Application of Targeted Next-Generation Sequencing for the Investigation of Thalassemia in a Developing Country: A Single Center Experience
Abstract
1. Introduction
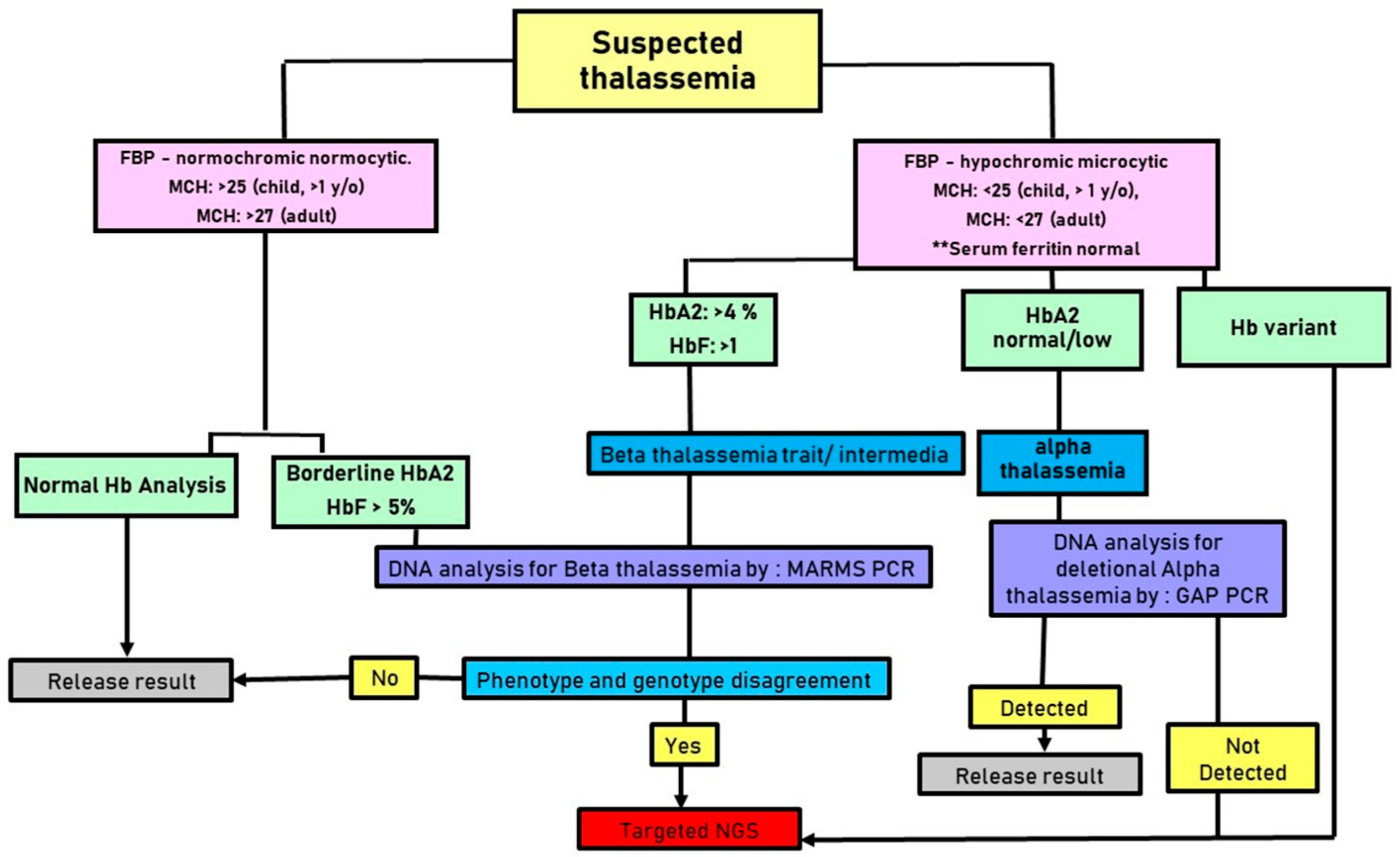
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weatherall, D.J. The Thalassemias: The Role of Molecular Genetics in an Evolving Global Health Problem. Am. J. Hum. Genet. 2004, 74, 385. [Google Scholar] [CrossRef] [PubMed]
- Cao, A.; Kan, Y.W. The Prevention of Thalassemia. Cold Spring Harb. Perspect. Med. 2013, 3, a011775. [Google Scholar] [CrossRef]
- Ngim, C.F.; Ibrahim, H.; Lai, N.M.; Ng, C.S. A single centre study on birth of children with transfusion-dependent thalassaemia in Malaysia and reasons for ineffective prevention. Prenat. Diagn. 2015, 35, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M. Malaysian thalassaemia registry report 2018. Minist. Health Malays. 2019, 19, 203–226. [Google Scholar]
- Ismail, A.; Campbell, M.J.; Ibrahim, H.M.; Jones, G.L. Health Related Quality of Life in Malaysian children with thalassaemia. Health Qual. Life Outcomes 2006, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, J.L.; Kim, H.-Y.; Thompson, A.A.; Quinn, C.T.; Mueller, B.U.; Odame, I.; Giardina, P.J.; Vichinsky, E.P.; Boudreaux, J.M.; Cohen, A.R.; et al. Chelation use and iron burden in North American and British thalassemia patients: A report from the Thalassemia Longitudinal Cohort. Blood 2012, 119, 2746. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Cho, S.I.; Park, S.S.; Seong, M.W. Molecular basis and diagnosis of thalassemia. Blood Res. 2021, 56, S39–S43. [Google Scholar] [CrossRef]
- Elizabeth, G.; Ann, T.J.A.M. Genotype-Phenotype Diversity of Beta-Thalassemia in Malaysia: Treatment Options and Emerging Therapies. Med. J. Malays. 2010, 65, 256–260. [Google Scholar]
- Azma, R.Z.; Ainoon, O.; Hafiza, A.; Azlin, I.; Farisah, A.R.N.; Hidayati, S.N.; Hamidah, H.N. Molecular characteristic of alpha thalassaemia among patients diagnosed in UKM Medical Centre. Malays. J. Pathol. 2014, 36, 27–32. [Google Scholar]
- Tan, J.-A.M.A.; Lee, P.-C.; Wee, Y.-C.; Tan, K.-L.; Mahali, N.F.; George, E.; Chua, K.-H. High prevalence of alpha- and beta-thalassemia in the kadazandusuns in east Malaysia: Challenges in providing effective health care for an indigenous group. J. Biomed. Biotechnol. 2010, 2010, 706872. [Google Scholar] [CrossRef]
- Lithanatudom, P.; Khampan, P.; Smith, D.R.; Svasti, S.; Fucharoen, S.; Kangwanpong, D.; Kampuansai, J. The prevalence of alpha-thalassemia amongst Tai and Mon-Khmer ethnic groups residing in northern Thailand: A population-based study. Hematology 2016, 21, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, W.; Chen, H.; Zhang, Y.; Wang, F.; Chen, H.; Zhou, Y.; Huang, Y.; Zhou, X.; Li, Q.; et al. Prevalence and molecular spectrum of α- and β-globin gene mutations in Hainan, China. Int. J. Hematol. 2021, 114, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Setianingsih, I.; Harahap, A.; Nainggolan, I.M. Alpha thalassaemia in Indonesia: Phenotypes and molecular defects. Adv. Exp. Med. Biol. 2003, 531, 47–56. [Google Scholar] [CrossRef] [PubMed]
