Prediction of the Topography of the Corticospinal Tract on T1-Weighted MR Images Using Deep-Learning-Based Segmentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Fiber Tracking and Coregistration
2.2. Generation of Label Images
2.3. Training of an Image Segmentation Model
2.4. Performance Evaluation
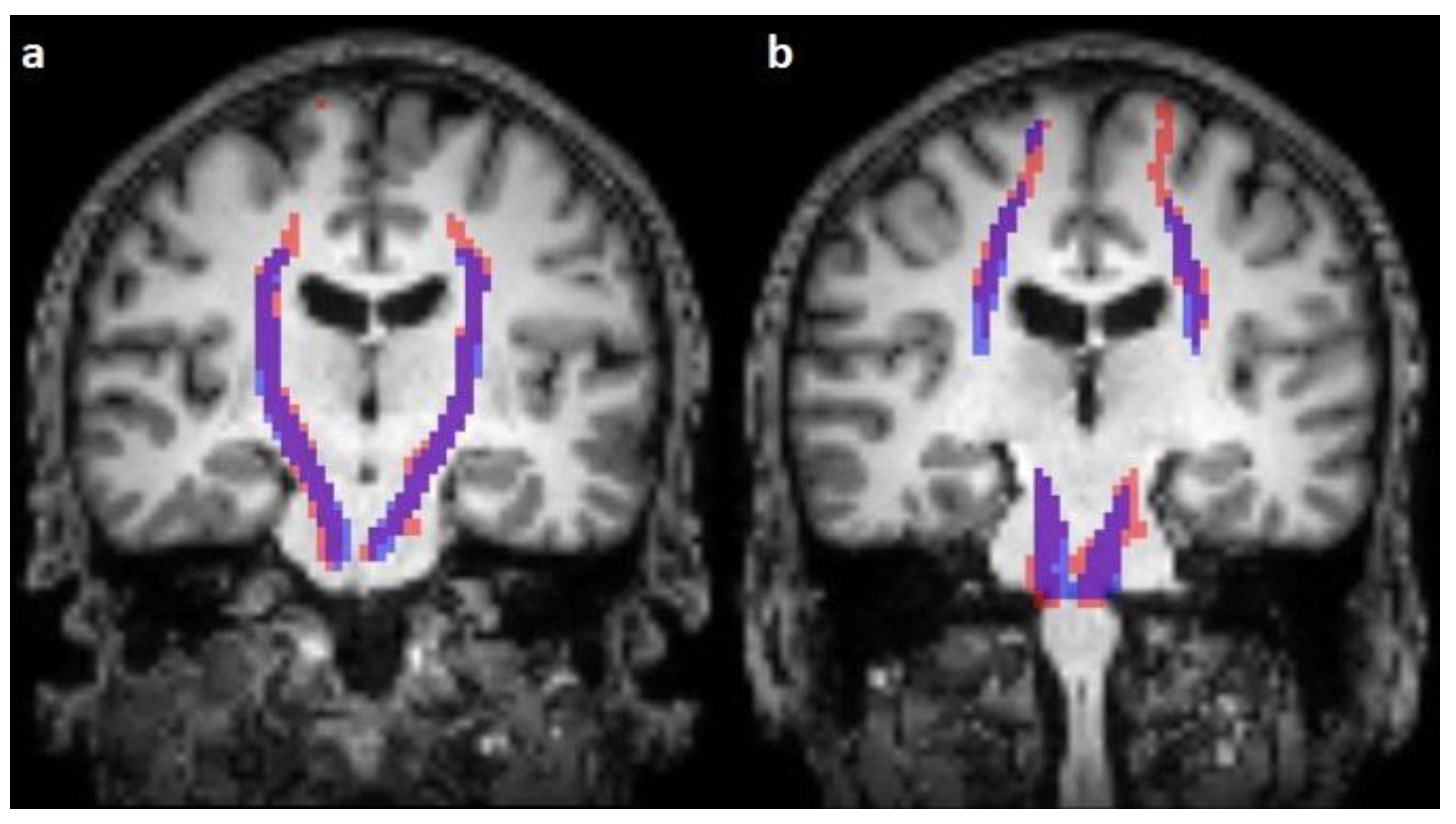
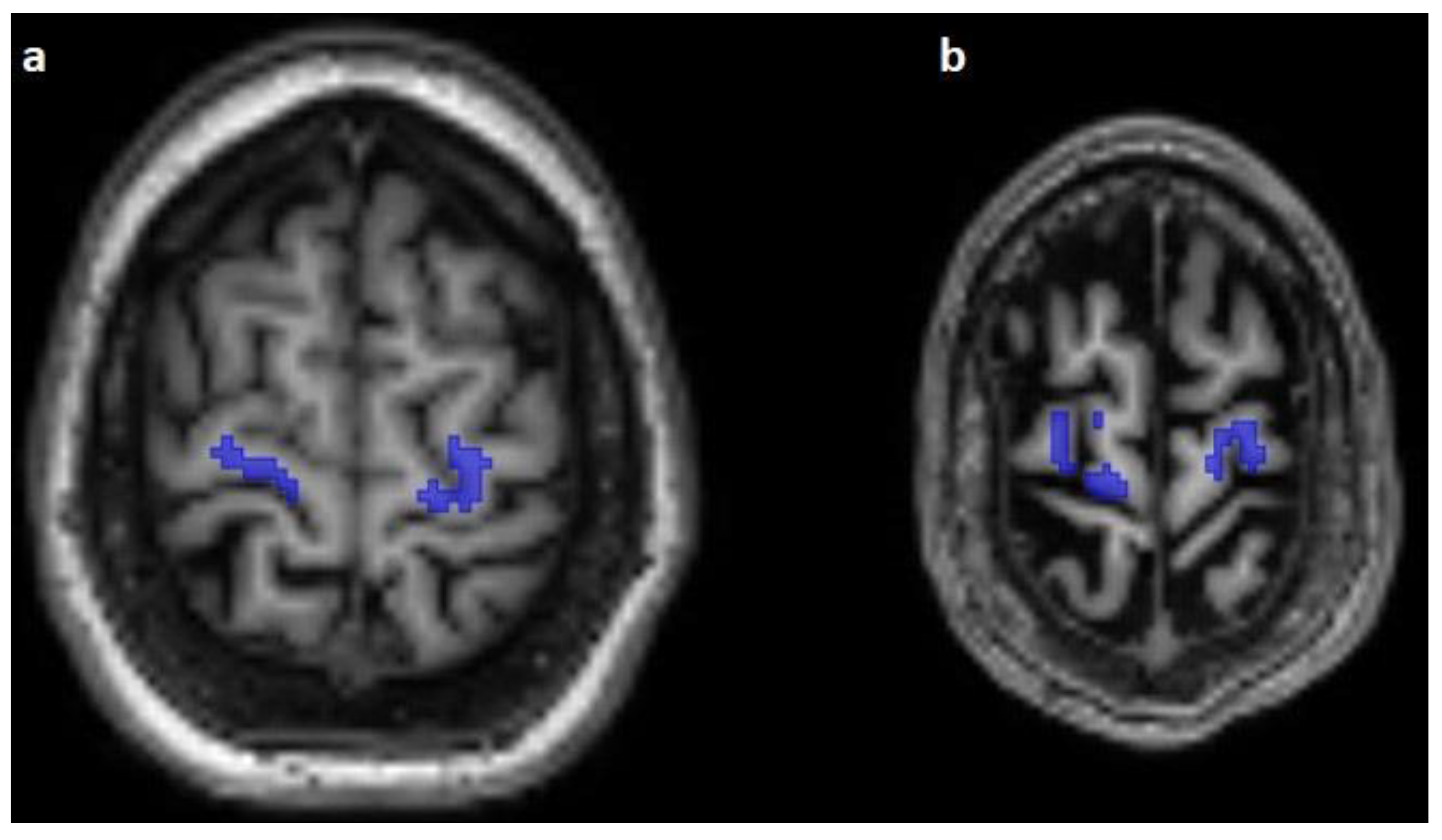
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berman, J.I.; Berger, M.S.; Chung, S.W.; Nagarajan, S.S.; Henry, R.G. Accuracy of Diffusion Tensor Magnetic Resonance Imaging Tractography Assessed Using Intraoperative Subcortical Stimulation Mapping and Magnetic Source Imaging. J. Neurosurg. 2007, 107, 488–494. [Google Scholar] [CrossRef]
- Berman, J.I.; Berger, M.S.; Mukherjee, P.; Henry, R.G. Diffusion-Tensor Imaging-Guided Tracking of Fibers of the Pyramidal Tract Combined with Intraoperative Cortical Stimulation Mapping in Patients with Gliomas. J. Neurosurg. 2004, 101, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.G.; Berman, J.I.; Nagarajan, S.S.; Mukherjee, P.; Berger, M.S. Subcortical Pathways Serving Cortical Language Sites: Initial Experience with Diffusion Tensor Imaging Fiber Tracking Combined with Intraoperative Language Mapping. Neuroimage 2004, 21, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Coenen, V.A.; Krings, T.; Mayfrank, L.; Polin, R.S.; Reinges, M.H.; Thron, A.; Gilsbach, J.M. Three-Dimensional Visualization of the Pyramidal Tract in a Neuronavigation System during Brain Tumor Surgery: First Experiences and Technical Note. Neurosurgery 2001, 49, 86–92; discussion 92–93. [Google Scholar] [CrossRef] [PubMed]
- Mikuni, N.; Okada, T.; Nishida, N.; Taki, J.; Enatsu, R.; Ikeda, A.; Miki, Y.; Hanakawa, T.; Fukuyama, H.; Hashimoto, N. Comparison between Motor Evoked Potential Recording and Fiber Tracking for Estimating Pyramidal Tracts near Brain Tumors. J. Neurosurg. 2007, 106, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Wieshmann, U.C.; Symms, M.R.; Parker, G.J.; Clark, C.A.; Lemieux, L.; Barker, G.J.; Shorvon, S.D. Diffusion Tensor Imaging Demonstrates Deviation of Fibres in Normal Appearing White Matter Adjacent to a Brain Tumour. J. Neurol. Neurosurg. Psychiatry 2000, 68, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Witwer, B.P.; Moftakhar, R.; Hasan, K.M.; Deshmukh, P.; Haughton, V.; Field, A.; Arfanakis, K.; Noyes, J.; Moritz, C.H.; Meyerand, M.E.; et al. Diffusion-Tensor Imaging of White Matter Tracts in Patients with Cerebral Neoplasm. J. Neurosurg. 2002, 97, 568–575. [Google Scholar] [CrossRef]
