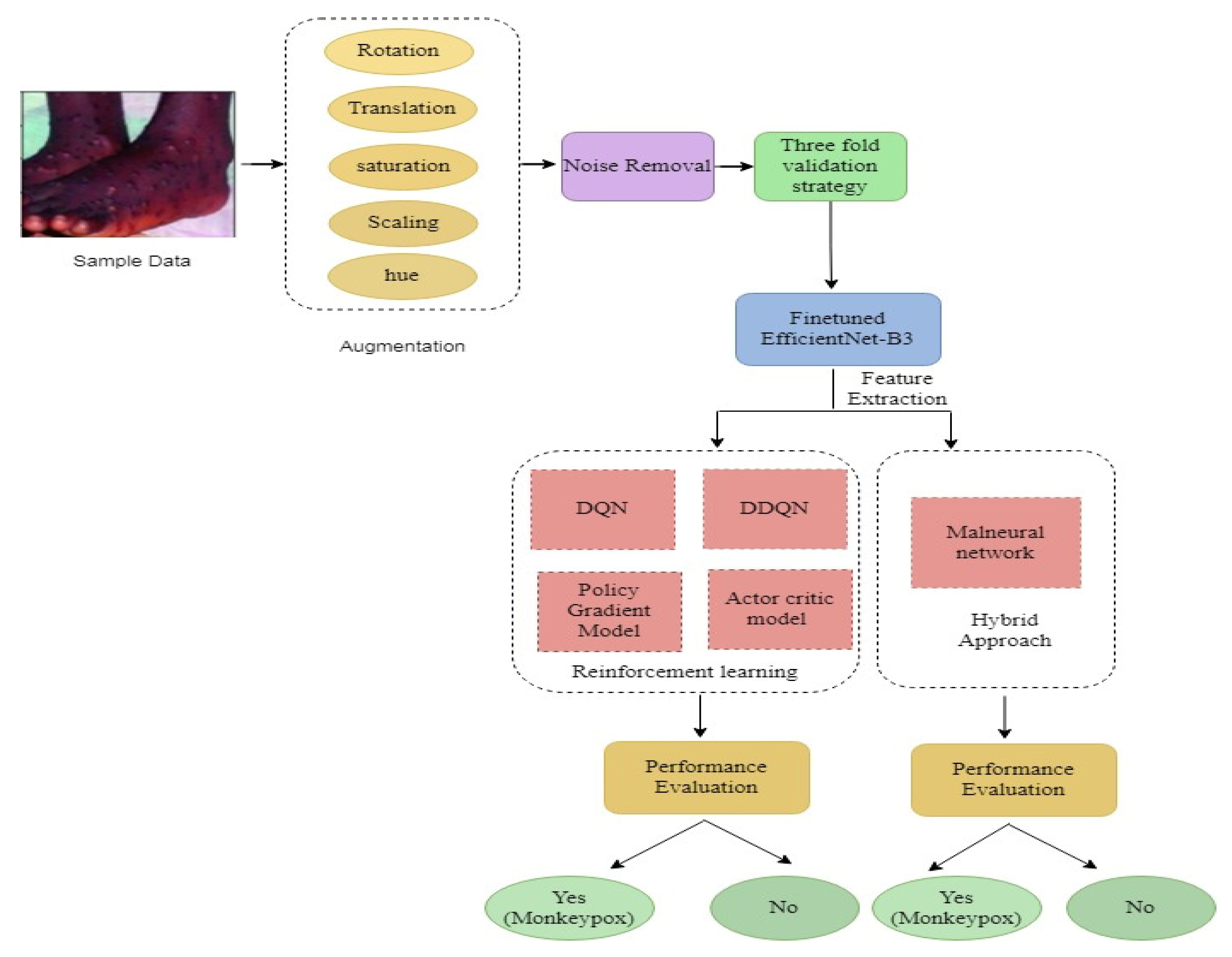
3.2. Data Preprocessing
3.2.1. Augmented Images
3.2.2. Fold1
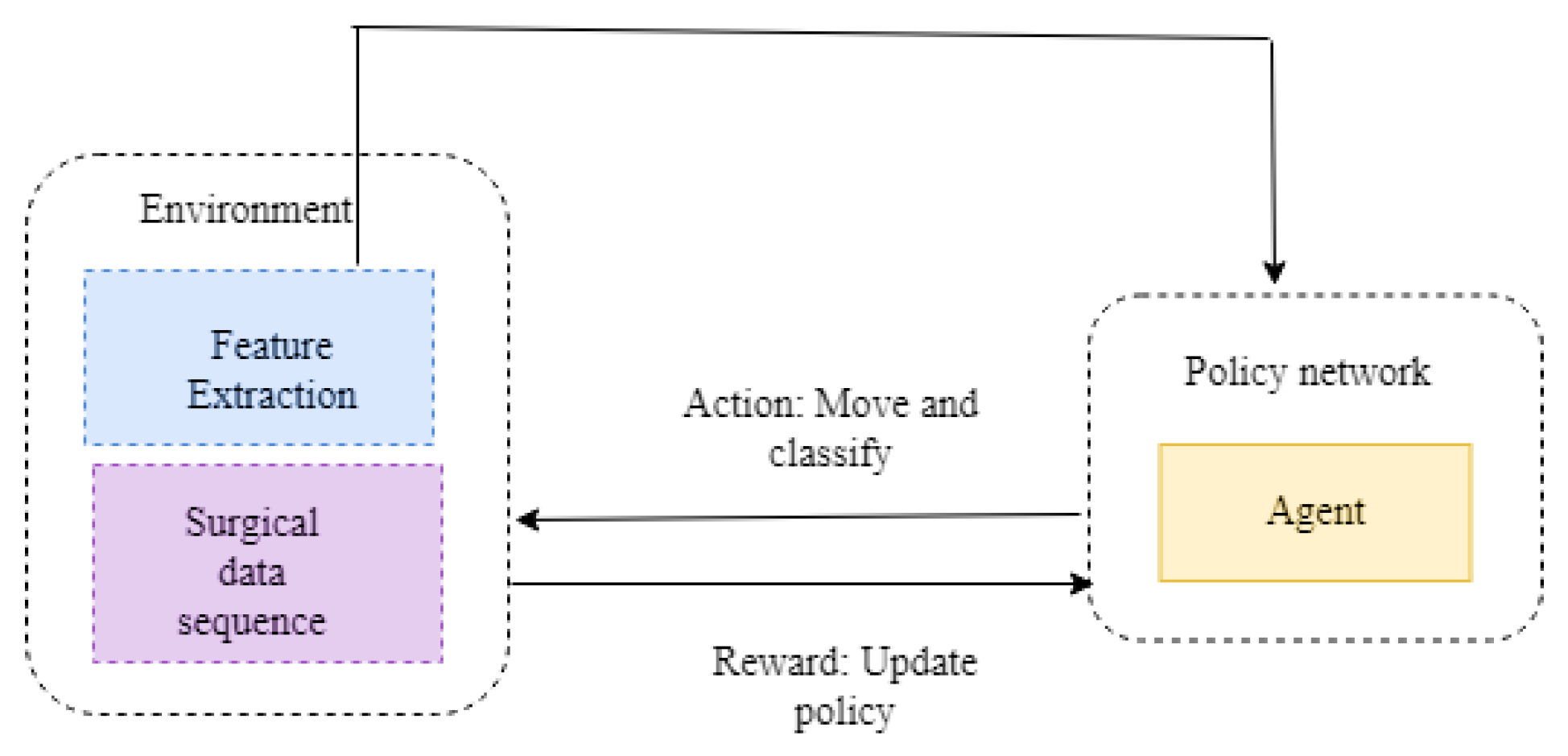
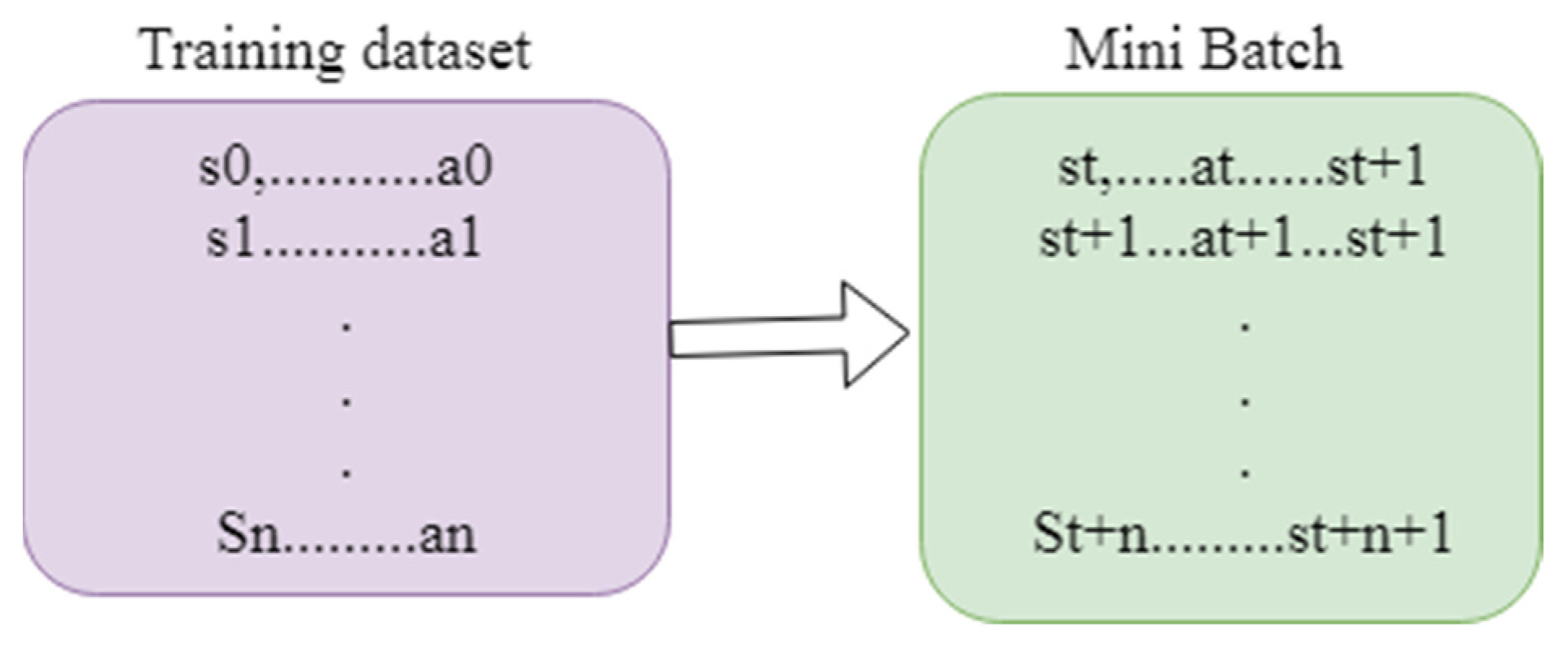
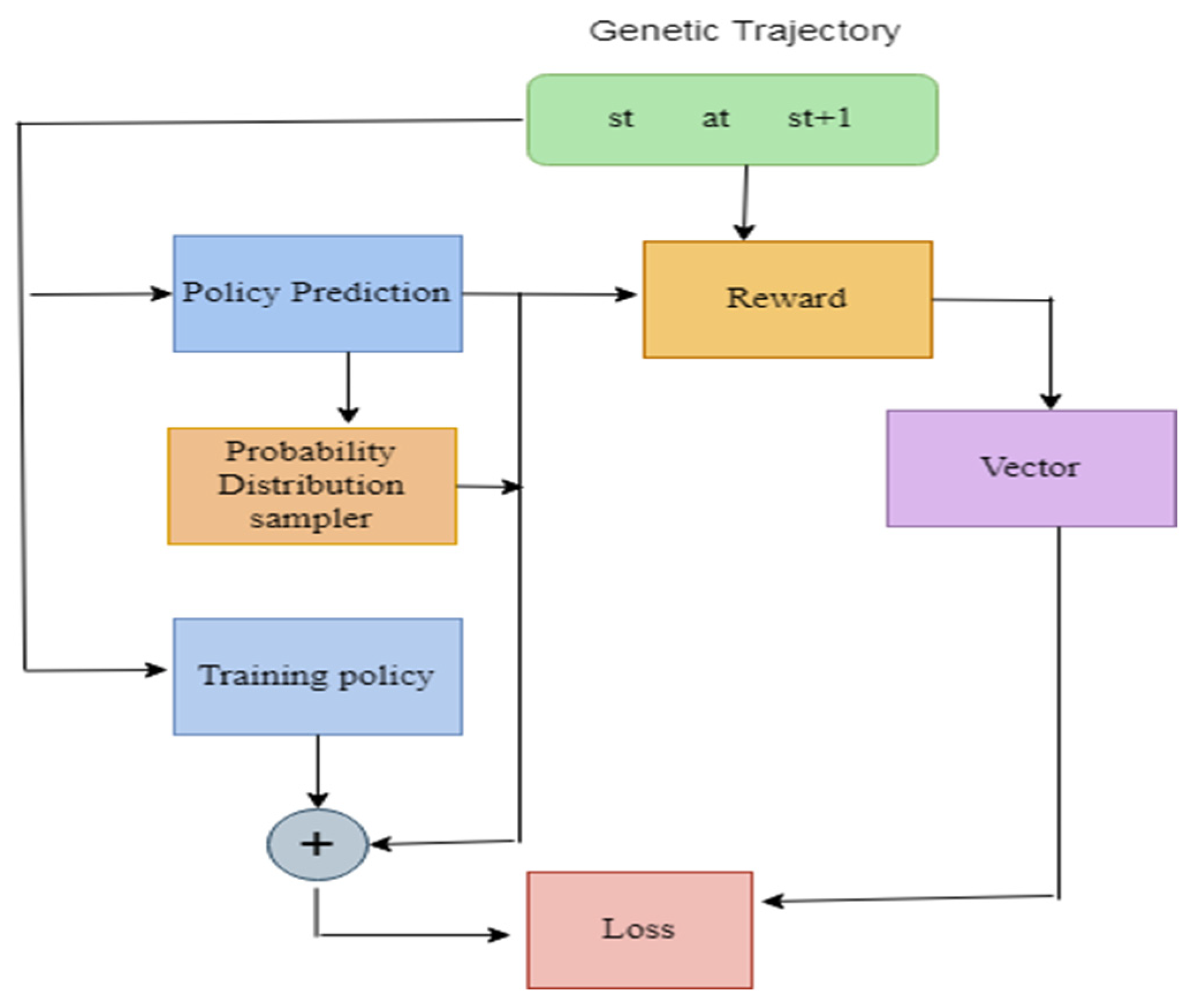
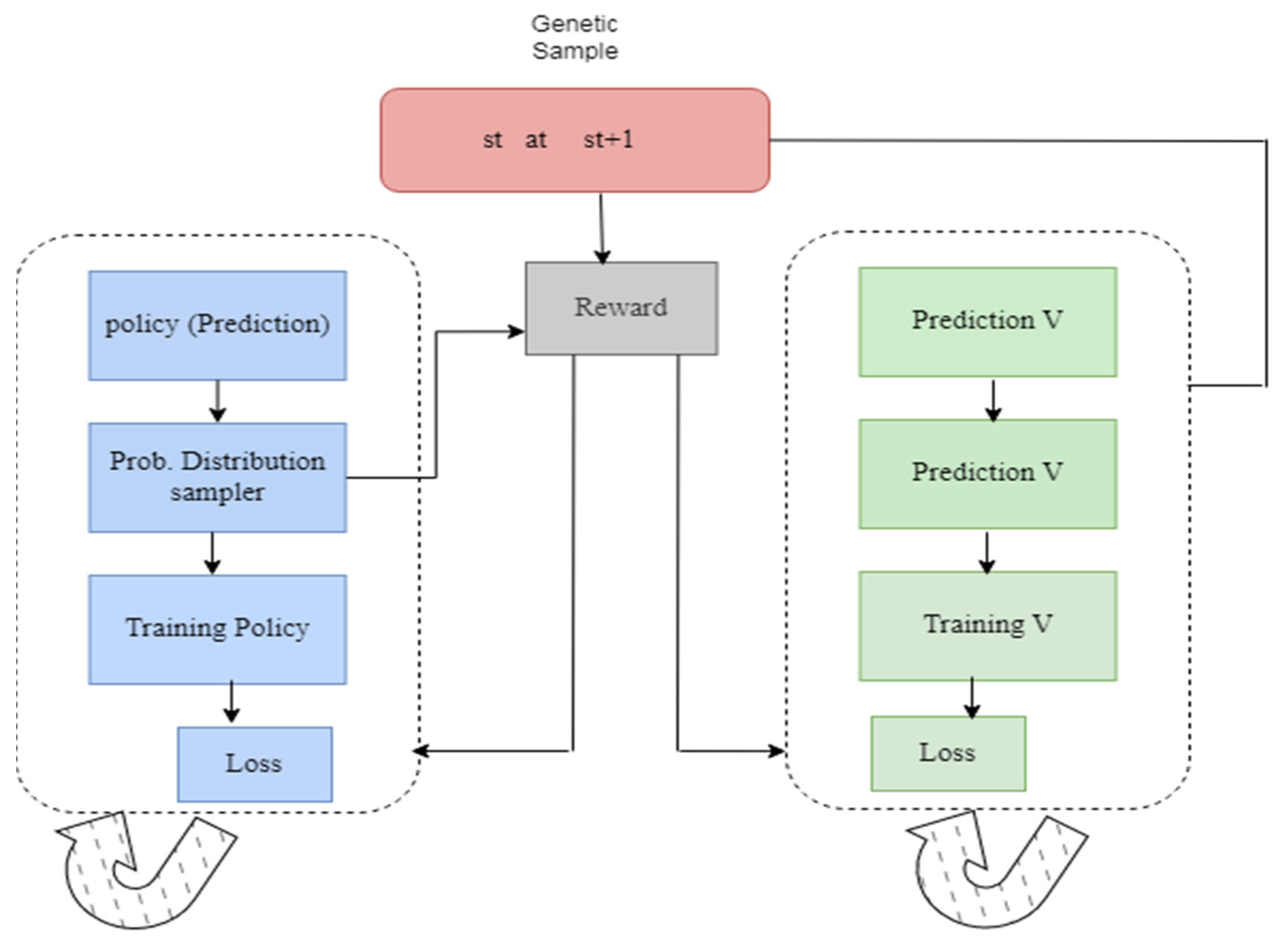
3.2.3. Reinforcement Learning
3.3. Proposed Methodology
3.3.1. System Model
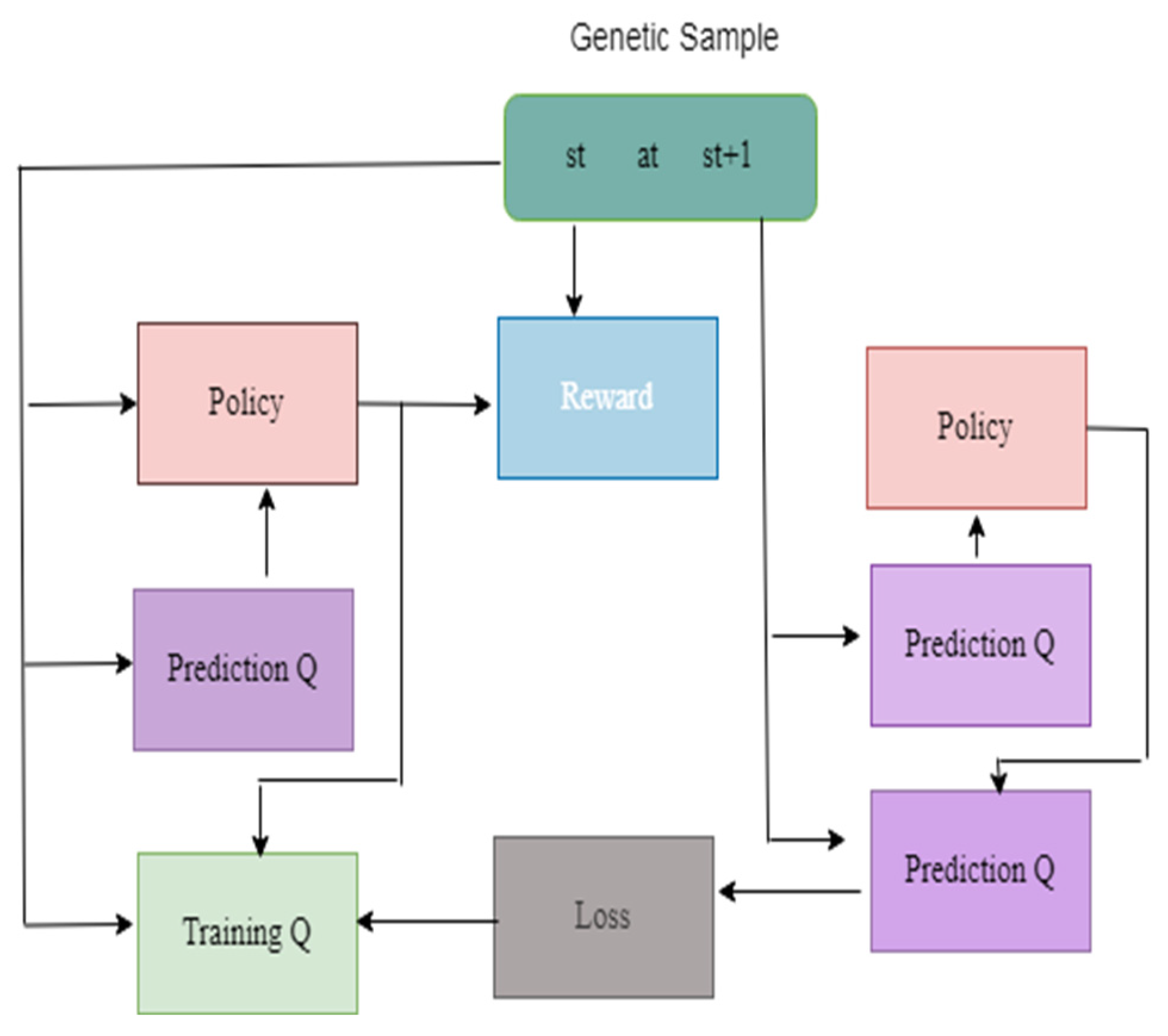
DQN
DDQN
Policy Gradient
Actor–Critic
| Algorithm 1: Malsneural algorithm |
|
1. Procedure Augmentation(image, pro) 2. prob pro: 3. 4. prob pro: 5. 6. prob pro: 7. 8. prob pro: 9. 10. prob pro: 11. 12. Return image 13. Adaptive median filter 14. Level 1: 15. 16. 17. If image1 > 0 and image 2 < 0 go to the next level 18. Else the size of the window increased 19. If windoe size <= size max redo the level 1 20. Else return zxy 21. Level 2: 22. 23. 24. If image 3 > 0 and image 4 < 0 return zxy 25. Else return zmedian 26. End if 27. Load replay memory M to the capacity C 28. Load the function action Q along with arbitrary weight W 29. Load destination value function Q along with weight W- = W 30. For iteration = 1,N do 31. Load sequence t = {y1} and preprocessed ϕ1 = ϕ(t1) 32. For q = 1, Q do 33. The random action choosen bQ 34. Orelse choose bq = argmaxb P(ϕ(tq),b;W) 35. Compile bq in emulator and notice reward rq and yq + 1 of input 36. Set t q + 1 = tq, bq, y q + 1 and process ϕq + 1 = ϕ(tq + 1) 37. Save the transition (ϕq, bq,rq,ϕq + 1) in M 38. Minibatch (ϕi, bi, fi,ϕi + 1 ) from M 39. If it stops at i + 1 40. Initialise fj 41. Else 42. Yj = {fi + ϑmax d P(ϕi + 1,bq,W) 43. Execute gradient descent by updating the gradient value (yi-P(ϕi,bi; W))2 44. Reset ό = P 45. End for 46. End for |
4. Experiment and Analysis
4.1. Experimental Setup
4.1.1. Precision
4.1.2. Recall
4.1.3. F1 Score
4.1.4. Recall and F1 Score Are given Equal Weighted values
5. Conclusions and Future Scope