- George, E.; Teh, L.K.; Rosli, R.; Lai, M.I.; Tan, J. Beta Thalassaemia Mutations in Malays: A Simplified Cost-effective Strategy to Identify the Mutations. Malays. J. Med. Health Sci. 2012, 8, 1–8. [Google Scholar]
- Boonyawat, B.; Monsereenusorn, C.; Traivaree, C. Molecular analysis of beta-globin gene mutations among Thai beta-thalassemia children: Results from a single center study. Appl. Clin. Genet. 2014, 7, 253–258. [Google Scholar] [CrossRef]
- Rujito, L.; Basalamah, M.; Mulatsih, S.; Sofro, A.S.M. Molecular Scanning of β-Thalassemia in the Southern Region of Central Java, Indonesia; a Step Towards a Local Prevention Program. Hemoglobin 2015, 39, 330–333. [Google Scholar]
- Leckngam, P.; Limweeraprajak, E.; Kiewkarnkha, T.; Tatu, T. The Hb E (HBB: C.79G>A), Mean Corpuscular Volume, Mean Corpuscular Hemoglobin Cutoff Points in Double Heterozygous Hb E/– –SEA α-Thalassemia-1 Carriers are Dependent on Hemoglobin Levels. Hemoglobin 2017, 41, 38–43. [Google Scholar] [CrossRef]
- Yokoyama, A.; Nakamaki, T.; Yamada, K.; Koike, M.; Tomoyasu, S.; Hirayama, N.; Tsuruoka, N.; Harano, T. Beta 0-thalassemia trait (IVS-I-1 G-->T) in a Japanese family. Intern. Med. 1993, 32, 865–868. [Google Scholar] [CrossRef]
- Wisedpanichkij, R.; Jindadamrongwech, S.; Butthep, P. Identification of Hb Constant Spring (HBA2: C.427T > C) by an Automated High Performance Liquid Chromatography Method. Hemoglobin 2015, 39, 190–195. [Google Scholar] [CrossRef]
- Alwi, Z.B.; Syed-Hassan, S.N.R.K. Thalassemia in Malaysia. Hemoglobin 2022, 46, 45–52. [Google Scholar] [CrossRef]
- Murad, H.; Moassas, F.; Fakseh, N.A.L. A rare gene variation cap +1 (A>C) (HBB: c. -50A>C) associated with codon 5 (-CT) (HBB: c.17_18delCT) mutation in Syrian family. Mol. Genet. Genom. Med. 2021, 9, e1602. [Google Scholar] [CrossRef]
- Fauzi, M.; Yusoff, M.; Rahayu, E.; Tohit, M.; Hashim, H.; Seman, Z. Evaluation of Dichlorophenolindophenol (DCIP) Test for Haemoglobin E (Hb E) in Normal Red Cell Indices Individuals. Malays. J. Med. Health Sci. 2021, 17, 10–13. [Google Scholar]
- Farmakis, D.; Porter, J.; Taher, A.; Cappellini, M.D.; Angastiniotis, M.; Eleftheriou, A. 2021 Thalassaemia International Federation Guidelines for the Management of Transfusion-dependent Thalassemia. HemaSphere 2022, 6, e732. [Google Scholar] [CrossRef]
- Wong, S.L.; Ng, H.P.; Tam, P.Y.; Hung, L.C.; Muda, Z. Report Management of Thalassaemia. Health Technology Assessment Unit Medical Development Division Ministry of Health Malaysia. 2003. Available online: https://www.moh.gov.my/moh/resources/autodownloadimages/587f136ce4807.pdf (accessed on 25 February 2023).
