A Foreground Prototype-Based One-Shot Segmentation of Brain Tumors
Abstract
1. Introduction
- Propose a one-shot learning segmentation model by considering the foreground prototypes for brain tumor detection on brain MRI that transcends the current deep learning-based tumor segmentation models in regard to the size of the training data.
- Adopt the VGG-16-based encoder with pre-trained weights from Resnet101 trained on MS COCO datasets as our few-shot learning model.
- Experiment with different N-shot K-way methods to show the effectiveness of the proposed single prototype for modeling the foreground classes based on its uniformity.
- Compare the performance of the proposed approach to architectures that are used for brain tumor segmentation.
- Evaluate the performance of our methods by using the dice similarity coefficient and intersection over union.
2. Related Work
2.1. Deep Learning-Based Brain Tumor Segmentation
2.2. Self-Supervised Learning
2.3. Few-Shot Learning
2.4. Few-Shot Segmentation
3. Proposed Methodology
3.1. Problem Foundation
3.2. Few-Shot Learning Model
3.2.1. Preprocessing and the Extraction of Prototype Features
3.2.2. Similarity Feature-Based Segmentation
| Algorithm 1. Foreground prototypes based on few-shot segmentation |
| Require: Pre-processing of images, such as identifying and cropping out ROI regions of the image. |
| Require: Base feature encoder model . |
| 1. Sample and from pre-processed images |
| 2. Initialize {, } empty |
| 3. for Image set {, } in {, } do |
| 4. cur 0 |
| 5. while i = 0 to C x K do |
| 6. index i + curr |
| 7. Sample slice u, v from { [index], } |
| 8. //where is T2 image and is its corresponding label |
| 9. |
| 10. {, } {{ ∈ {} |
| 11. end while |
| 12. end for |
| 13. for in {, } do |
| 14. Query set |
| 15. Support set |
| 16. |
| 17. |
| 18. end for |
| 19. for k = 0 to len() do |
| 20. ℙ[k] // = masked average pooling function (MAP) |
| 21. end for |
| 22. for n = 0 to len( do |
| 23. Predictions[n] R(ℙ[n], ) |
| 24. Result[n] (Predictions[n]) // = Threshold function to eliminate small artifacts |
| 25. end for |
| Algorithm 2. |
| Pseudocode for training one-shot segmentation model. |
| Define the size and dimension of images in the dataset. |
| Initialization: |
| Support set , Query set Q = {, } contains image mask pair having a single slice. |
| Choose a backbone architecture as a feature encoder model with multi-class identification having θ parameters. |
| For epoch in S steps / I iterations. |