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- McCollum, A.M.; Damon, I.K. Human Monkey Pox. Clin. Infect. Dis. 2014, 58, 260–267. [Google Scholar] [CrossRef]
- Alakunle, E.; Moens, U.; Nchinda, G.; Okeke, M.I. Monkey Pox Virus in Nigeria: Infection Biology, Epidemiology, and Evolution. Viruses 2020, 12, 1257. [Google Scholar] [CrossRef]
- Moore, M.J.; Rathish, B.; Zahra, F. Monkey Pox. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Nolen, L.D.; Osadebe, L.; Katomba, J.; Likofata, J.; Mukadi, D.; Monroe, B.; Doty, J.; Hughes, C.M.; Kabamba, J.; Malekani, J.; et al. Extended Human-to-Human Transmission during a Monkey Pox Outbreak in the Democratic Republic of the Congo. Emerg. Infect. Dis. 2016, 22, 1014–1021. [Google Scholar] [CrossRef]
- Nguyen, P.Y.; Ajisegiri, W.S.; Costantino, V.; Chughtai, A.A.; MacIntyre, C.R. Reemergence of Human Monkey pox and Declining Population Immunity in the Context of Urbanization, Nigeria, 2017–2020. Emerg. Infect. Dis. 2021, 27, 1007. [Google Scholar] [CrossRef]
- El-kenawy, E.S.M.; Albalawi, F.; Ward, S.A.; Ghoneim, S.S.M.; Eid, M.M.; Abdelhamid, A.A.; Bailek, N.; Ibrahim, A. Feature Selection and Classification of Transformer Faults Based on Novel Meta-Heuristic Algorithm. Mathematics 2022, 10, 3144. [Google Scholar] [CrossRef]
- Ali Salamai, A.; El-kenawy, E.S.M.; Abdelhameed, I. Dynamic Voting Classifier for Risk Identification in Supply Chain 4. Comput. Mater. Contin. 2021, 69, 3749–3766. [Google Scholar] [CrossRef]
- Ali, S.; Ahmed, M.; Paul, J.; Jahan, T.; Sani, S.; Noor, N.; Hasan, T. Monkey Pox Skin Lesion Detection Using Deep Learning Models: A Feasibility Study. arXiv 2022, arXiv:2207.03342. [Google Scholar]
- Adler, H.; Gould, S.; Hine, P.; Snell, L.B.; Wong, W.E.A. Clinical features and management of human monkey pox: A retrospective observational study in the U.K. Lancet Infect. Dis. 2022, 22, 1153–1162. [Google Scholar] [CrossRef]