- Traeger-Synodinos, J.; Harteveld, C.L.; Old, J.M.; Petrou, M.; Galanello, R.; Giordano, P.; Angastioniotis, M.; De la Salle, B.; Henderson, S.; May, A.; et al. EMQN Best Practice Guidelines for molecular and haematology methods for carrier identification and prenatal diagnosis of the haemoglobinopathies. Eur. J. Hum. Genet. 2015, 23, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Bahar, R.; Johan, M.F.; Hashim, E.K.M.; Abdullah, W.Z.; Esa, E.; Hamid, F.S.A.; Zulkafli, Z. Next-Generation Sequencing (NGS) and Third-Generation Sequencing (TGS) for the Diagnosis of Thalassemia. Diagnostics 2023, 13, 373. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Song, W.; Yang, J.; Lu, S.; Yuan, Y.; Guo, J.; Zhang, J.; Ye, K.; Yang, F.; Long, F.; et al. Next-generation sequencing improves thalassemia carrier screening among premarital adults in a high prevalence population: The Dai nationality, China. Genet. Med. 2017, 19, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- SSuhaimi, A.; Zulkipli, I.N.; Ghani, H.; Abdul-Hamid, M.R.W. Applications of next generation sequencing in the screening and diagnosis of thalassemia: A mini-review. Front. Pediatr. 2022, 10, 1015769. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Peng, Z.; Ye, Y.; Asan; Zhang, X.; Chen, Y.; Zhu, B.; Cai, W.; Chen, S.; Cai, R.; et al. Rapid Targeted Next-Generation Sequencing Platform for Molecular Screening and Clinical Genotyping in Subjects with Hemoglobinopathies. EBioMedicine 2017, 23, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, C.; Li, J.; Hou, S.; Chen, D.; Yan, H.; Chen, S.; Liu, S.; Yin, Z.; Yang, X.; et al. Next-generation sequencing improves molecular epidemiological characterization of thalassemia in Chenzhou Region, P.R. China. J. Clin. Lab. Anal. 2019, 33, e22845. [Google Scholar] [CrossRef]
- Kwaifa, I.K.; Lai, M.I.; Noor, S.M. Non-deletional alpha thalassaemia: A review. Orphanet J. Rare Dis. 2020, 15, 166. [Google Scholar] [CrossRef]
- Jaing, T.H.; Chang, T.Y.; Chen, S.H.; Lin, C.W.; Wen, Y.C.; Chiu, C.C. Molecular genetics of β-thalassemia: A narrative review. Medicine 2021, 100, E27522. [Google Scholar] [CrossRef] [PubMed]