| For each image label pair in the train dataset. |
| Divide image label pairs into query and support sets containing random slices from the image. Both the query and support set contain one slice and its corresponding label image. |
| Pass both sets through the feature encoder model to extract their features. |
| Calculate support prototypes using masked average pooling of the support features set. |
| Perform segmentation based on similarities found between query features and support prototypes. |
| Update total loss consisting of segmentation and PAR loss. |
| End for |
| End for |
4. Experimental Settings
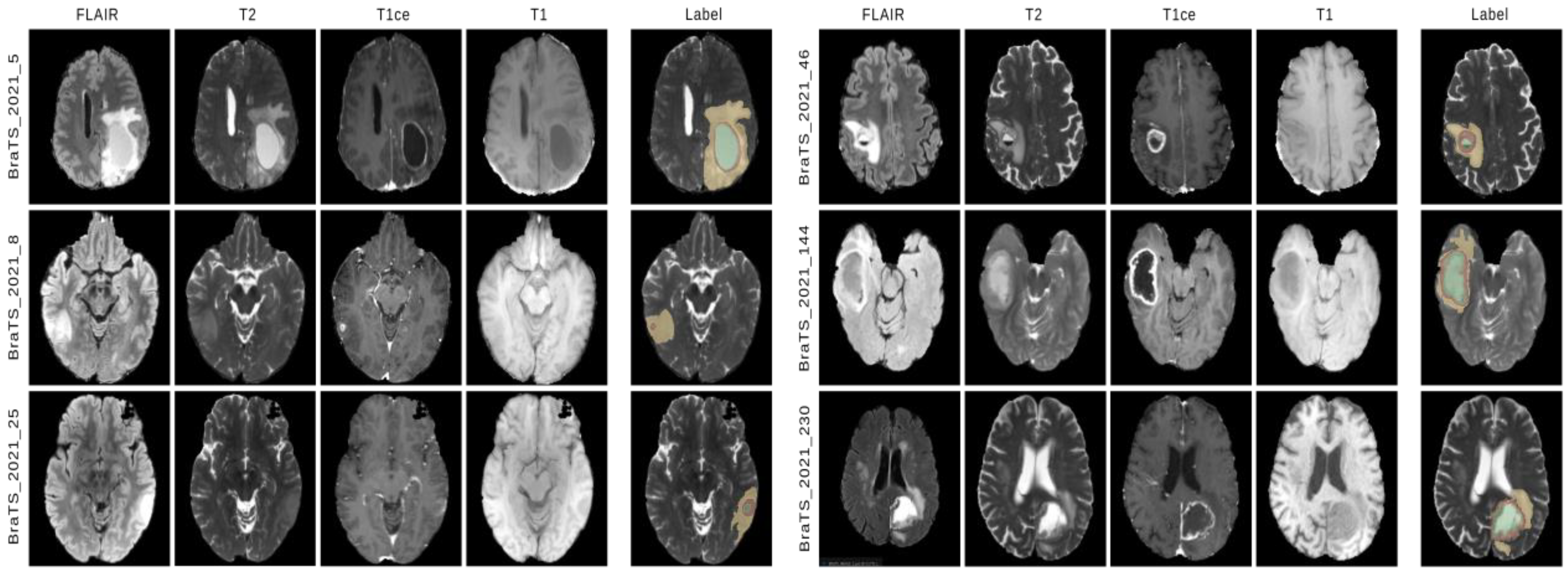
4.1. Dataset
4.2. Parameter Settings
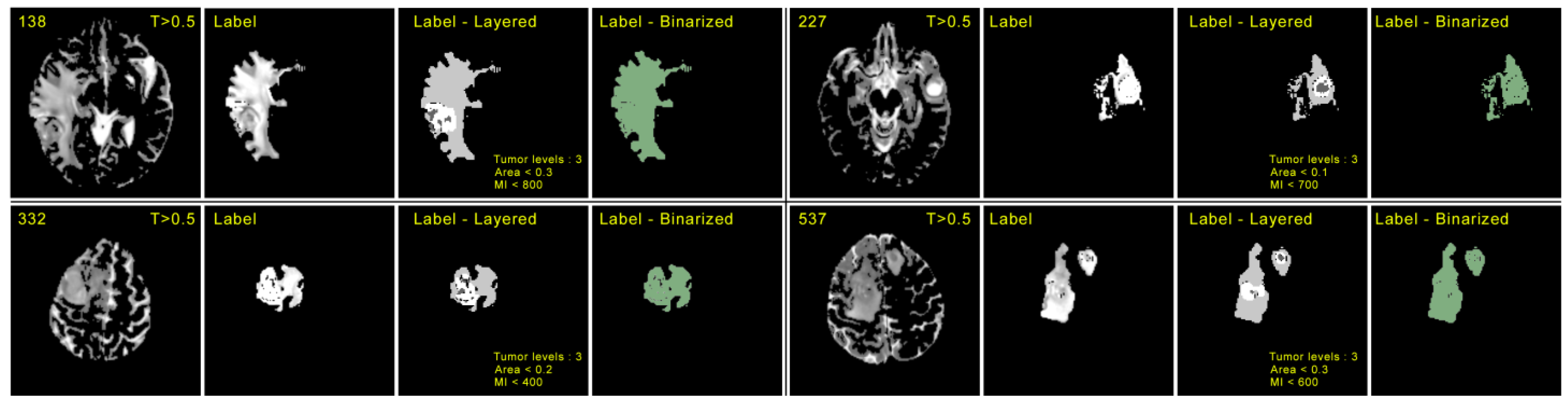
4.3. Evaluation Metrics
4.4. Evaluation Protocols
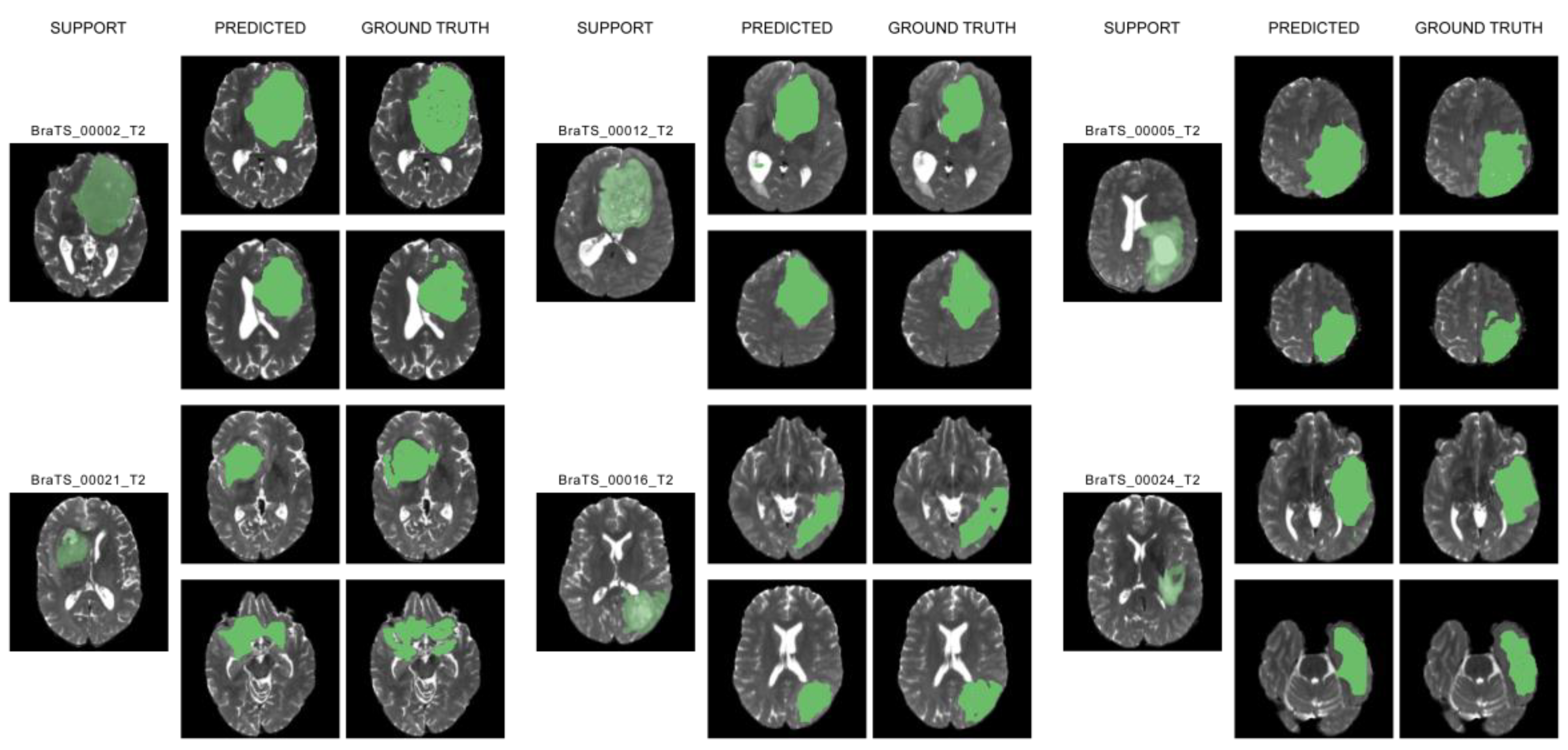
5. Results and Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dasgupta, A.; Gupta, T.; Jalali, R. Indian data on central nervous tumors: A summary of published work. S. Asian J. Cancer 2016, 5, 147–153. [Google Scholar]
- Wrensch, M.; Minn, Y.; Chew, T.; Bondy, M.; Berger, M.S. Epidemiology of primary brain tumors: Current concepts and review of the literature. Neuro-Oncology 2002, 4, 278–299. [Google Scholar] [CrossRef] [PubMed]
- Hicham, M.; Bouchaib, C.; Lhoussain, B. Convolutional Neural Networks for Multimodal Brain MRI Images Segmentation: A Comparative Study. In Proceedings of the Smart Applications and Data Analysis: Third International Conference, SADASC 2020, Marrakesh, Morocco, 25–26 June 2020; Springer International Publishing: New York, NY, USA, 2020; Volume 1207, pp. 329–338. [Google Scholar]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2016, 36, 61–78. [Google Scholar] [CrossRef]
- Zhou, C.; Ding, C.; Wang, X.; Lu, Z.; Tao, D. One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans. Image Process. 2020, 29, 4516–4529. [Google Scholar] [CrossRef]
- Li, S.; Sui, X.; Luo, X.; Xu, X.; Liu, Y.; Goh, R. Medical image segmentation using squeeze-and-expansion transformers. arXiv 2021, arXiv:2105.09511. [Google Scholar]
- Myronenko, A. 3D MRI brain tumor segmentation using Autoencoder regularization. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 4th International Workshop, Granada, Spain, 16 September 2018; Springer International Publishing: New York, NY, USA, 2019; pp. 311–320. [Google Scholar]
- Hansen, S.; Gautam, S.; Jenssen, R.; Kampffmeyer, M. Anomaly detection-inspired few-shot medical image segmentation through self-supervision with supervoxels. Med. Image Anal. 2022, 78, 102385. [Google Scholar] [CrossRef]