- Akin, K.D.; Gurkan, C.; Budak, A.; Karataş, H. Classification of Monkey Pox Skin Lesion Using the Explainable Artificial Intelligence Assisted Convolutional Neural Networks. Avrupa Bilim Ve Teknol. Derg. 2022, 40, 106–110. [Google Scholar]
- Ibrahim, A.; Mirjalili, S.; El-Said, M.; Ghoneim, S.S.M.; Al-Harthi, M.M.; Ibrahim, T.F.; El-Kenawy, E.S.M. Wind Speed Ensemble Forecasting Based on Deep Learning Using Adaptive Dynamic Optimization Algorithm. IEEE Access 2021, 9, 125787–125804. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Gupta, K.D.; Islam, M.M.; Sen, S.; Rahman, M.L.; Shakhawat Hossain, M. COVID-19 Symptoms Detection Based on NasNetMobile with Explainable A.I. Using Various Imaging Modalities. Mach. Learn. Knowl. Extr. 2020, 2, 490–504. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Alam, T.E.; Trafalis, T.; Huebner, P. Deep MLP-CNN Model Using Mixed-Data to Distinguish between COVID-19 and Non-COVID-19 Patients. Symmetry 2020, 12, 1526. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Ahad, M.T.; Soma, F.A.; Paul, S.; Chowdhury, A.; Luna, S.A.; Yazdan, M.M.S.; Rahman, A.; Siddique, Z.; Huebner, P. Detecting SARS-CoV-2 from Chest X-Ray Using Artificial Intelligence. IEEE Access 2021, 9, 35501–35513. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Nazim, R.; Siddique, Z.; Huebner, P. Detection of COVID-19 Patients from CT Scan and Chest X-ray Data Using Modified MobileNetV2 and LIME. Healthcare 2021, 9, 1099. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Siddique, Z. Machine learning-based heart disease diagnosis: A systematic literature review. Artif. Intell. Med. 2022, 128, 102289. [Google Scholar] [CrossRef] [PubMed]