- Panyasai, S.; Permsripong, N.; Pornprasert, S. Hemoglobin J-Singapore [α79(EF8)Ala→Gly, GCG>GGG] in a pregnant Thai woman. J. Med. Assoc. Thail. 2018, 101, 275–278. [Google Scholar]
- Blackwell, R.Q.; Boon, W.H.; Liu, C.-S.; Weng, M.-I. Hemoglobin J Singapore: α78 Asn → Asp; α79 Ala → Gly. Biochim. Biophys. Acta Protein Struct. 1972, 278, 482–490. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, D.A.; Clark, B.; Rai, D.K. Alpha78(EF7)Asn-->Asp is a posttranslational modification in Hb J-Singapore [alpha78(EF7)Asn-->Asp;alpha79(EF8)Ala-->Gly]. Hemoglobin 2007, 31, 483–489. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, F.; Li, D.Z. Hematological Characteristics of β-Globin Gene Mutation -50 (G>A) (HBB: c.-100G>A) Carriers in Mainland China. Hemoglobin 2020, 44, 240–243. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, X.M.; Wu, L.S.; Chen, J.; Chen, Y. Prevalence and clinical phenotype of the triplicated α-globin genes and its ethnic and geographical distribution in Guizhou of China. BMC Med. Genom. 2021, 14, 97. [Google Scholar] [CrossRef]
- Rameli, N.; Ramli, M.; Zulkafli, Z.; Hassan, M.N.; Yusoff, S.M.; Noor, N.H.M.; Hussin, S.; Kamarudin, N.K.M.; Yusoff, Y.M.; Bahar, R. Challenges in diagnosis of Beta Thalassemia syndrome: The Importance of Molecular Diagnosis. Oman Med. J. 2022, 37, e331. [Google Scholar] [CrossRef]
- Steinberg-Shemer, O.; Ulirsch, J.; Noy-Lotan, S.; Krasnov, T.; Attias, D.; Dgany, O.; Laor, R.; Sankaran, V.G.; Tamary, H. Whole-exome sequencing identifies an α-globin cluster triplication resulting in increased clinical severity of β-thalassemia. Cold Spring Harb. Mol. Case Stud. 2017, 3, a001941. [Google Scholar] [CrossRef]
- Yang, K.G.; Kutlar, F.; George, E.; Wilson, J.B.; Kutlar, A.; Stoming, T.A.; Redondo, J.M.G.; Huisman, T.H.J. Molecular characterization of β-globin gene mutations in Malay patients with Hb E-β-thalassaemia and thalassaemia major. Br. J. Haematol. 1989, 72, 73–80. [Google Scholar] [CrossRef]