- Henderson, F.; Abdullah, K.G.; Verma, R.; Brem, S. Tractography and the Connectome in Neurosurgical Treatment of Gliomas: The Premise, the Progress, and the Potential. Neurosurg. Focus 2020, 48, E6. [Google Scholar] [CrossRef]
- Farquharson, S.; Tournier, J.-D.; Calamante, F.; Fabinyi, G.; Schneider-Kolsky, M.; Jackson, G.D.; Connelly, A. White Matter Fiber Tractography: Why We Need to Move beyond DTI. J. Neurosurg. 2013, 118, 1367–1377. [Google Scholar] [CrossRef]
- Pujol, S.; Wells, W.; Pierpaoli, C.; Brun, C.; Gee, J.; Cheng, G.; Vemuri, B.; Commowick, O.; Prima, S.; Stamm, A.; et al. The DTI Challenge: Toward Standardized Evaluation of Diffusion Tensor Imaging Tractography for Neurosurgery. J. Neuroimaging 2015, 25, 875–882. [Google Scholar] [CrossRef]
- Panesar, S.S.; Abhinav, K.; Yeh, F.-C.; Jacquesson, T.; Collins, M.; Fernandez-Miranda, J. Tractography for Surgical Neuro-Oncology Planning: Towards a Gold Standard. Neurotherapeutics 2019, 16, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Essayed, W.I.; Zhang, F.; Unadkat, P.; Cosgrove, G.R.; Golby, A.J.; O’Donnell, L.J. White Matter Tractography for Neurosurgical Planning: A Topography-Based Review of the Current State of the Art. Neuroimage Clin. 2017, 15, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.-M.; Yeh, C.-H.; Poupon, C.; Calamante, F. Diffusion MRI Tractography for Neurosurgery: The Basics, Current State, Technical Reliability and Challenges. Phys. Med. Biol. 2021, 66, 15TR01. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.A.; Barrick, T.R.; Murphy, M.M.; Bell, B.A. White Matter Fiber Tracking in Patients with Space-Occupying Lesions of the Brain: A New Technique for Neurosurgical Planning? Neuroimage 2003, 20, 1601–1608. [Google Scholar] [CrossRef]
- Szmuda, T.; Kierońska, S.; Ali, S.; Słoniewski, P.; Pacholski, M.; Dzierżanowski, J.; Sabisz, A.; Szurowska, E. Tractography-Guided Surgery of Brain Tumours: What Is the Best Method to Outline the Corticospinal Tract? Folia Morphol. 2021, 80, 40–46. [Google Scholar] [CrossRef]
- Castellano, A.; Bello, L.; Michelozzi, C.; Gallucci, M.; Fava, E.; Iadanza, A.; Riva, M.; Casaceli, G.; Falini, A. Role of Diffusion Tensor Magnetic Resonance Tractography in Predicting the Extent of Resection in Glioma Surgery. Neuro Oncol. 2012, 14, 192–202. [Google Scholar] [CrossRef]
- O’Donnell, L.J.; Suter, Y.; Rigolo, L.; Kahali, P.; Zhang, F.; Norton, I.; Albi, A.; Olubiyi, O.; Meola, A.; Essayed, W.I.; et al. Automated White Matter Fiber Tract Identification in Patients with Brain Tumors. Neuroimage Clin. 2017, 13, 138–153. [Google Scholar] [CrossRef]