- Bendou, Y.; Hu, Y.; Lafargue, R.; Lioi, G.; Pasdeloup, B.; Pateux, S.; Gripon, V. Easy—Ensemble Augmented-Shot-Y-Shaped Learning: State-of-The-Art Few-Shot Classification with Simple Components. J. Imaging 2022, 8, 179. [Google Scholar] [CrossRef]
- Jie, C.; Luming, Z.; Naijie, G.; Xiaoci, Z.; Minquan, Y.; Rongzhang, Y.; Meng, Q. A mix-pooling CNN architecture with FCRF for brain tumor segmentation. J. Vis. Commun. Image Represent. 2019, 58, 316–322. [Google Scholar]
- Lucas, F.; Wenqi, L.; Luis, C.G.P.H.; Jinendra, E.; Neil, K.; Sebastian, O.; Tom, V. Scalable Multimodal Convolutional Networks for Brain Tumour Segmentation; Medical Image Computing and Computer Assisted Inter-vention—MICCAI 2017; Springer: Cham, Switzerland, 2017; Volume 10435. [Google Scholar]
- ZainEldin, H.; Gamel, S.A.; El-Kenawy, E.-S.M.; Alharbi, A.H.; Khafaga, D.S.; Ibrahim, A.; Talaat, F.M. Brain Tumor Detection and Classification Using Deep Learning and Sine-Cosine Fitness Grey Wolf Optimization. Bioengineering 2023, 10, 18. [Google Scholar]
- Feng, X.; Tustison, N.J.; Patel, S.H.; Meyer, C.H. Brain tumor segmentation using an ensemble of 3d u-nets and overall survival prediction using radiomic features. Front. Comput. Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef]
- Yogananda, C.G.B.; Shah, B.R.; Vejdani-Jahromi, M.; Nalawade, S.S.; Murugesan, G.K.; Yu, F.F.; Pinho, M.C.; Wagner, B.C.; Emblem, K.E.; Bjørnerud, A.; et al. A fully automated deep learning network for brain tumor segmentation. Tomography 2020, 6, 186–193. [Google Scholar] [CrossRef]
- Madhupriya, G.; Guru, N.M.; Praveen, S.; Nivetha, B. Brain tumor segmentation with deep learning technique. In Proceedings of the 2019 3rd International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 23–25 April 2019; pp. 758–763. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Third International Workshop, BrainLes 2017, Proceedings of the Conjunction with MICCAI 2017, Quebec City, QC, Canada, 14 September 2017, Revised Selected Papers 3; Springer International Publishing: New York, NY, USA, 2018; pp. 178–190. [Google Scholar]
- Liu, J.; Li, M.; Wang, J.; Wu, F.; Liu, T.; Pan, Y. A survey of MRI-based brain tumor segmentation methods. Tsinghua Sci. Technol. 2014, 19, 578–595. [Google Scholar]
- Wang, Y.; Yao, Q.; Kwok, J.T.; Ni, L.M. Generalizing from a few examples: A survey on few-shot learning. ACM Comput. Surv. (Csur) 2020, 53, 1–34. [Google Scholar] [CrossRef]