- Ma, S.K.; Chow, E.Y.; Chan, A.Y.; Kung, N.N.; Waye, J.S.; Chan, L.C.; Chui, D.H. Thalassemia Intermedia Caused by Compound Heterozygosity for Hb Malay (Codon 19 AAC→AGC; Asn→Ser) and Codons 41/42 (-CTTT) 0-Thalassemia Mutation. J. Hematol. 2000, 64, 206–209. [Google Scholar] [CrossRef]
- Shahida, N.S.; Nazri, M.H.; Haslina, N.M.; Zaidah, W.A. The Diagnosis of Beta Thalassemia with Borderline HbA2 Level among Kelantan Population. J. Blood Disord. Transfus. 2017, 8, 2. [Google Scholar] [CrossRef]
- Amran, H.S.; Sarijan, N.; Sathar, J.S.; Noor, S.M. Case Series of Homozygous and Compound Heterozygosity of Hb Malay, the Diagnostic Features and Transfusion Requirements. J. Biomed. Clin. Sci. 2018, 2, 23–25. [Google Scholar]
- Pervez, M.T.; Hasnain, M.J.U.; Abbas, S.H.; Moustafa, M.F.; Aslam, N.; Shah, S.S.M. A Comprehensive Review of Performance of Next-Generation Sequencing Platforms. BioMed Res. Int. 2022, 2022, 3457806. [Google Scholar] [CrossRef] [PubMed]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child. Educ. Pract. Ed. 2013, 98, 236. [Google Scholar] [CrossRef] [PubMed]
- Sabath, D.E. Molecular Diagnosis of Thalassemias and HemoglobinopathiesAn ACLPS Critical Review. Am. J. Clin. Pathol. 2017, 148, 6–15. [Google Scholar] [CrossRef]
- Achour, A.; Koopmann, T.T.; Baas, F.; Harteveld, C.L. The Evolving Role of Next-Generation Sequencing in Screening and Diagnosis of Hemoglobinopathies. Front. Physiol. 2021, 12, 686689. [Google Scholar] [CrossRef]
- Synthesis (SBS) Technology. Sequencing Technology|Sequencing by Synthesis. Available online: https://sapac.illumina.com/science/technology/next-generation-sequencing/sequencing-technology.html (accessed on 29 January 2023).
- Vijian, D.; Rahman, W.S.W.A.; Ponnuraj, K.T.; Zulkafli, Z.; Noor, N.H.M. Molecular Detection of Alpha Thalassemia: A Review of Prevalent Techniques. Medeni. Med. J. 2021, 36, 257. [Google Scholar] [CrossRef]
- Clark, B.E.; Shooter, C.; Smith, F.; Brawand, D.; Thein, S.L. Next-generation sequencing as a tool for breakpoint analysis in rearrangements of the globin gene clusters. Int. J. Lab. Hematol. 2017, 39, 111–120. [Google Scholar] [CrossRef]
| Alpha-Thalassemia | |||
|---|---|---|---|
| Countries | Genotype Variants | Incidence | References |