- Miranda, G.H.B.; Felipe, J.C. Computer-aided diagnosis system based on fuzzy logic for breast cancer categorization. Comput. Biol. Med. 2015, 64, 334–346. [Google Scholar] [CrossRef]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using C.T. images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Sandeep, R.; Vishal, K.P.; Shamanth, M.S.; Chethan, K. Diagnosis of Visible Diseases Using CNNs. In Proceedings of the International Conference on Communication and Artificial Intelligence; Goyal, V., Gupta, M., Mirjalili, S., Trivedi, A., Eds.; Lecture Notes in Networks and Systems; Springer Nature: Singapore, 2022; pp. 459–468. [Google Scholar]
- Roy, K.; Chaudhuri, S.S.; Ghosh, S.; Dutta, S.K.; Chakraborty, P.; Sarkar, R. Skin Disease detection based on different Segmentation Techniques. In Proceedings of the 2019 International Conference on Opto-Electronics and Applied Optics (Optronix), Kolkata, India, 18–20 March 2019; pp. 1–5. [Google Scholar]
- Chandra, M.M.G. Effective Heart Disease Prediction Using Hybrid Machine Learning Techniques. Int. J. Sci. Res. Eng. Manag. 2022, 6, 81542–81554. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, X.; Adeli, E.; Zhu, Y.; Nie, F.; Munsell, B.; Wu, G. Multi-modal classification of neurodegenerative disease by progressive graph-based transductive learning. Med. Image Anal. 2017, 39, 218–230. [Google Scholar] [CrossRef]
- Roman, R.C.; Precup, R.E.; Petriu, E.M. Hybrid data-driven fuzzy active disturbance rejection control for tower crane systems. Eur. J. Control 2021, 58, 373–387. [Google Scholar] [CrossRef]
- Chi, R.; Li, H.; Shen, D.; Hou, Z.; Huang, B. Enhanced P-type Control: Indirect Adaptive Learning from Set-Point Updates. IEEE Trans. Autom. Control. 2022, 1, 1600–1613. [Google Scholar] [CrossRef]