- Bai, W.; Chen, C.; Tarroni, G.; Duan, J.; Guitton, F.; Petersen, S.E.; Guo, Y.; Matthews, P.M.; Rueckert, D. Self-supervised learning for cardiac mr image segmentation by anatomical position prediction. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part II 22 2019; Springer International Publishing: New York, NY, USA, 2019; pp. 541–549. [Google Scholar]
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. Self-supervised learning for medical image analysis using image context restoration. Med. Image Anal. 2019, 58, 101539. [Google Scholar] [CrossRef]
- Tong, T.; Wolz, R.; Wang, Z.; Gao, Q.; Misawa, K.; Fujiwara, M.; Mori, K.; Hajnal, J.V.; Rueckert, D. Discriminative dictionary learning for abdominal multi-organ segmentation. Med. Image Anal. 2015, 23, 92–104. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, G.; Liu, F.; Yao, R.; Shen, C. Canet: Class-Agnostic Segmentation Networks with Iterative Refinement and Attentive Few-Shot Learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2019, Long Beach, CA, USA, 15–20 June 2019; pp. 5217–5226. [Google Scholar]
- Yu, M.; Liang, L.; Biqing, H.; Xiu, L. Few-shot RUL estimation based on model-agnostic meta-learning. J. Intell. Manuf. 2022, 1572–8145. [Google Scholar]
- Finn, C.; Abbeel, P.; Levine, S. Model-agnostic meta-learning for fast adaptation of deep networks. In Proceedings of the 34th International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 1126–1135. [Google Scholar]
- Vinyals, O.; Blundell, C.; Lillicrap, T.; Wierstra, D. Matching networks for one shot learning. In Proceedings of the Advances in Neural Information PROCESSING systems 29, Annual Conference on Neural Information Processing Systems 2016, Barcelona, Spain, 5–10 December 2016; Morgan Kaufmann Publishers Inc.: Burlington, MA, USA, 2016. [Google Scholar]
- Snell, J.; Swersky, K.; Zemel, R. Prototypical networks for few-shot learning. In Proceedings of the Advances in Neural Information Processing Systems 30, Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Fan, Q.; Pei, W.; Tai, Y.-W.; Tang, C.-K. Self-Support Few-Shot Semantic Segmentation. arXiv 2022, arXiv:2207.11549. [Google Scholar]
- Shi, X.; Wei, D.; Zhang, Y.; Lu, D.; Ning, M.; Chen, J.; Ma, K.; Zheng, Y. Dense cross-query-and-support attention weighted mask aggregation for few-shot segmentation. In Proceedings of the Computer Vision–ECCV 2022: 17th European Conference, Tel Aviv, Israel, 23–27 October 2022; Springer Nature: Cham, Switzerland, 2022; pp. 151–168. [Google Scholar]