| Malaysia | |||
| Malay | αα/-SEA | 44.3% | Azma et al. [9] |
| αα/-α3.7 | 33.5% | ||
| -α3.7/-SEA | 7.1% | ||
| αα/ααCS | 6.2% | ||
| Chinese | αα/-SEA | 83.4% | |
| αα/-α3.7 | 4.9% | ||
| -α3.7/-SEA | 7.2% | ||
| αα/ααCS | 0.5% | ||
| -SEA/-SEA | 2.0% | ||
| Indian | αα/-SEA | 5% | |
| αα/-α3.7 | 90% | ||
| Kadazandusun | -α3.7/αα | 33.6% (42/125) | Tan et al. [10] |
| Thailand | αα/-α3.7 | 17.7% | Lithanatudom et al. [11] |
| αα/-SEA | 3.5% | ||
| _αCS/αα | 2.1% | ||
| China | -SEA/αα | 31.53% | Wang et al. [12] |
| -α4.2/αα | 11.15% | ||
| -α3.7/αα | 11.02% | ||
| -αWS/αα | 5.48% | ||
| Indonesia | SEA | 16.2% | Setianingsih et al. [13] |
| -α3.7 | 14.1% | ||
| Cd 59 (GGC>GAC) | 3.0% | ||
| Cd 22 (GGC>GGT) | 1.0% | ||
| Hb Constant Spring | 1.0% | ||
| Beta-thalassemia | |||
| Countries | Genotype variants | Incidence | References |
| Malaysia | |||
| Malay | IVS I-5 | 15.3% | George et al. [14] |
| CD 26 | 32.1% | ||
| IVS I-1 | 8.0% | ||
| CD 19 | 7.3% | ||
| CD 41/42 | 3.6% | ||
| CD 17 | 1.5% | ||
| -29 | 1.5% | ||
| CD 8/9 | 0.7 | ||
| CAP + 1 | 0% | ||
| Homozygous CD 19/CD 19 | 0.7% | ||
| Homozygous CD 71/72 | 0.7% | ||
| IVS I-1/CD 19 | 0.7% | ||
| CD 26/CD 26 | 3.6% | ||
| CD 26/IVS I-5 | 9.5% | ||
| CD 26/IVS I-1 | 5.1% | ||
| CD 26/CD 19 | 3.6% | ||
| Kadazandusun | 45 kb Filipino β-deletion | 12.8% (16/125) | Tan et al. [10] |
| Thailand | Codon 41/42 (-TCTT) | 37.5% | Traivaree et al. [15] |
| Codon 17 (A>T) | 26.1% | ||
| IVS-I-5 (G>C) | 8.0% | ||
| IVS-II-654 (C>T) | 6.8% | ||
| IVS-I-I (G>T) | 4.5% | ||
| Codon 71/72 (+A) | 2.3% | ||
| Codon 35 (C > A) | 4.5% | ||
| Initiation codon mutation (c.2T > G) | 1.1% | ||
| Codon 15 (G>A) | 1.1% | ||
| Codon 19 (A>G) | 1.1% | ||
| Codon 27/28 (+C) | 1.1% | ||
| Codon 123/124/125 | 1.1% | ||
| (-ACCCCACC) | |||
| 3.2 kb deletion | 4.5% | ||
| China | CD41/42 | 30.27% | Wang et al. [12] |
| -28 | 2.56% | ||
| IVS-II-654 | 1.02% | ||
| Indonesia | IVS-I-5 (G>C) | 43.5% | Rujito et al. [16] |
| Codon 26 (G>A) | 28.2% | ||
| IVS-I-1 (G>A) | 5.0% | ||
| Codon 15 (TGG>TAG) | 3.8% | ||
| IVS-I-1(G>T) | 3.1% | ||
| Codon 35 (-C) | 2.4% | ||
