Advances in the Microbiological Diagnosis of Prosthetic Joint Infections
Abstract
1. Introduction
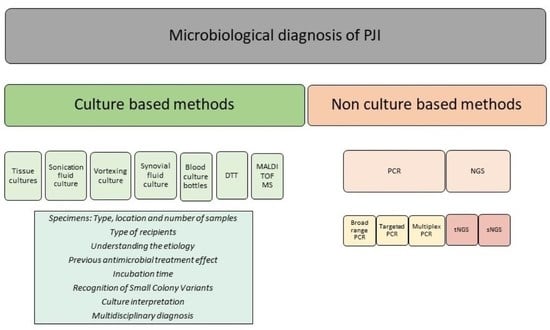
2. Key Aspects about the Microbiological Diagnosis of PJI
2.1. Specimens: Type, Location and Number of Samples
2.2. Type of Recipients
2.3. Etiopathogenesis
2.4. Previous Antimicrobial Treatment
2.5. Incubation Time
2.6. Recognition of Small Colony Variants (SCV)
2.7. Culture Interpretation
2.8. Multidisciplinary Diagnosis
2.9. Non-Culture-Based Methods
2.9.1. Polymerase Chain Reaction Techniques (PCR)
2.9.2. Next-Generation Sequencing Methods (NGS)
3. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jamari, J.; Ammarullah, M.I.; Santoso, G.; Sugiharto, S.; Supriyono, T.; Permana, M.S.; Winarni, T.I.; van der Heide, E. Adopted walking condition for computational simulation approach on bearing of hip joint prosthesis: Review over the past 30 years. Heliyon 2022, 8, e12050. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. Periprosthetic Joint Infection. N. Engl. J. Med. 2023, 388, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.M.; Ong, K.L.; Schmier, J.; Mowat, F.; Saleh, K.; Dybvik, E.; Karrholm, J.; Garellick, G.; Havelin, L.I.; Furnes, O.; et al. Future clinical and economic impact of revision total hip and knee arthroplasty. J. Bone Jt. Surg. 2007, 89 (Suppl. 3), 144–151. [Google Scholar]
- Kurtz, S.; Ong, K.; Lau, E.; Mowat, F.; Halpern, M. Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J. Bone Jt. Surg. 2007, 89, 780–785. [Google Scholar] [CrossRef]
- Del Pozo, J.L.; Tran, N.V.; Petty, P.M.; Johnson, C.H.; Walsh, M.F.; Bite, U.; Clay, R.P.; Mandrekar, J.N.; Piper, K.E.; Steckelberg, J.M.; et al. Pilot study of association of bacteria on breast implants with capsular contracture. J. Clin. Microbiol. 2009, 47, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Pannu, T.S.; Villa, J.M.; Higuera, C.A. Diagnosis and management of infected arthroplasty. SICOT-J 2021, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Trampuz, A.; Piper, K.E.; Jacobson, M.J.; Hanssen, A.D.; Unni, K.K.; Osmon, D.R.; Mandrekar, J.N.; Cockerill, F.R.; Steckelberg, J.M.; Greenleaf, J.F.; et al. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 2007, 357, 654–663. [Google Scholar] [CrossRef]
- Berbari, E.F.; Marculescu, C.; Sia, I.; Lahr, B.D.; Hanssen, A.D.; Steckelberg, J.M.; Gullerud, R.; Osmon, D.R. Culture-negative prosthetic joint infection. Clin. Infect. Dis. 2007, 45, 1113–1119. [Google Scholar] [CrossRef]
- Bellova, P.; Knop-Hammad, V.; Königshausen, M.; Mempel, E.; Frieler, S.; Gessmann, J.; Schildhauer, T.A.; Baecker, H. Sonication of retrieved implants improves sensitivity in the diagnosis of periprosthetic joint infection. BMC Musculoskelet. Disord. 2019, 20, 623. [Google Scholar] [CrossRef]
- Baron, E.J.; Miller, J.M.; Weinstein, M.P.; Richter, S.S.; Gilligan, P.H.; Thomson, R.B.; Bourbeau, P.; Carroll, K.C.; Kehl, S.C.; Dunne, W.M.; et al. Executive summary: A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM)(a). Clin. Infect. Dis. 2013, 57, 485–488. [Google Scholar] [CrossRef]