- Boudiaf, M.; Kervadec, H.; Masud, Z.I.; Piantanida, P.; Ben Ayed, I.; Dolz, J. Few-shot segmentation without meta-learning: A good transductive inference is all you need? In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13979–13988. [Google Scholar]
- Cui, H.; Wei, D.; Ma, K.; Gu, S.; Zheng, Y. A unified framework for generalized low-shot medical image segmentation with scarce data. IEEE Trans. Med. Imaging 2020, 40, 2656–2671. [Google Scholar] [CrossRef]
- Voulodimos, A.; Protopapadakis, E.; Katsamenis, I.; Doulamis, A.; Doulamis, N. A few-shot U-net deep learning model for COVID-19 infected area segmentation in CT images. Sensors 2021, 21, 2215. [Google Scholar] [CrossRef]
- Wang, K.; Liew, J.H.; Zou, Y.; Zhou, D.; Feng, J. Panet: Few-shot image semantic segmentation with prototype alignment. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9197–9206. [Google Scholar]
- Li, S.; Liu, J.; Song, Z. Brain tumor segmentation based on region of interest-aided localization and segmentation U-Net. Int. J. Mach. Learn. Cybern. 2022, 13, 2435–2445. [Google Scholar] [CrossRef]
- Ouyang, C.; Biffi, C.; Chen, C.; Kart, T.; Qiu, H.; Rueckert, D. Self-Supervision with Superpixels: Training Few-shot Medical Image Segmentation without Annotation. arXiv 2020, arXiv:2007.09886. [Google Scholar]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollar, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the 13th European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; Springer International Publishing: New York, NY, USA, 2014; pp. 740–755. [Google Scholar]
- Han, Z.; Hao, Y.; Dong, L.; Sun, Y.; Wei, F. Prototypical Calibration for Few-shot Learning of Language Models. arXiv 2022, arXiv:2205.10183. [Google Scholar]
- Lang, C.; Cheng, G.; Tu, B.; Han, J. Learning what not to segment: A new perspective on few-shot segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Hu, S.X.; Li, D.; Stuhmer, J.; Kim, M.; Hospedales, T.M. Pushing the limits of simple pipelines for few-shot learning: External data and fine-tuning make a difference. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Schick, T.; Schütze, H. Few-shot text generation with pattern-exploiting training. arXiv 2020, arXiv:2012.11926. [Google Scholar]
- Huo, Y.; Xu, Z.; Xiong, Y.; Aboud, K.; Parvathaneni, P.; Bao, S.; Bermudez, C.; Resnick, S.M.; Cutting, L.E.; Landman, B.A. 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage 2019, 94, 105–119. [Google Scholar] [CrossRef]

| Method | Input Size (MB) | Forward/Backward Pass Size (MB) | Estimated Total Size (MB) | Encoding Shape |