- Mancini, M.; Vos, S.B.; Vakharia, V.N.; O’Keeffe, A.G.; Trimmel, K.; Barkhof, F.; Dorfer, C.; Soman, S.; Winston, G.P.; Wu, C.; et al. Automated Fiber Tract Reconstruction for Surgery Planning: Extensive Validation in Language-Related White Matter Tracts. Neuroimage Clin. 2019, 23, 101883. [Google Scholar] [CrossRef]
- Conti Nibali, M.; Rossi, M.; Sciortino, T.; Riva, M.; Gay, L.G.; Pessina, F.; Bello, L. Preoperative Surgical Planning of Glioma: Limitations and Reliability of FMRI and DTI Tractography. J. Neurosurg. Sci. 2019, 63, 127–134. [Google Scholar] [CrossRef]
- Umana, G.E.; Scalia, G.; Graziano, F.; Maugeri, R.; Alberio, N.; Barone, F.; Crea, A.; Fagone, S.; Giammalva, G.R.; Brunasso, L.; et al. Navigated Transcranial Magnetic Stimulation Motor Mapping Usefulness in the Surgical Management of Patients Affected by Brain Tumors in Eloquent Areas: A Systematic Review and Meta-Analysis. Front Neurol. 2021, 12, 644198. [Google Scholar] [CrossRef]
- Mandelli, M.L.; Berger, M.S.; Bucci, M.; Berman, J.I.; Amirbekian, B.; Henry, R.G. Quantifying Accuracy and Precision of Diffusion MR Tractography of the Corticospinal Tract in Brain Tumors. J. Neurosurg. 2014, 121, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Wassermann, D.; Makris, N.; Rathi, Y.; Shenton, M.; Kikinis, R.; Kubicki, M.; Westin, C.-F. The White Matter Query Language: A Novel Approach for Describing Human White Matter Anatomy. Brain Struct. Funct. 2016, 221, 4705–4721. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, D.; Duffau, H.; Delmaire, C.; Capelle, L.; Gatignol, P.; Ducros, M.; Chiras, J.; Lehéricy, S. Comparison of Diffusion Tensor Imaging Tractography of Language Tracts and Intraoperative Subcortical Stimulations. J. Neurosurg. 2010, 112, 503–511. [Google Scholar] [CrossRef]
- Kinoshita, M.; Yamada, K.; Hashimoto, N.; Kato, A.; Izumoto, S.; Baba, T.; Maruno, M.; Nishimura, T.; Yoshimine, T. Fiber-Tracking Does Not Accurately Estimate Size of Fiber Bundle in Pathological Condition: Initial Neurosurgical Experience Using Neuronavigation and Subcortical White Matter Stimulation. Neuroimage 2005, 25, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-W.; Chou, M.-C.; Chen, C.-Y. Principles and Limitations of Computational Algorithms in Clinical Diffusion Tensor MR Tractography. AJNR Am. J. Neuroradiol. 2011, 32, 3–13. [Google Scholar] [CrossRef]
- Bello, L.; Castellano, A.; Fava, E.; Casaceli, G.; Riva, M.; Scotti, G.; Gaini, S.M.; Falini, A. Intraoperative Use of Diffusion Tensor Imaging Fiber Tractography and Subcortical Mapping for Resection of Gliomas: Technical Considerations. Neurosurg. Focus 2010, 28, E6. [Google Scholar] [CrossRef]