- Corvec, S.; Portillo, M.E.; Pasticci, B.M.; Borens, O.; Trampuz, A. Epidemiology and new developments in the diagnosis of prosthetic joint infection. Int. J. Artif. Organs 2012, 35, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Renz, N.; Trampuz, A.; Ojeda-Thies, C. Twenty common errors in the diagnosis and treatment of periprosthetic joint infection. Int. Orthop. 2020, 44, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Goswami, K.; Parvizi, J.; Maxwell Courtney, P. Current Recommendations for the Diagnosis of Acute and Chronic PJI for Hip and Knee-Cell Counts, Alpha-Defensin, Leukocyte Esterase, Next-generation Sequencing. Curr. Rev. Musculoskelet. Med. 2018, 11, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.; Gómez-Barrena, E. An update about molecular biology techniques to detect orthopaedic implant-related infections. EFORT Open Rev. 2021, 6, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.E.; Salvado, M.; Alier, A.; Martinez, S.; Sorli, L.; Horcajada, J.P.; Puig, L. Advantages of sonication fluid culture for the diagnosis of prosthetic joint infection. J. Infect. 2014, 69, 35–41. [Google Scholar] [CrossRef]
- Atkins, B.L.; Athanasou, N.; Deeks, J.J.; Crook, D.W.; Simpson, H.; Peto, T.E.; McLardy-Smith, P.; Berendt, A.R. Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. The OSIRIS Collaborative Study Group. J. Clin. Microbiol. 1998, 36, 2932–2939. [Google Scholar] [CrossRef]
- Spangehl, M.J.; Masri, B.A.; O’Connell, J.X.; Duncan, C.P. Prospective analysis of preoperative and intraoperative investigations for the diagnosis of infection at the sites of two hundred and two revision total hip arthroplasties. J. Bone Jt. Surg. 1999, 81, 672–683. [Google Scholar] [CrossRef]
- Schindler, M.; Christofilopoulos, P.; Wyssa, B.; Belaieff, W.; Garzoni, C.; Bernard, L.; Lew, D.; Hoffmeyer, P.; Uckay, I. Poor performance of microbiological sampling in the prediction of recurrent arthroplasty infection. Int. Orthop. 2011, 35, 647–654. [Google Scholar] [CrossRef]
- Hughes, J.G.; Vetter, E.A.; Patel, R.; Schleck, C.D.; Harmsen, S.; Turgeant, L.T.; Cockerill, F.R. Culture with BACTEC Peds Plus/F bottle compared with conventional methods for detection of bacteria in synovial fluid. J. Clin. Microbiol. 2001, 39, 4468–4471. [Google Scholar] [CrossRef]
- Font-Vizcarra, L.; Garcia, S.; Martinez-Pastor, J.C.; Sierra, J.M.; Soriano, A. Blood culture flasks for culturing synovial fluid in prosthetic joint infections. Clin. Orthop. Relat. Res. 2010, 468, 2238–2243. [Google Scholar] [CrossRef]
- Osmon, D.R.; Berbari, E.F.; Berendt, A.R.; Lew, D.; Zimmerli, W.; Steckelberg, J.M.; Rao, N.; Hanssen, A.; Wilson, W.R. Diagnosis and management of prosthetic joint infection: Clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 2013, 56, e1–e25. [Google Scholar] [CrossRef] [PubMed]
- Corvec, S.; Portillo, M.; Vossen, A.; Trampuz, A.; Haar, P. Diagnostics. In Principles of Orthopedic Infection Management. AOTrauma; Georg Thieme Verlag: Stuttgart, Germany, 2016; pp. 91–122. [Google Scholar]
- Ascione, T.; Barrack, R.; Benito, N.; Blevins, K.; Brause, B.; Cornu, O.; Frommelt, L.; Gant, V.; Goswami, K.; Hu, R.; et al. General Assembly, Diagnosis, Pathogen Isolation-Culture Matters: Proceedings of International Consensus on Orthopedic Infections. J. Arthroplast. 2019, 34, S197–S206. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, M.; Barrett, L.; Figtree, M.; Scarborough, M.; Watanabe, M.; Newnham, R.; Wallis, R.; Oakley, S.; Kendrick, B.; Stubbs, D.; et al. Sonication versus tissue sampling for diagnosis of prosthetic joint and other orthopedic device-related infections. J. Clin. Microbiol. 2018, 56, e00688-18. [Google Scholar] [CrossRef] [PubMed]
- Izakovicova, P.; Borens, O.; Trampuz, A. Periprosthetic joint infection: Current concepts and outlook. EFORT Open Rev. 2019, 4, 482–494. [Google Scholar] [CrossRef]