| Codon 41/42 (-TTCT) | 1.0% | ||
| Codon 8/9 (+G) | 1.0% | ||
| Codon 19 (AAC > AGC) | 0.7% | ||
| Codon123/124/125 | 0.5% | ||
| (-ACCCCACC) | |||
| Cases | FBC | Hb Analysis (HPLC) | Molecular | Devyser NGS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RBC | Hb (g/L) | MCV (80-100 fl) | MCH (pg/Cell) <25, Child <27, Adult | HbA | HbA2 | HbF | MARMS (Beta-Thalassemia) | GAP-PCR (Alpha-Thalassemia) | |||
| 1 | 3-year-old female | 3.06 | 6.5 | 66.0 | 21.2 | 5.9 | 46.1 | 48 | CD 26 (G>A) IVS I-1 (G>T) | Not performed | CD 26 (HBB:c.79G>A) IVS I-1 (HBB:c.92 + 1G>T) |
| 2 | 27-year-old female | 3.98 | 8.6 | 66.1 | 21.6 | 51.6 | 43.6 | 4.8 | CD 26 (G>A) Poly A (A>G) | Not performed | CD 26 (HBB:c.79G>A) Poly A (HBB:c.*112A>G) |
| 3 | 3-year-old female | 5.00 | 12.5 | 72.4 | 25 | 71.7 | 24.7 | 3.6 | Not performed | aa/a4.2 | aa/a4.2 CD 26 (HBB:c.79G>A) |
| 4 | 4-year-old male | 5.59 | 9.7 | 55.5 | 17.4 | 95.9 | 2.9 | 1.2 | Not performed | SEA deletion | SEA deletion |
| 5 | 23-year-old male | 6.02 | 13.4 | 69.6 | 22.3 | 97.1 | 2.6 | 0.3 | Not performed | THAI deletion | THAI deletion |
| 6 | 36-year-old female | 6.06 | 14.2 | 76.2 | 23.4 | 96 | 2.8 | 1.2 | Not performed | aa/a3.7 | aa/a3.7 |
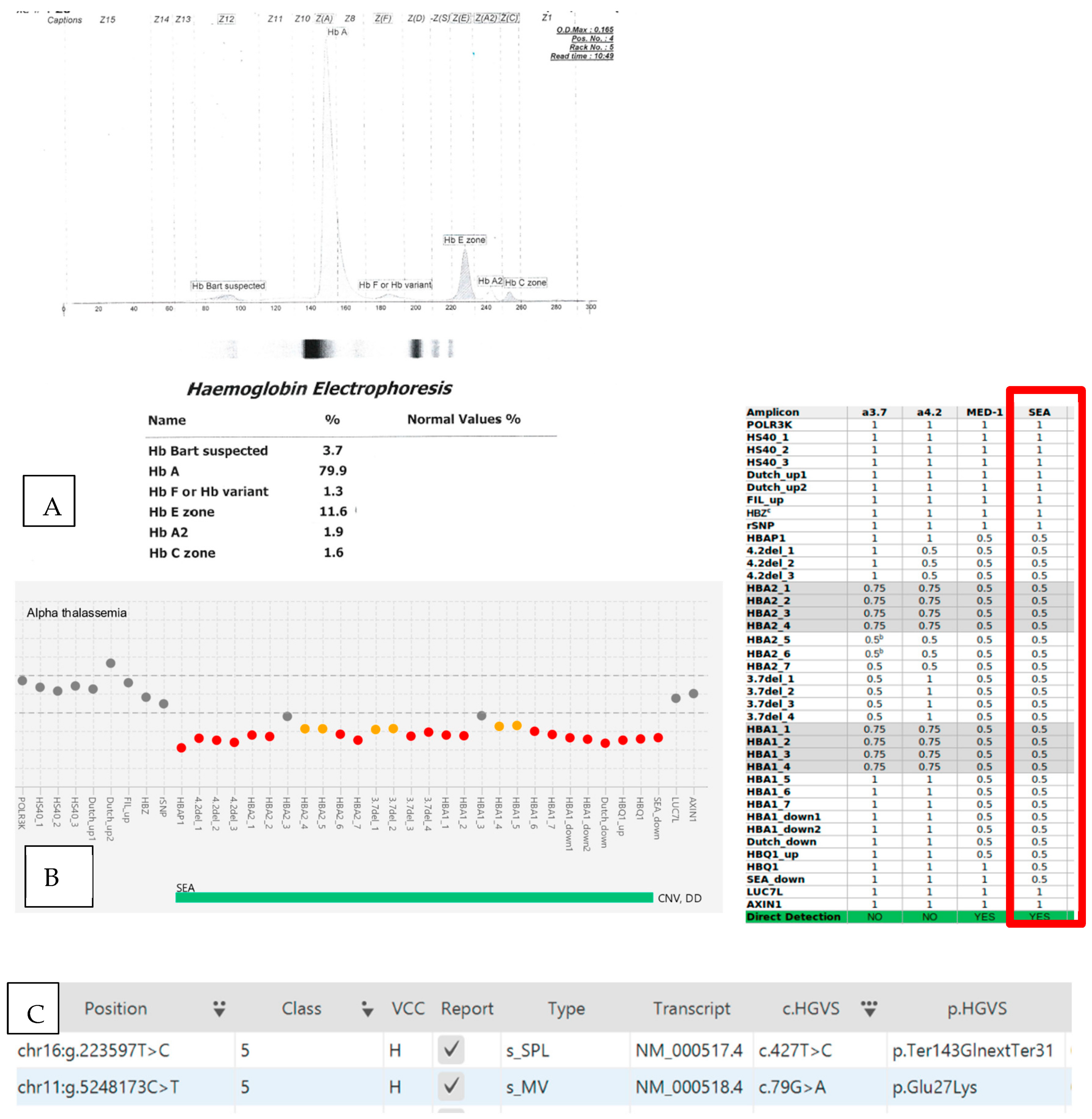
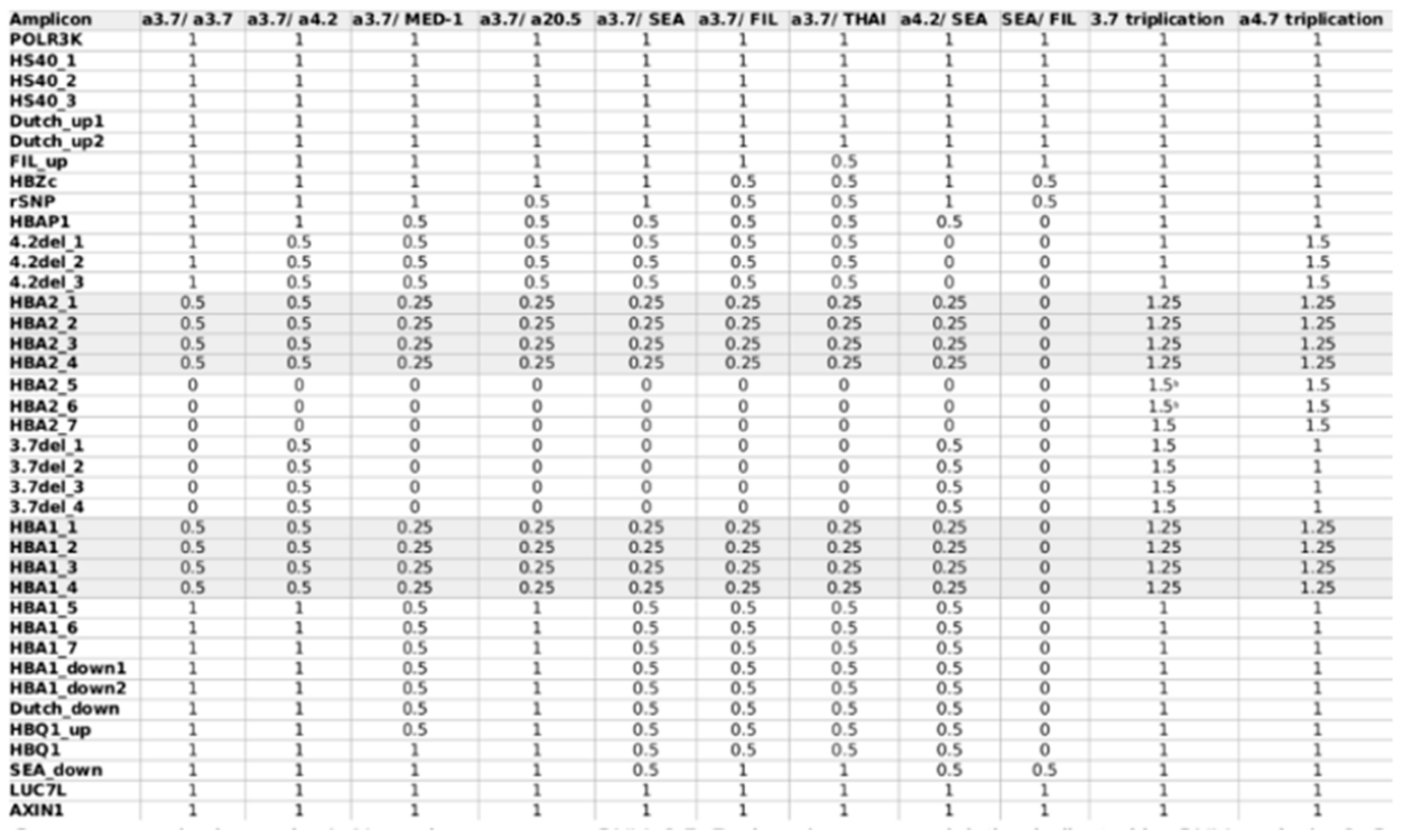
| 7 | 27-year-old female | 4.98 | 7.5 | 55 | 15.1 | 81.9 | 14.6 | 3.5 | CD 26 (HBB:c.79G>A) | SEA deletion | SEA deletion CD 26 (HBB:c.79G>A) CD 142 (HBA2:c.427T>C) |
| Hb Bart’s disease. HbE trait with Hb Constant Spring | |||||||||||
| 8 | 29-year-old female | 5.23 | 10.7 | 65 | 20.5 | 92.1 | 5.5 | 2.4 | Heterozygous IVS I-5(G>C) | Not performed | IVS I-5 (HBB:c.92 + 5G>C) |
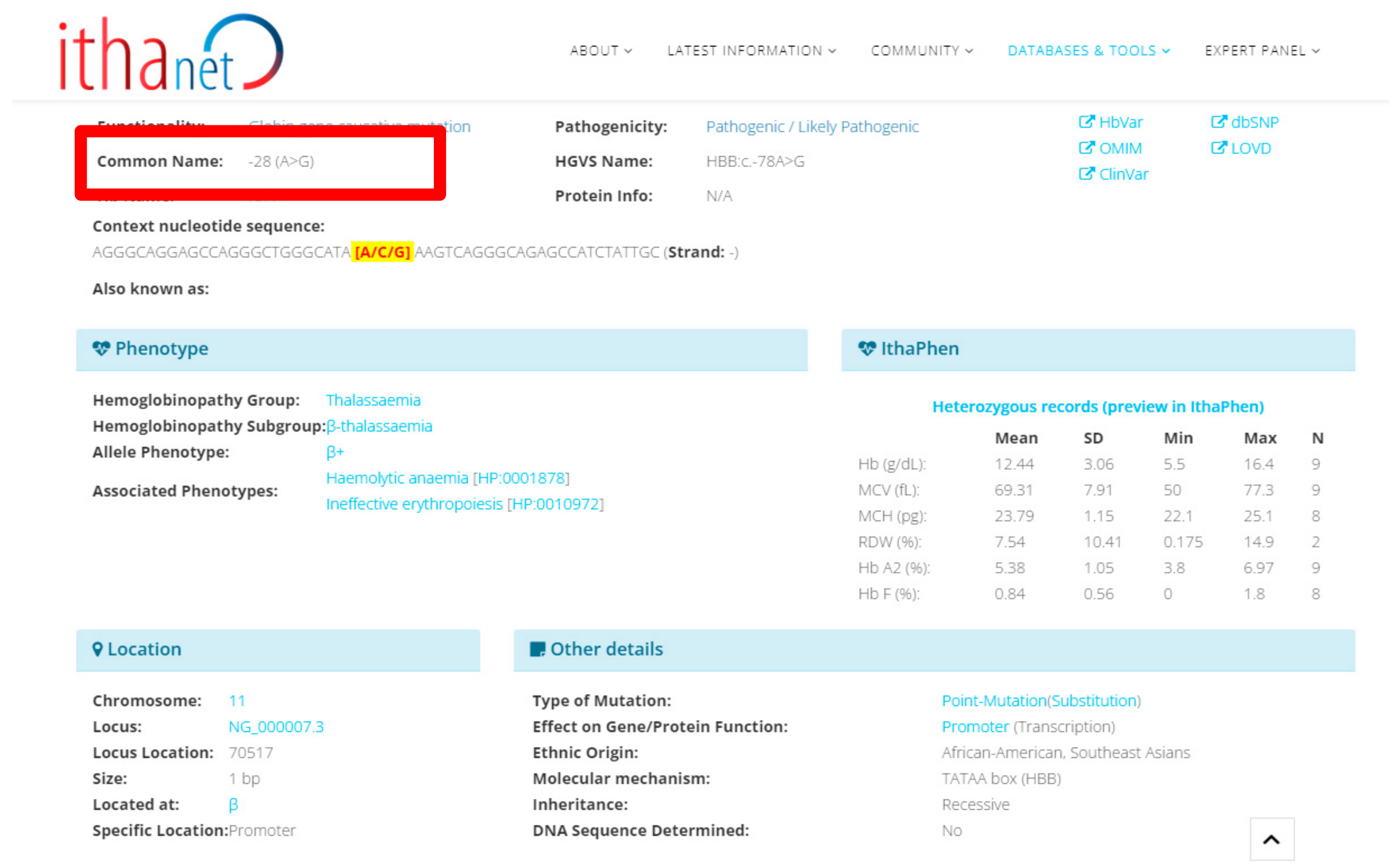
| 9 | 30-year-old female | 3.37 | 8.0 | 73 | 23.7 | Not available (Refer by other hospital) | Homozygous -28 (A>G) | Not performed | -28 (A>G) (HBB:c.-78A>G) | ||
| 10 | 18-year-old female | 5.72 | 8.8 | 53.5 | 15.4 | 90.3 | 4.3 | 5.4 | Heterozygous CD 41/42 | Not performed | CD 41/42; (HBB:c.126_129del) CD 79 (HBA2:c.239C>G) |
| Abnormal band (Presence of fast band at Hb Bart’s region with prominent A2/E band) on Alkaline gel electrophoresis | |||||||||||