- Wu, C.C.; Yeh, W.C.; Hsu, W.D.; Islam, M.M.; Nguyen, P.A.A.; Poly, T.N.; Wang, Y.C.; Yang, H.C.; Li, Y.C.J. Prediction of fatty liver disease using machine learning algorithms. Comput. Methods Programs Biomed. 2019, 170, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Chen, L.; Liu, Y.; Liu, C.; Feng, C.; Chen, B. A Machine Learning Methodology for Diagnosing Chronic Kidney Disease. IEEE Access 2020, 8, 20991–21002. [Google Scholar] [CrossRef]
- Khan, M.M.R.; Arif, R.B.; Siddique, M.A.B.; Oishe, M.R. Study and Observation of the Variation of Accuracies of KNN, SVM, LMNN, ENN Algorithms on Eleven Different Datasets from UCI Machine Learning Repository. In Proceedings of the 2018 4th International Conference on Electrical Engineering and Information & Communication Technology (iCEEiCT), Dhaka, Bangladesh, 13–15 September 2018; pp. 124–129. [Google Scholar]
- Abdar, M.; Ksiązek, W.; Acharya, U.R.; Tan, R.S.; Makarenkov, V.; Pławiak, P. A new machine learning technique for an accurate diagnosis of coronary artery disease. Comput. Methods Programs Biomed. 2019, 179, 104992. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yang, Z.; Li, P.; Yang, Z.; You, Y. Work-in-Progress: On the Feasibility of Lightweight Scheme of Real-Time Atrial Fibrillation Detection Using Deep Learning. In Proceedings of the 2019 IEEE Real-Time Systems Symposium (RTSS), Hong Kong, China, 3–6 December 2019; pp. 552–555. [Google Scholar]
- Hayward, G. Remarks on Smallpox, Cowpox and Varioloid. Boston Med. Surg. J. 1860, 62, 173–177. [Google Scholar] [CrossRef]
- Islam, T.; Hussain, M.A.; Chowdhury, F.U.H.; Islam, B.M.R. A Web-scraped Skin Image Database of Monkeypox, Chickenpox, Smallpox, Cowpox, and Measles; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2022. [Google Scholar] [CrossRef]
- Chen, J.; Wu, L.; Zhang, J.; Zhang, L.; Gong, D.; Zhao, Y.; Chen, Q.; Huang, S.; Yang, M.; Yang, X.; et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020, 10, 19196. [Google Scholar] [CrossRef]