- Trampuz, A.; Piper, K.E.; Hanssen, A.D.; Osmon, D.R.; Cockerill, F.R.; Steckelberg, J.M.; Patel, R. Sonication of explanted prosthetic components in bags for diagnosis of prosthetic joint infection is associated with risk of contamination. J. Clin. Microbiol. 2006, 44, 628–631. [Google Scholar] [CrossRef]
- Zimmerli, W.; Trampuz, A.; Ochsner, P.E. Prosthetic-joint infections. N. Engl. J. Med. 2004, 351, 1645–1654. [Google Scholar] [CrossRef]
- Portillo, M.E.; Salvado, M.; Alier, A.; Sorli, L.; Martinez, S.; Horcajada, J.P.; Puig, L. Prosthesis failure within 2 years of implantation is highly predictive of infection. Clin. Orthop. Relat. Res. 2013, 471, 3672–3678. [Google Scholar] [CrossRef]
- O’Toole, P.; Osmon, D.; Soriano, A.; Berdal, J.E.; Bostrum, M.; Franco-Cendejas, R.; Huang, D.; Nelson, C.; Nishisaka, F.; Roslund, B.; et al. Oral antibiotic therapy. J. Arthroplast. 2014, 29, 115–118. [Google Scholar] [CrossRef]
- Burnett, R.S.; Aggarwal, A.; Givens, S.A.; McClure, J.T.; Morgan, P.M.; Barrack, R.L. Prophylactic antibiotics do not affect cultures in the treatment of an infected TKA: A prospective trial. Clin. Orthop. Relat. Res. 2010, 468, 127–134. [Google Scholar] [CrossRef]
- Ghanem, E.; Parvizi, J.; Clohisy, J.; Burnett, S.; Sharkey, P.F.; Barrack, R. Perioperative antibiotics should not be withheld in proven cases of periprosthetic infection. Clin. Orthop. Relat. Res. 2007, 461, 44–47. [Google Scholar] [CrossRef]
- Pérez-Prieto, D.; Portillo, M.E.; Puig-Verdié, L.; Alier, A.; Gamba, C.; Guirro, P.; Martínez-Díaz, S.; Horcajada, J.P.J.P.; Trampuz, A.; Monllau, J.C.J.C. Preoperative antibiotic prophylaxis in prosthetic joint infections: Not a concern for intraoperative cultures. Diagn. Microbiol. Infect. Dis. 2016, 86, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Wouthuyzen-Bakker, M.; Benito, N.; Soriano, A. The effect of preoperative antimicrobial prophylaxis on intraoperative culture results in patients with a suspected or confirmed prosthetic joint infection: A systematic review. J. Clin. Microbiol. 2017, 55, 2765–2774. [Google Scholar] [CrossRef]
- Parsek, M.R.; Singh, P.K. Bacterial biofilms: An emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003, 57, 677–701. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.; Alvarez-Alvarez, B.; Blanco, A.; Fernandez-Roblas, R.; Gadea, I.; Garcia-Canete, J.; Sandoval, E.; Valdazo, M. Prolonged incubation time does not increase sensitivity for the diagnosis of implant-related infection using samples prepared by sonication of the implants. Bone Jt. J. 2013, 95, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.E.; Salvadó, M.; Trampuz, A.; Plasencia, V.; Rodriguez-Villasante, M.; Sorli, L.; Puig, L.; Horcajada, J.P. Sonication versus Vortexing of Implants for Diagnosis of Prosthetic Joint Infection. J. Clin. Microbiol. 2013, 51, 591–594. [Google Scholar] [CrossRef]
- Portillo, M.E.; Salvadó, M.; Trampuz, A.; Siverio, A.; Alier, A.; Sorli, L.; Martínez, S.; Pérez-Prieto, D.; Horcajada, J.P.J.P.; Puig-Verdie, L. Improved diagnosis of orthopedic implant-associated infection by inoculation of sonication fluid into blood culture bottles. J. Clin. Microbiol. 2015, 53, 1622–1627. [Google Scholar] [CrossRef] [PubMed]
- van den Bijllaardt, W.; van der Jagt, O.P.; Peijs, M.; Janssens, M.; Buiting, A.G.; Reuwer, A.Q. Culturing periprosthetic tissue in blood culture bottles results in isolation of additional microorganisms. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 245–252. [Google Scholar] [CrossRef]