- Bello, L.; Gambini, A.; Castellano, A.; Carrabba, G.; Acerbi, F.; Fava, E.; Giussani, C.; Cadioli, M.; Blasi, V.; Casarotti, A.; et al. Motor and Language DTI Fiber Tracking Combined with Intraoperative Subcortical Mapping for Surgical Removal of Gliomas. Neuroimage 2008, 39, 369–382. [Google Scholar] [CrossRef]
- Kamada, K.; Todo, T.; Masutani, Y.; Aoki, S.; Ino, K.; Takano, T.; Kirino, T.; Kawahara, N.; Morita, A. Combined Use of Tractography-Integrated Functional Neuronavigation and Direct Fiber Stimulation. J. Neurosurg. 2005, 102, 664–672. [Google Scholar] [CrossRef]
- Kamada, K.; Todo, T.; Ota, T.; Ino, K.; Masutani, Y.; Aoki, S.; Takeuchi, F.; Kawai, K.; Saito, N. The Motor-Evoked Potential Threshold Evaluated by Tractography and Electrical Stimulation. J. Neurosurg. 2009, 111, 785–795. [Google Scholar] [CrossRef]
- Maesawa, S.; Fujii, M.; Nakahara, N.; Watanabe, T.; Wakabayashi, T.; Yoshida, J. Intraoperative Tractography and Motor Evoked Potential (MEP) Monitoring in Surgery for Gliomas around the Corticospinal Tract. World Neurosurg. 2010, 74, 153–161. [Google Scholar] [CrossRef]
- Nossek, E.; Korn, A.; Shahar, T.; Kanner, A.A.; Yaffe, H.; Marcovici, D.; Ben-Harosh, C.; ben Ami, H.; Weinstein, M.; Shapira-Lichter, I.; et al. Intraoperative Mapping and Monitoring of the Corticospinal Tracts with Neurophysiological Assessment and 3-Dimensional Ultrasonography-Based Navigation. Clinical Article. J. Neurosurg. 2011, 114, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, S.S.; Gasco, J.; Tummala, S.; Weinberg, J.S.; Rao, G. Intraoperative Magnetic Resonance Imaging-Guided Tractography with Integrated Monopolar Subcortical Functional Mapping for Resection of Brain Tumors. Clinical Article. J. Neurosurg. 2011, 114, 719–726. [Google Scholar] [CrossRef]
- Yamaguchi, F.; Takahashi, H.; Teramoto, A. Navigation-Assisted Subcortical Mapping: Intraoperative Motor Tract Detection by Bipolar Needle Electrode in Combination with Neuronavigation System. J. NeuroOncol. 2009, 93, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Bonney, P.A.; Conner, A.K.; Boettcher, L.B.; Cheema, A.A.; Glenn, C.A.; Smitherman, A.D.; Pittman, N.A.; Sughrue, M.E. A Simplified Method of Accurate Postprocessing of Diffusion Tensor Imaging for Use in Brain Tumor Resection. Oper. Neurosurg. 2017, 13, 47–59. [Google Scholar] [CrossRef]
- Ohue, S.; Kohno, S.; Inoue, A.; Yamashita, D.; Harada, H.; Kumon, Y.; Kikuchi, K.; Miki, H.; Ohnishi, T. Accuracy of Diffusion Tensor Magnetic Resonance Imaging-Based Tractography for Surgery of Gliomas near the Pyramidal Tract: A Significant Correlation between Subcortical Electrical Stimulation and Postoperative Tractography. Neurosurgery 2012, 70, 283–293; discussion 294. [Google Scholar] [CrossRef] [PubMed]