| 11 | 7-year-old female | 5.53 | 10.8 | 62 | 19.5 | 55.2 | 42.6 | 2.2 | CD 26 (G>A) CAP + 1 (A>C) | Not performed | CD 26 (HBB:c.79G>A) CAP + 1 (HBB:c.-50A>C); CD 142 (HBA2:c.427T>C) |
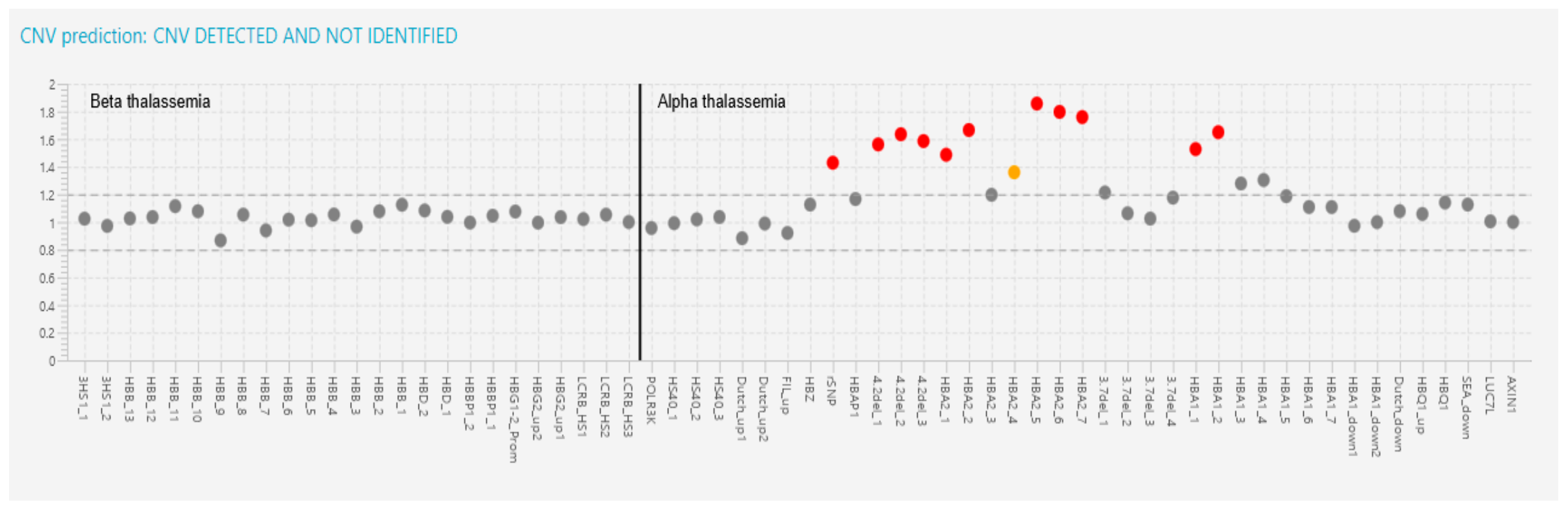
| 12 | 35-year-old female | 3.3 | 6.8 | 66.4 | 20.6 | 92.1 | 5.8 | 2.1 | Heterozygous CD 8/9 (+G) | Not performed | CD 8/9 (HBB:c.27dup); -50 G>A (HBB:c.-100G>A) 4.2 triplication |
| Recently transfuse sample | |||||||||||
| 13 | 8-year-old male | 5.93 | 11.2 | 62.1 | 18.9 | 93.9 | 4.7 | 1.4 | Heterozygous CD19 (A>G) | Not performed | CD19 (HBB:c.59A>G) |
| 14 | 5-month-old male | 1.87 | 4.6 | 71.1 | 24.6 | 1.9 | 2.4 | 95.7 | CD17 (A>T) IVS II-654 (C>T) | Not performed | CD17 (HBB:c.52A>T) IVS II-654 (HBB:c.316-197C>T) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zulkeflee, R.H.; Bahar, R.; Abdullah, M.; Mohd Radzi, M.A.R.; Md Fauzi, A.; Hassan, R. Application of Targeted Next-Generation Sequencing for the Investigation of Thalassemia in a Developing Country: A Single Center Experience. Diagnostics 2023, 13, 1379. https://doi.org/10.3390/diagnostics13081379
Zulkeflee RH, Bahar R, Abdullah M, Mohd Radzi MAR, Md Fauzi A, Hassan R. Application of Targeted Next-Generation Sequencing for the Investigation of Thalassemia in a Developing Country: A Single Center Experience. Diagnostics. 2023; 13(8):1379. https://doi.org/10.3390/diagnostics13081379
Chicago/Turabian StyleZulkeflee, Razan Hayati, Rosnah Bahar, Marne Abdullah, Muhammad Amiro Rasheeq Mohd Radzi, Alina Md Fauzi, and Rosline Hassan. 2023. "Application of Targeted Next-Generation Sequencing for the Investigation of Thalassemia in a Developing Country: A Single Center Experience" Diagnostics 13, no. 8: 1379. https://doi.org/10.3390/diagnostics13081379
APA StyleZulkeflee, R. H., Bahar, R., Abdullah, M., Mohd Radzi, M. A. R., Md Fauzi, A., & Hassan, R. (2023). Application of Targeted Next-Generation Sequencing for the Investigation of Thalassemia in a Developing Country: A Single Center Experience. Diagnostics, 13(8), 1379. https://doi.org/10.3390/diagnostics13081379