- Li, C.; Renz, N.; Thies, C.O.; Trampuz, A. Meta-analysis of sonicate fluid in blood culture bottles for diagnosing periprosthetic joint infection. J. Bone Jt. Infect. 2018, 3, 273–279. [Google Scholar] [CrossRef]
- Minassian, A.M.; Newnham, R.; Kalimeris, E.; Bejon, P.; Atkins, B.L.; Bowler, I.C. Use of an automated blood culture system (BD BACTEC) for diagnosis of prosthetic joint infections: Easy and fast. BMC Infect. Dis. 2014, 14, 233. [Google Scholar] [CrossRef]
- Godec, M.; Kocijan, A.; Dolinar, D.; Mandrino, D.; Jenko, M.; Antolic, V. An investigation of the aseptic loosening of an AISI 316L stainless steel hip prosthesis. Biomed. Mater. 2010, 5, 45012. [Google Scholar] [CrossRef]
- Drago, L.; Clerici, P.; Morelli, I.; Ashok, J.; Benzakour, T.; Bozhkova, S.; Alizadeh, C.; Del Sel, H.; Sharma, H.K.; Peel, T.; et al. The World Association against Infection in Orthopaedics and Trauma (WAIOT) procedures for Microbiological Sampling and Processing for Periprosthetic Joint Infections (PJIs) and other Implant-Related Infections. J. Clin. Med. 2019, 8, 933. [Google Scholar] [CrossRef]
- Sendi, P.; Frei, R.; Maurer, T.B.; Trampuz, A.; Zimmerli, W.; Graber, P. Escherichia coli variants in periprosthetic joint infection: Diagnostic challenges with sessile bacteria and sonication. J. Clin. Microbiol. 2010, 48, 1720–1725. [Google Scholar] [CrossRef] [PubMed]
- Sendi, P.; Proctor, R.A. Staphylococcus aureus as an intracellular pathogen: The role of small colony variants. Trends Microbiol. 2009, 17, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Rao, Y.; Li, J.; Huang, Q.; Rao, X. Staphylococcus aureus small-colony variants: Formation, infection, and treatment. Microbiol. Res. 2022, 260, 127040. [Google Scholar] [CrossRef]
- Manasherob, R.; Mooney, J.A.; Lowenberg, D.W.; Bollyky, P.L.; Amanatullah, D.F. Tolerant Small-colony Variants Form Prior to Resistance Within a Staphylococcus aureus Biofilm Based on Antibiotic Selective Pressure. Clin. Orthop. Relat. Res. 2021, 479, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Rodríguez, D.; Colín-Castro, C.A.; Hernández-Durán, M.; López-Jácome, L.E.; Franco-Cendejas, R. Staphylococcus epidermidis small colony variants, clinically significant quiescent threats for patients with prosthetic joint infection. Microbes Infect. 2021, 23, 104854. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.; Chadfield, M.S.; Christensen, J.P.; Christensen, H.; Bisgaard, M. Characterization of small-colony variants of Enterococcus faecalis isolated from chickens with amyloid arthropathy. J. Clin. Microbiol. 2008, 46, 2686–2691. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Tong, Y.; Cheng, J.; Abbas, Z.; Li, Z.; Wang, J.; Zhou, Y.; Si, D.; Zhang, R. Biofilm and Small Colony Variants-An Update on Staphylococcus aureus Strategies toward Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1241. [Google Scholar] [CrossRef] [PubMed]
- Neut, D.; Van Der Mei, H.C.; Bulstra, S.K.; Busscher, H.J. The role of small-colony variants in failure to diagnose and treat biofilm infections in orthopedics. Acta Orthop. 2007, 78, 299–308. [Google Scholar] [CrossRef]