- Bucci, M.; Mandelli, M.L.; Berman, J.I.; Amirbekian, B.; Nguyen, C.; Berger, M.S.; Henry, R.G. Quantifying Diffusion MRI Tractography of the Corticospinal Tract in Brain Tumors with Deterministic and Probabilistic Methods. Neuroimage Clin. 2013, 3, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Mikuni, N.; Miki, Y.; Kikuta, K.-I.; Urayama, S.-I.; Hanakawa, T.; Fushimi, Y.; Yamamoto, A.; Kanagaki, M.; Fukuyama, H.; et al. Corticospinal Tract Localization: Integration of Diffusion-Tensor Tractography at 3-T MR Imaging with Intraoperative White Matter Stimulation Mapping--Preliminary Results. Radiology 2006, 240, 849–857. [Google Scholar] [CrossRef]
- Mikuni, N.; Okada, T.; Enatsu, R.; Miki, Y.; Hanakawa, T.; Urayama, S.; Kikuta, K.; Takahashi, J.A.; Nozaki, K.; Fukuyama, H.; et al. Clinical Impact of Integrated Functional Neuronavigation and Subcortical Electrical Stimulation to Preserve Motor Function during Resection of Brain Tumors. J. Neurosurg. 2007, 106, 593–598. [Google Scholar] [CrossRef]
- Wakana, S.; Caprihan, A.; Panzenboeck, M.M.; Fallon, J.H.; Perry, M.; Gollub, R.L.; Hua, K.; Zhang, J.; Jiang, H.; Dubey, P.; et al. Reproducibility of Quantitative Tractography Methods Applied to Cerebral White Matter. Neuroimage 2007, 36, 630–644. [Google Scholar] [CrossRef]
- Colon-Perez, L.M.; Triplett, W.; Bohsali, A.; Corti, M.; Nguyen, P.T.; Patten, C.; Mareci, T.H.; Price, C.C. A Majority Rule Approach for Region-of-Interest-Guided Streamline Fiber Tractography. Brain Imaging Behav. 2016, 10, 1137–1147. [Google Scholar] [CrossRef]
- Wasserthal, J.; Neher, P.; Maier-Hein, K.H. TractSeg—Fast and Accurate White Matter Tract Segmentation. Neuroimage 2018, 183, 239–253. [Google Scholar] [CrossRef]
- Garyfallidis, E.; Côté, M.-A.; Rheault, F.; Sidhu, J.; Hau, J.; Petit, L.; Fortin, D.; Cunanne, S.; Descoteaux, M. Recognition of White Matter Bundles Using Local and Global Streamline-Based Registration and Clustering. Neuroimage 2018, 170, 283–295. [Google Scholar] [CrossRef]
- Yendiki, A.; Panneck, P.; Srinivasan, P.; Stevens, A.; Zöllei, L.; Augustinack, J.; Wang, R.; Salat, D.; Ehrlich, S.; Behrens, T.; et al. Automated Probabilistic Reconstruction of White-Matter Pathways in Health and Disease Using an Atlas of the Underlying Anatomy. Front. Neuroinf. 2011, 5, 23. [Google Scholar] [CrossRef]
- O’Donnell, L.J.; Wells, W.M.; Golby, A.J.; Westin, C.-F. Unbiased Groupwise Registration of White Matter Tractography. Med. Image Comput. Comput. Assist. Interv. 2012, 15, 123–130. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, Y.; Norton, I.; Rathi, Y.; Golby, A.J.; O’Donnell, L.J. Test-Retest Reproducibility of White Matter Parcellation Using Diffusion MRI Tractography Fiber Clustering. Hum. Brain Mapp. 2019, 40, 3041–3057. [Google Scholar] [CrossRef]