- Reynolds, M.; Emerson, G.; Pukuta, E.; Karhemere, S.; Muyembe, J.; Bikin-dou, A.; McCollum, A.; Moses, C.; Wilkins, K.; Zhao, H.; et al. Detection of human monkeypox in the republic of the congo following intensive community education. Am. J. Trop. Med. Hygiene 2013, 88, 982. [Google Scholar] [CrossRef]
- Sitaula, C.; Shahi, T.B. Monkeypox virus detection using pre-trained deep learning-based approaches. J. Med. Syst. 2022, 46, 1–9. [Google Scholar] [CrossRef]
- Haque, M.; Ahmed, M.; Nila, R.S.; Islam, S. Classification of human monkeypox disease using deep learning models and attention mechanisms. arXiv 2022, arXiv:2211.15459. [Google Scholar]
- Shoieb, D.A.; Youssef, S.M.; Aly, W.M. Computer-Aided Model for Skin Diagnosis Using Deep Learning. J. Image Graph. 2016, 4, 122–129. [Google Scholar] [CrossRef]
- Abdelhamid, A.A.; El-Kenawy, E.S.M.; Khodadadi, N.; Mirjalili, S.; Khafaga, D.S.; Alharbi, A.H.; Saber, M. Classification of monkeypox images based on transfer learning and the Al-Biruni Earth Radius Optimization algorithm. Mathematics 2022, 10, 3614. [Google Scholar] [CrossRef]




| Reference | Techniques | Datasetcount | Recall (%) | Accuracy (%) | F1 Score (%) | Precision (%) |
|---|---|---|---|---|---|---|
| [34] | VGG-16 | 1428 | 81.0 | 81.48 | 83.01 | 85.01 |
| [34] | ResNet 50 | 1428 | 83.0 | 82.96 | 84.01 | 87 |
| [34] | Inception v3 | 1428 | 81.0 | 74.07 | 78 | 74.10 |
| [35] | DenseNet-169 | 1784 | 83.00 | 84.24 | 83.83 | 83.12 |
| Reference | Techniques | Datasetcount | Precision (%) | Accuracy (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|---|---|
| [34] | DQN | 1428 | 84 | 79.26 | 79.0 | 81.1 |
| [36] | DDQN | 1000 | 79.2 | 84.0 | 79.0 | 81.0 |
| [37] | Policy Gradient | 1200 | 80.8 | 85.1 | 91.1 | 76.5 |
| [38] | Actor–Critic | 89.0 | 63.0 | 92.0 | 74.0 | 90.0 |
| Parameters | Values |
|---|---|
| Optimizer | Adam |
| Learning rate | 0.001 |
| Loss | Binary_crossentropy |
| epoch | 200 |