|---|---|---|---|---|
| 1-shot 1-way | 17.56 | 21,038.63 | 21,228.29 | (22,256,32,32) |
| 1-shot 5-way | 21.76 | 24,863.83 | 25,057.69 | (26,256,32,32) |
| 5-shot 1-way | 21.76 | 24,863.83 | 25,057.69 | (26,256,32,32) |
| 5-shot 5-way | 42.73 | 43,989.76 | 44,204.69 | (46,256,32,32) |
| 10-shot 1-way | 27.00 | 29,645.34 | 29,844.44 | (31,256,32,32) |
| 10-shot 5-way | 68.95 | 67,897.39 | 68,138.44 | (71,256,32,32) |
| Method | Training Data | Dice (%) | Dataset |
|---|---|---|---|
| OM-Net + CGap | 559 | 91.59 | BraTS 2015 + BraTS 2018 |
| 3DCNN + CRF | 338 | 90.10 | BraTS 2015 + ISLES 2015 |
| Segtran | 335 | 81.70 | BraTS 2019 |
| NVDLMED | 285 | 87.04 | BraTS 2018 |
| AFN-6 | 274 | 89.30 | BraTS 2015 |
| CNN + 3D filters | 274 | 80.00 | BraTS 2015 |
| Few-shot-based | 60 | 83.42 | BraTS 2021 |
| Method | Score Metrics | ||||
|---|---|---|---|---|---|
| Avg. Dice | Max. Dice | Avg. IOU | Max. IOU | Precision | |
| 1-shot 1-way | 83.42 ± 0.35 | 83.85 ± 1.14 | 80.97 ± 1.12 | 81.57 ± 1.23 | 61.03 ± 0.38 |
| 1-shot 5-way | 83.78 ± 1.14 | 82.67 ± 2.81 | 80.99 ± 1.16 | 81.39 ± 1.61 | 60.78 ±0.82 |
| 5-shot 1-way | 83.28 ± 2.27 | 82.50 ± 3.13 | 79.44 ± 2.29 | 80.97 ± 0.25 | 60.45 ± 1.05 |
| 5-shot 5-way | 83.41 ± 2.30 | 81.23 ± 3.43 | 79.64 ± 2.44 | 80.22 ± 1.68 | 60.61 ± 1.75 |
| 10-shot 1-way | 82.89 ± 2.78 | 81.45 ± 2.25 | 79.59 ± 2.54 | 80.98 ± 3.02 | 79.62 ± 1.68 |
| 10-shot 5-way | 83.40 ± 3.28 | 80.23 ± 4.17 | 80.02 ± 2.69 | 80.82 ± 2.93 | 59.88 ± 2.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balasundaram, A.; Kavitha, M.S.; Pratheepan, Y.; Akshat, D.; Kaushik, M.V. A Foreground Prototype-Based One-Shot Segmentation of Brain Tumors. Diagnostics 2023, 13, 1282. https://doi.org/10.3390/diagnostics13071282
Balasundaram A, Kavitha MS, Pratheepan Y, Akshat D, Kaushik MV. A Foreground Prototype-Based One-Shot Segmentation of Brain Tumors. Diagnostics. 2023; 13(7):1282. https://doi.org/10.3390/diagnostics13071282
Chicago/Turabian StyleBalasundaram, Ananthakrishnan, Muthu Subash Kavitha, Yogarajah Pratheepan, Dhamale Akshat, and Maddirala Venkata Kaushik. 2023. "A Foreground Prototype-Based One-Shot Segmentation of Brain Tumors" Diagnostics 13, no. 7: 1282. https://doi.org/10.3390/diagnostics13071282
APA StyleBalasundaram, A., Kavitha, M. S., Pratheepan, Y., Akshat, D., & Kaushik, M. V. (2023). A Foreground Prototype-Based One-Shot Segmentation of Brain Tumors. Diagnostics, 13(7), 1282. https://doi.org/10.3390/diagnostics13071282