- O’Donnell, L.J.; Westin, C.-F. Automatic Tractography Segmentation Using a High-Dimensional White Matter Atlas. IEEE Trans. Med. Imaging 2007, 26, 1562–1575. [Google Scholar] [CrossRef]
- Sydnor, V.J.; Rivas-Grajales, A.M.; Lyall, A.E.; Zhang, F.; Bouix, S.; Karmacharya, S.; Shenton, M.E.; Westin, C.-F.; Makris, N.; Wassermann, D.; et al. A Comparison of Three Fiber Tract Delineation Methods and Their Impact on White Matter Analysis. Neuroimage 2018, 178, 318–331. [Google Scholar] [CrossRef]
- Poulin, P.; Jörgens, D.; Jodoin, P.-M.; Descoteaux, M. Tractography and Machine Learning: Current State and Open Challenges. Magn. Reson. Imaging 2019, 64, 37–48. [Google Scholar] [CrossRef]
- Yang, Q.; Hansen, C.B.; Cai, L.Y.; Rheault, F.; Lee, H.H.; Bao, S.; Chandio, B.Q.; Williams, O.; Resnick, S.M.; Garyfallidis, E.; et al. Learning White Matter Subject-specific Segmentation from Structural MRI. Med. Phys. 2022, 49, 2502–2513. [Google Scholar] [CrossRef]
- Snoek, L.; van der Miesen, M.M.; Beemsterboer, T.; van der Leij, A.; Eigenhuis, A.; Steven Scholte, H. The Amsterdam Open MRI Collection, a Set of Multimodal MRI Datasets for Individual Difference Analyses. Sci. Data 2021, 8, 85. [Google Scholar] [CrossRef]
- Yan, C.; Gong, G.; Wang, J.; Wang, D.; Liu, D.; Zhu, C.; Chen, Z.J.; Evans, A.; Zang, Y.; He, Y. Sex- and Brain Size–Related Small-World Structural Cortical Networks in Young Adults: A DTI Tractography Study. Cereb. Cortex 2011, 21, 449–458. [Google Scholar] [CrossRef]
- Lloyd, W.K.; Morriss, J.; Macdonald, B.; Joanknecht, K.; Nihouarn, J.; van Reekum, C.M. Longitudinal Change in Executive Function Is Associated with Impaired Top-down Frontolimbic Regulation during Reappraisal in Older Adults. Neuroimage 2021, 225, 117488. [Google Scholar] [CrossRef]
- Hanke, M.; Baumgartner, F.J.; Ibe, P.; Kaule, F.R.; Pollmann, S.; Speck, O.; Zinke, W.; Stadler, J. A High-Resolution 7-Tesla FMRI Dataset from Complex Natural Stimulation with an Audio Movie. Sci. Data 2014, 1, 140003. [Google Scholar] [CrossRef]
- Available online: https://openneuro.org/datasets/ds001771 (accessed on 29 January 2023).
- Boekel, W.; Forstmann, B.U.; Keuken, M.C. A Test-Retest Reliability Analysis of Diffusion Measures of White Matter Tracts Relevant for Cognitive Control. Psychophysiology 2017, 54, 24–33. [Google Scholar] [CrossRef]
- Available online: https://Dsi-Studio.Labsolver.Org/ (accessed on 29 January 2023).