| Batch size | 32 |
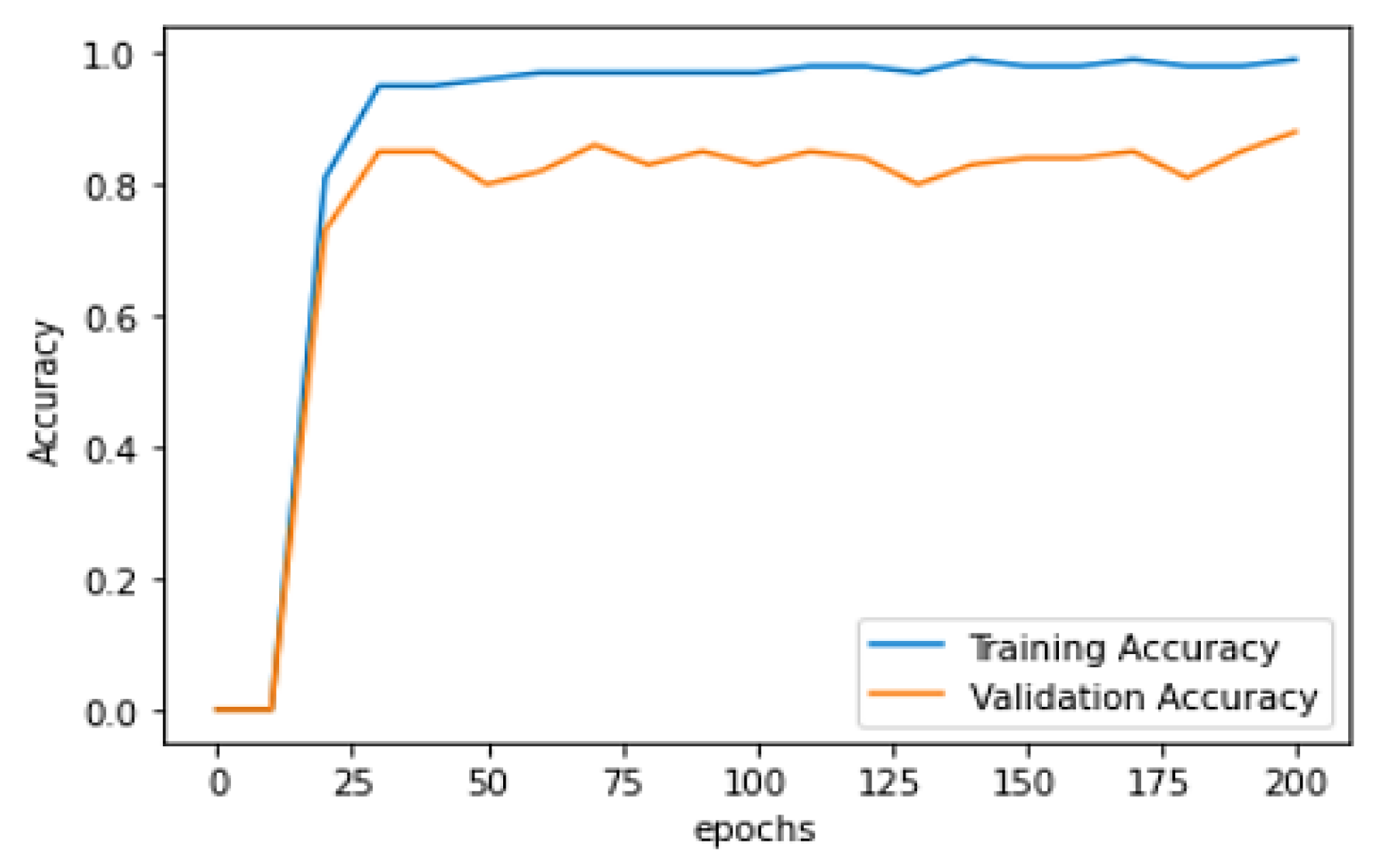
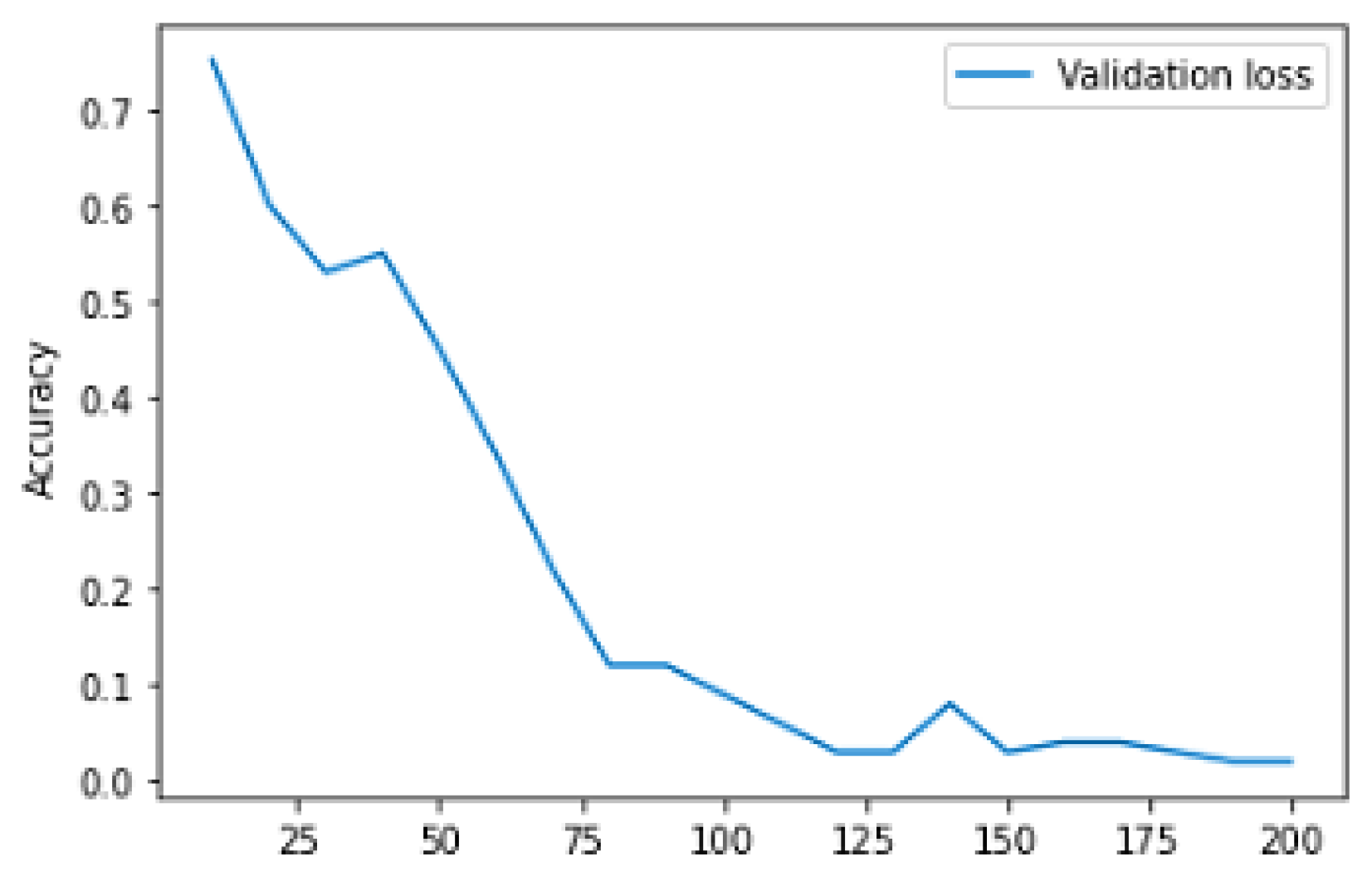
| Epoch | Training Accuracy | Training Loss | Validation Accuracy | Validation Loss |
|---|---|---|---|---|
| 10 | 0.8179 | 1.0625 | 0.7381 | 0.8807 |
| 20 | 0.9519 | 0.3025 | 0.8524 | 0.6117 |
| 30 | 0.9534 | 0.2310 | 0.8095 | 1.0032 |
| 40 | 0.9631 | 0.1515 | 0.7881 | 1.4253 |
| 50 | 0.9701 | 0.1502 | 0.7238 | 1.7884 |
| 60 | 0.9720 | 0.1743 | 0.9626 | 0.8833 |
| 70 | 0.9757 | 0.1239 | 0.8381 | 1.1028 |
| 80 | 0.9753 | 0.1242 | 0.8595 | 0.9159 |
| 90 | 0.9795 | 0.1258 | 0.8310 | 1.3588 |
| 100 | 0.9823 | 0.0971 | 0.8524 | 0.9706 |
| 110 | 0.9841 | 0.0676 | 0.8405 | 1.5409 |
| 120 | 0.9771 | 0.1351 | 0.8095 | 1.4022 |
| 140 | 0.9925 | 0.0320 | 0.8310 | 1.3769 |
| 150 | 0.9851 | 0.0806 | 0.8429 | 1.2375 |
| 160 | 0.9893 | 0.0393 | 0.8405 | 1.0522 |
| 170 | 0.9916 | 0.0394 | 0.8476 | 1.1742 |
| 180 | 0.9874 | 0.0470 | 0.8571 | 1.5278 |
| 190 | 0.9897 | 0.0476 | 0.8119 | 1.1780 |
| 200 | 0.9907 | 0.0528 | 0.8571 | 0.9906 |
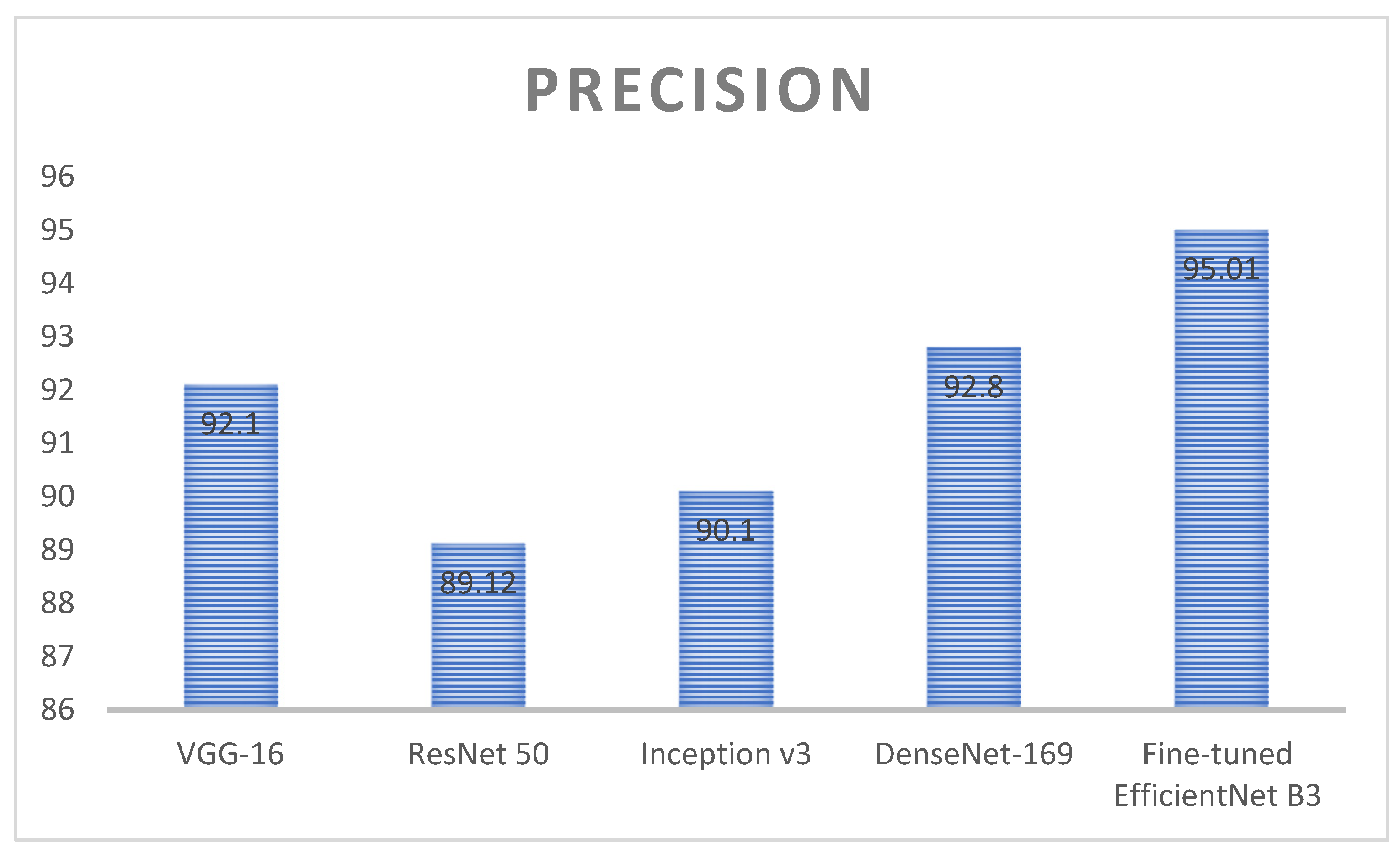
| Techniques | Precision (%) | Dataset Count |
|---|---|---|
| VGG-16 | 92.1 | 3192 |
| ResNet 50 | 89.12 | |
| Inception v3 | 90.1 | |
| DenseNet-169 | 92.8 | |
| Proposed model (fine-tuned EfficientNet B3) | 95.01 |
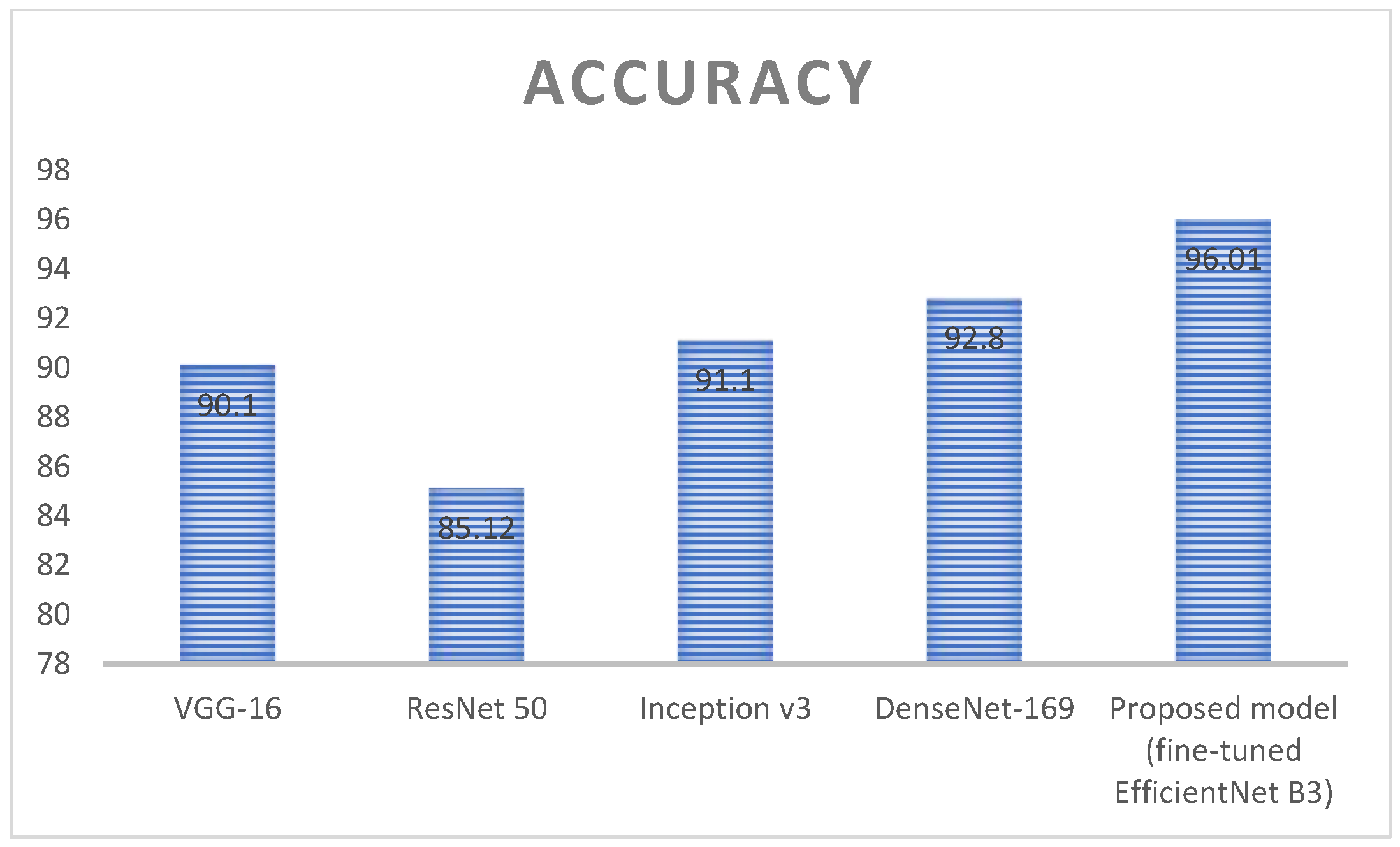
| Techniques | Accuracy (%) | Dataset Count |
|---|---|---|
| VGG-16 | 90.1 | 3192 |
| ResNet 50 | 85.12 | |
| Inception v3 | 91.1 | |
| DenseNet-169 | 92.8 | |
| Proposed model (fine-tuned EfficientNet B3) | 96.01 |
| Techniques | Recall (%) | Dataset Count |
|---|---|---|
| VGG-16 | 85.1 | 3192 |
| ResNet 50 | 85.12 | |
| Inception v3 | 84.1 | |
| DenseNet-169 | 90.8 | |
| Proposed model (fine-tuned EfficientNet B3) | 96.01 |
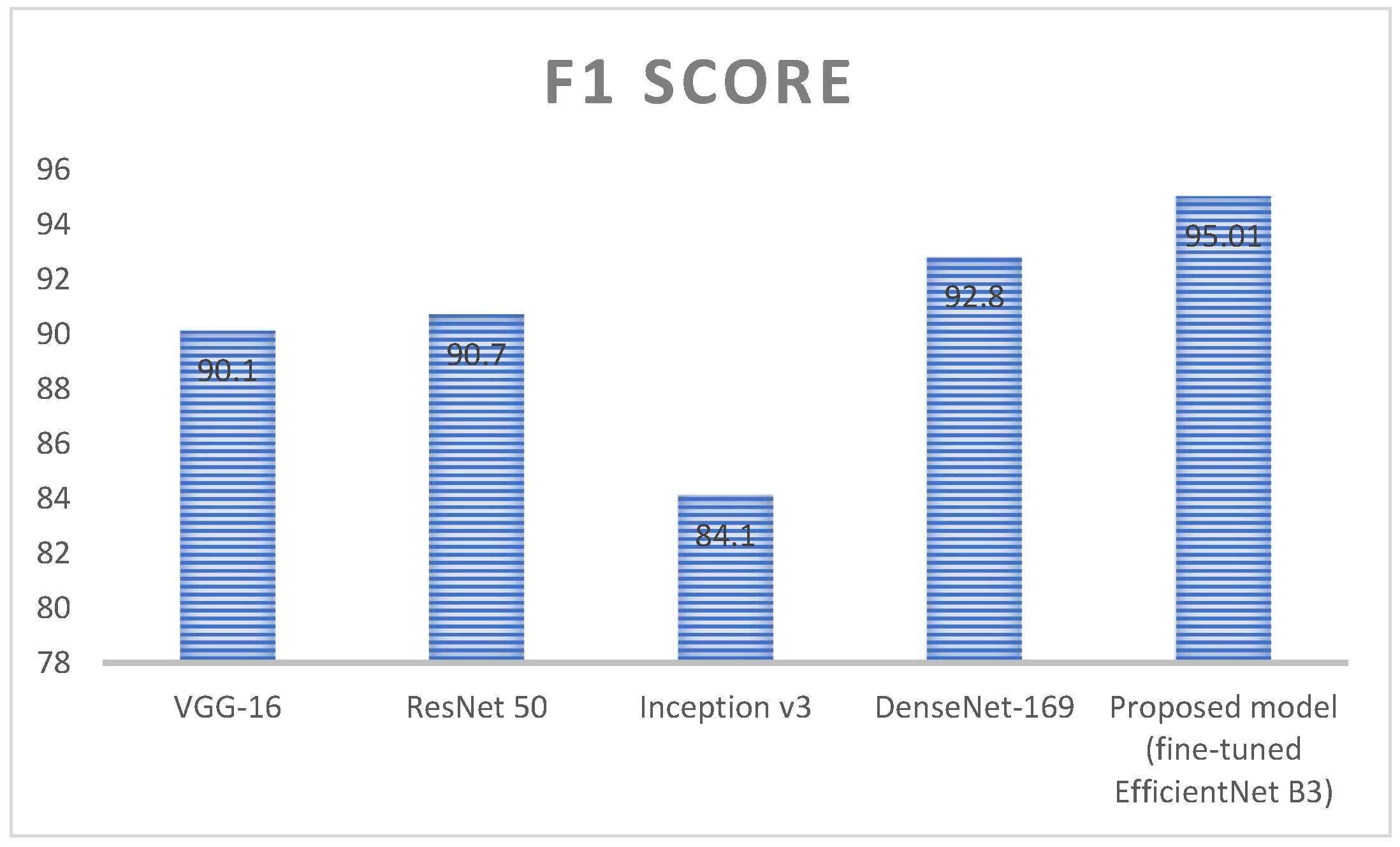
| Techniques | F1 Score (%) | Dataset Count |
|---|---|---|