- Yeh, F.-C.; Panesar, S.; Fernandes, D.; Meola, A.; Yoshino, M.; Fernandez-Miranda, J.C.; Vettel, J.M.; Verstynen, T. Population-Averaged Atlas of the Macroscale Human Structural Connectome and Its Network Topology. Neuroimage 2018, 178, 57–68. [Google Scholar] [CrossRef]
- Yeh, F.-C.; Panesar, S.; Barrios, J.; Fernandes, D.; Abhinav, K.; Meola, A.; Fernandez-Miranda, J.C. Automatic Removal of False Connections in Diffusion MRI Tractography Using Topology-InforMed. Pruning (TIP). Neurotherapeutics 2019, 16, 52–58. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn. Reson Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy; Jolesz, F., Ed.; Springer: New York, NY, USA, 2004; Available online: http://Www.Slicer.Org (accessed on 29 January 2023). [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. NnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Ouyang, M.; Dubois, J.; Yu, Q.; Mukherjee, P.; Huang, H. Delineation of Early Brain Development from Fetuses to Infants with Diffusion MRI and Beyond. Neuroimage 2019, 185, 836–850. [Google Scholar] [CrossRef]
- Zhang, F.; Daducci, A.; He, Y.; Schiavi, S.; Seguin, C.; Smith, R.E.; Yeh, C.-H.; Zhao, T.; O’Donnell, L.J. Quantitative Mapping of the Brain’s Structural Connectivity Using Diffusion MRI Tractography: A Review. Neuroimage 2022, 249, 118870. [Google Scholar] [CrossRef] [PubMed]
- Raja, R.; Rosenberg, G.; Caprihan, A. Review of Diffusion MRI Studies in Chronic White Matter Diseases. NeuroSci. Lett. 2019, 694, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Penadés, R.; Franck, N.; González-Vallespí, L.; Dekerle, M. Neuroimaging Studies of Cognitive Function in Schizophrenia. Adv. Exp. Med. Biol. 2019, 1118, 117–134. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, G.; Familiari, P.; di Lauro, A.; Angelini, A.; Sessa, G. Safe Resection of Gliomas of the Dominant Angular Gyrus Availing of Preoperative FMRI and Intraoperative DTI: Preliminary Series and Surgical Technique. World Neurosurg. 2016, 87, 627–639. [Google Scholar] [CrossRef]
- Bürgel, U.; Mädler, B.; Honey, C.R.; Thron, A.; Gilsbach, J.; Coenen, V.A. Fiber Tracking with Distinct Software Tools Results in a Clear Diversity in Anatomical Fiber Tract Portrayal. Cent. Eur. Neurosurg. 2009, 70, 27–35. [Google Scholar] [CrossRef]
- Itoh, D.; Aoki, S.; Maruyama, K.; Masutani, Y.; Mori, H.; Masumoto, T.; Abe, O.; Hayashi, N.; Okubo, T.; Ohtomo, K. Corticospinal Tracts by Diffusion Tensor Tractography in Patients with Arteriovenous Malformations. J. Comput. Assist. Tomogr. 2006, 30, 618–623. [Google Scholar] [CrossRef]
- Okada, T.; Miki, Y.; Kikuta, K.; Mikuni, N.; Urayama, S.; Fushimi, Y.; Yamamoto, A.; Mori, N.; Fukuyama, H.; Hashimoto, N.; et al. Diffusion Tensor Fiber Tractography for Arteriovenous Malformations: Quantitative Analyses to Evaluate the Corticospinal Tract and Optic Radiation. AJNR Am. J. Neuroradiol. 2007, 28, 1107–1113. [Google Scholar] [CrossRef]
- Yamada, K.; Kizu, O.; Ito, H.; Nishimura, T. Tractography for an Arteriovenous Malformation. Neurology 2004, 62, 669. [Google Scholar] [CrossRef]
- Chen, Z.; Tie, Y.; Olubiyi, O.; Rigolo, L.; Mehrtash, A.; Norton, I.; Pasternak, O.; Rathi, Y.; Golby, A.J.; O’Donnell, L.J. Reconstruction of the Arcuate Fasciculus for Surgical Planning in the Setting of Peritumoral Edema Using Two-Tensor Unscented Kalman Filter Tractography. Neuroimage Clin. 2015, 7, 815–822. [Google Scholar] [CrossRef]
- Caverzasi, E.; Papinutto, N.; Amirbekian, B.; Berger, M.S.; Henry, R.G. Q-Ball of Inferior Fronto-Occipital Fasciculus and Beyond. PLoS ONE 2014, 9, e100274. [Google Scholar] [CrossRef]
- Côté, M.-A.; Girard, G.; Boré, A.; Garyfallidis, E.; Houde, J.-C.; Descoteaux, M. Tractometer: Towards Validation of Tractography Pipelines. Med. Image Anal. 2013, 17, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Schilling, K.G.; Daducci, A.; Maier-Hein, K.; Poupon, C.; Houde, J.-C.; Nath, V.; Anderson, A.W.; Landman, B.A.; Descoteaux, M. Challenges in Diffusion MRI Tractography—Lessons Learned from International Benchmark Competitions. Magn. Reson Imaging 2019, 57, 194–209. [Google Scholar] [CrossRef]