| VGG-16 | 90.1 | 3192 |
| ResNet 50 | 91.12 | |
| Inception v3 | 84.1 | |
| DenseNet-169 | 90.7 | |
| Proposed model (fine-tuned EfficientNet B3) | 95.01 |
| Reinforcement Learning | Model | Accuracy |
| DQN | 96.5 | |
| DDQN | 89.7 | |
| Policy Gradient | 78.7 | |
| Actor–Critic | 80.7 | |
| Malneural | 97.7 |
| Reinforcement Learning | Model | F1 Score |
| DQN | 97.4 | |
| DDQN | 91.2 | |
| Policy Gradient | 79.0 | |
| Actor–Critic | 81.1 | |
| Malneural | 98.1 |
| Reinforcement Learning | Model | Precision |
| DQN | 94.3 | |
| DDQN | 89.4 | |
| Policy Gradient | 89.8 | |
| Actor–Critic | 92.0 | |
| Malneural | 96.1 |
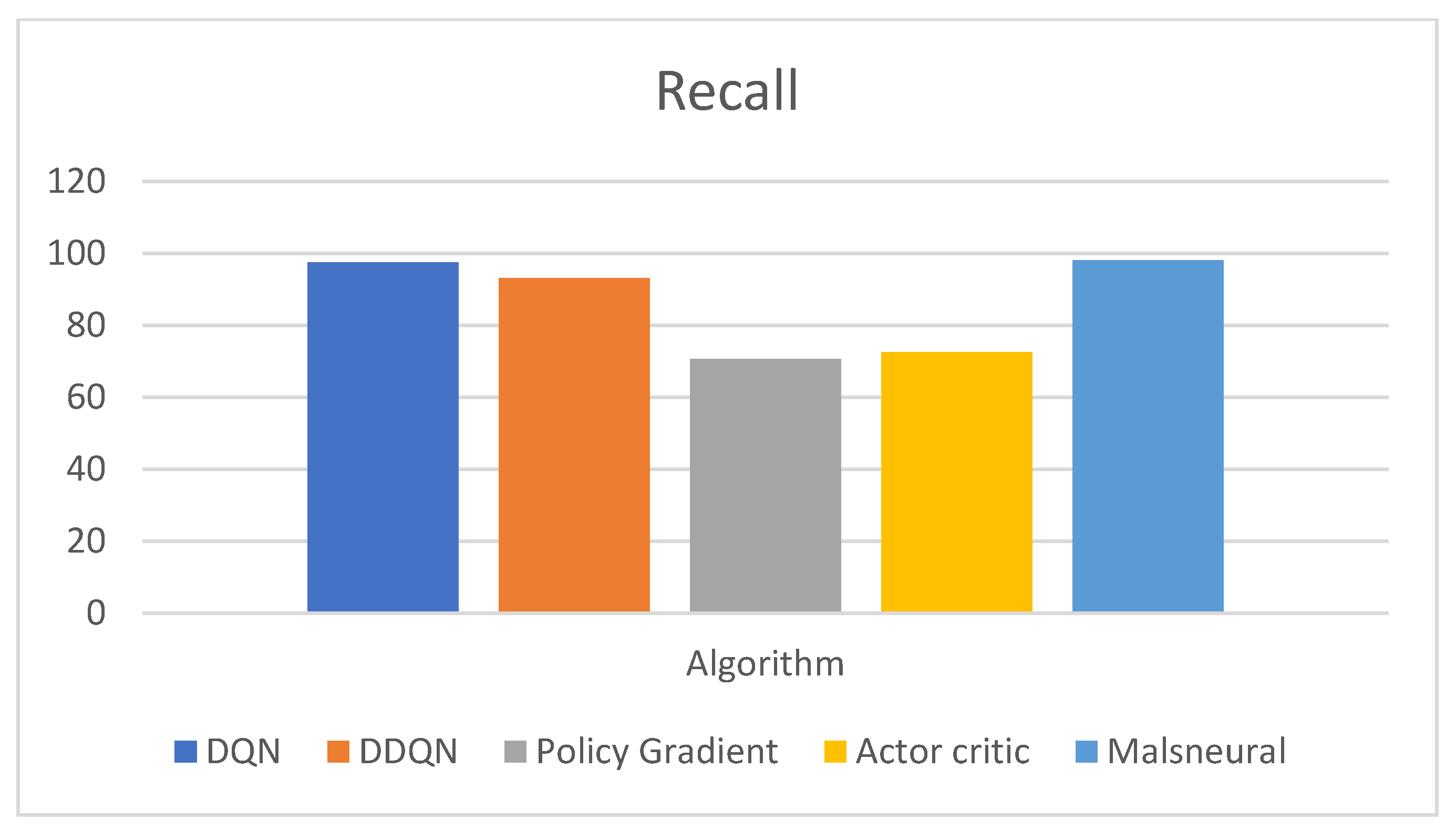
| Reinforcement Learning | Model | Recall |
| DQN | 97.4 | |
| DDQN | 93.0 | |
| Policy Gradient | 70.6 | |
| Actor–Critic | 72.5 | |
| Malneural | 98.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velu, M.; Dhanaraj, R.K.; Balusamy, B.; Kadry, S.; Yu, Y.; Nadeem, A.; Rauf, H.T. Human Pathogenic Monkeypox Disease Recognition Using Q-Learning Approach. Diagnostics 2023, 13, 1491. https://doi.org/10.3390/diagnostics13081491
Velu M, Dhanaraj RK, Balusamy B, Kadry S, Yu Y, Nadeem A, Rauf HT. Human Pathogenic Monkeypox Disease Recognition Using Q-Learning Approach. Diagnostics. 2023; 13(8):1491. https://doi.org/10.3390/diagnostics13081491
Chicago/Turabian StyleVelu, Malathi, Rajesh Kumar Dhanaraj, Balamurugan Balusamy, Seifedine Kadry, Yang Yu, Ahmed Nadeem, and Hafiz Tayyab Rauf. 2023. "Human Pathogenic Monkeypox Disease Recognition Using Q-Learning Approach" Diagnostics 13, no. 8: 1491. https://doi.org/10.3390/diagnostics13081491
APA StyleVelu, M., Dhanaraj, R. K., Balusamy, B., Kadry, S., Yu, Y., Nadeem, A., & Rauf, H. T. (2023). Human Pathogenic Monkeypox Disease Recognition Using Q-Learning Approach. Diagnostics, 13(8), 1491. https://doi.org/10.3390/diagnostics13081491