- Deletis, V. Intraoperative Monitoring of the Functional Integrity of the Motor Pathways. Adv. Neurol. 1993, 63, 201–214. [Google Scholar] [PubMed]
- Maier-Hein, K.H.; Neher, P.F.; Houde, J.-C.; Côté, M.-A.; Garyfallidis, E.; Zhong, J.; Chamberland, M.; Yeh, F.-C.; Lin, Y.-C.; Ji, Q.; et al. The Challenge of Mapping the Human Connectome Based on Diffusion Tractography. Nat. Commun. 2017, 8, 1349. [Google Scholar] [CrossRef] [PubMed]
- de Witt Hamer, P.C.; Robles, S.G.; Zwinderman, A.H.; Duffau, H.; Berger, M.S. Impact of Intraoperative Stimulation Brain Mapping on Glioma Surgery Outcome: A Meta-Analysis. J. Clin. Oncol. 2012, 30, 2559–2565. [Google Scholar] [CrossRef] [PubMed]
- Southwell, D.G.; Birk, H.S.; Han, S.J.; Li, J.; Sall, J.W.; Berger, M.S. Resection of Gliomas DeeMed. Inoperable by Neurosurgeons Based on Preoperative Imaging Studies. J. Neurosurg. 2018, 129, 567–575. [Google Scholar] [CrossRef]
- Sanai, N.; Berger, M.S. Mapping the Horizon: Techniques to Optimize Tumor Resection before and during Surgery. Clin. Neurosurg. 2008, 55, 14–19. [Google Scholar]
- Hervey-Jumper, S.L.; Li, J.; Lau, D.; Molinaro, A.M.; Perry, D.W.; Meng, L.; Berger, M.S. Awake Craniotomy to Maximize Glioma Resection: Methods and Technical Nuances over a 27-Year Period. J. Neurosurg. 2015, 123, 325–339. [Google Scholar] [CrossRef]
- Feigl, G.C.; Hiergeist, W.; Fellner, C.; Schebesch, K.-M.M.; Doenitz, C.; Finkenzeller, T.; Brawanski, A.; Schlaier, J. Magnetic Resonance Imaging Diffusion Tensor Tractography: Evaluation of Anatomic Accuracy of Different Fiber Tracking Software Packages. World Neurosurg. 2014, 81, 144–150. [Google Scholar] [CrossRef]
- Duffau, H. The Dangers of Magnetic Resonance Imaging Diffusion Tensor Tractography in Brain Surgery. World Neurosurg. 2014, 81, 56–58. [Google Scholar] [CrossRef]
- Schilling, K.G.; Tax, C.M.W.; Rheault, F.; Hansen, C.; Yang, Q.; Yeh, F.-C.; Cai, L.; Anderson, A.W.; Landman, B.A. Fiber Tractography Bundle Segmentation Depends on Scanner Effects, Vendor Effects, Acquisition Resolution, Diffusion Sampling Scheme, Diffusion Sensitization, and Bundle Segmentation Workflow. Neuroimage 2021, 242, 118451. [Google Scholar] [CrossRef] [PubMed]
- Rheault, F.; de Benedictis, A.; Daducci, A.; Maffei, C.; Tax, C.M.W.; Romascano, D.; Caverzasi, E.; Morency, F.C.; Corrivetti, F.; Pestilli, F.; et al. Tractostorm: The What, Why, and How of Tractography Dissection Reproducibility. Hum. Brain Mapp. 2020, 41, 1859–1874. [Google Scholar] [CrossRef] [PubMed]


| Dataset Name | Voxel Size (mm) | B-Value | Nr of DWI | |
|---|---|---|---|---|
| T1 | DWI | |||
| AOMIC-PIOP2 | 1.0 × 1.0 × 1.0 | 2.0 × 2.0 × 2.0 | 1000 | 32 |
| Forrest Gump | 0.7 × 0.7 × 0.7 | 2.0 × 2.0 × 2.0 | 800 | 32 |
| interTVA | 0.8 × 0.8 × 0.8 | 1.8 × 1.8 × 1.8 | various | 102 |
| Emotion regulation | 1.0 × 1.0 × 1.0 | 2.0 × 2.0 × 2.0 | 1000 | 30 |
| Test-Retest | 1.0 × 1.0 × 1.0 | 2.0 × 2.0 × 2.0 | 1000 | 32 |
| Beijing | 1.3 × 1.0 × 1.0 | 2.0 × 2.0 × 2.0 | 1000 | 64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barany, L.; Hore, N.; Stadlbauer, A.; Buchfelder, M.; Brandner, S. Prediction of the Topography of the Corticospinal Tract on T1-Weighted MR Images Using Deep-Learning-Based Segmentation. Diagnostics 2023, 13, 911. https://doi.org/10.3390/diagnostics13050911
Barany L, Hore N, Stadlbauer A, Buchfelder M, Brandner S. Prediction of the Topography of the Corticospinal Tract on T1-Weighted MR Images Using Deep-Learning-Based Segmentation. Diagnostics. 2023; 13(5):911. https://doi.org/10.3390/diagnostics13050911
Chicago/Turabian StyleBarany, Laszlo, Nirjhar Hore, Andreas Stadlbauer, Michael Buchfelder, and Sebastian Brandner. 2023. "Prediction of the Topography of the Corticospinal Tract on T1-Weighted MR Images Using Deep-Learning-Based Segmentation" Diagnostics 13, no. 5: 911. https://doi.org/10.3390/diagnostics13050911
APA StyleBarany, L., Hore, N., Stadlbauer, A., Buchfelder, M., & Brandner, S. (2023). Prediction of the Topography of the Corticospinal Tract on T1-Weighted MR Images Using Deep-Learning-Based Segmentation. Diagnostics, 13(5), 911. https://doi.org/10.3390/diagnostics13050911