- Meléndez-Carmona, M.Á.; Muñoz-Gallego, I.; Viedma, E.; Lora-Tamayo, J.; Chaves, F. Intraosteoblastic activity of levofloxacin and rifampin alone and in combination against clinical isolates of meticillin-susceptible Staphylococcus aureus causing prosthetic joint infection. Int. J. Antimicrob. Agents 2019, 54, 356–360. [Google Scholar] [CrossRef]
- McNally, M.; Sousa, R.; Wouthuyzen-Bakker, M.; Chen, A.F.; Soriano, A.; Vogely, H.C.; Clauss, M.; Higuera, C.A.; Trebse, R. The EBJIS definition of periprosthetic joint infection: A practical guide for clinicians. Bone Jt. J. 2021, 103, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Oliva, A.; Miele, M.C.; Al Ismail, D.; Di Timoteo, F.; De Angelis, M.; Rosa, L.; Cutone, A.; Venditti, M.; Mascellino, M.T.; Valenti, P.; et al. Challenges in the Microbiological Diagnosis of Implant-Associated Infections: A Summary of the Current Knowledge. Front. Microbiol. 2021, 12, 750460. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Chen, S.; Zhang, C.; Zhao, X.; Huang, X.; Cai, Z. Effect of the Biofilm Age and Starvation on Acid Tolerance of Biofilm Formed by Streptococcus mutans Isolated from Caries-Active and Caries-Free Adults. Int. J. Mol. Sci. 2017, 18, 713. [Google Scholar] [CrossRef] [PubMed]
- Argenson, J.N.; Arndt, M.; Babis, G.; Battenberg, A.; Budhiparama, N.; Catani, F.; Chen, F.; de Beaubien, B.; Ebied, A.; Esposito, S.; et al. Hip and Knee Section, Treatment, Debridement and Retention of Implant: Proceedings of International Consensus on Orthopedic Infections. J. Arthroplast. 2019, 34, S399–S419. [Google Scholar] [CrossRef] [PubMed]
- Karbysheva, S.; Cabric, S.; Koliszak, A.; Bervar, M.; Kirschbaum, S.; Hardt, S.; Perka, C.; Trampuz, A. Clinical evaluation of dithiothreitol in comparison with sonication for biofilm dislodgement in the microbiological diagnosis of periprosthetic joint infection. Diagn. Microbiol. Infect. Dis. 2022, 103, 115679. [Google Scholar] [CrossRef]
- Lausmann, C.; Zahar, A.; Citak, M.; Brañes, J.; Schmidl, S.; Frommelt, L.; Gehrke, T.; Gebauer, M. Are There Benefits In Early Diagnosis Of Prosthetic Joint Infection With Multiplex Polymerase Chain Reaction? J. Bone Jt. Infect. 2017, 2, 175–183. [Google Scholar] [CrossRef]
- Pérez-Prieto, D.; Portillo, M.E.; Puig-Verdié, L.; Alier, A.; Martínez, S.; Sorlí, L.; Horcajada, J.P.; Monllau, J.C. C-reactive protein may misdiagnose prosthetic joint infections, particularly chronic and low-grade infections. Int. Orthop. 2017, 41, 1315–1319. [Google Scholar] [CrossRef]
- Portillo, M.E.; Corvec, S. Chapter 4. Identification of Pathogens in Bone and Joint Infections by Non-Culture Techniques. In Bone and Joint Infection, 2nd ed.; John Wiley and Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Gamie, Z.; Karthikappallil, D.; Gamie, E.; Stamiris, S.; Kenanidis, E.; Tsiridis, E. Molecular sequencing technologies in the diagnosis and management of prosthetic joint infections. Expert Rev. Mol. Diagn. 2022, 22, 603–624. [Google Scholar] [CrossRef]
- Portillo, M.E.; Salvado, M.; Sorli, L.; Alier, A.; Martinez, S.; Trampuz, A.; Gomez, J.; Puig, L.; Horcajada, J.P. Multiplex PCR of sonication fluid accurately differentiates between prosthetic joint infection and aseptic failure. J. Infect. 2012, 65, 541–548. [Google Scholar] [CrossRef]
- Esteban, J.; Alonso-Rodriguez, N.; Del-Prado, G.; Ortiz-Perez, A.; Molina-Manso, D.; Cordero-Ampuero, J.; Sandoval, E.; Fernandez-Roblas, R.; Gomez-Barrena, E. PCR-hybridization after sonication improves diagnosis of implant-related infection. Acta Orthop. 2012, 83, 299–304. [Google Scholar] [CrossRef]
- Curtoni, A.; Cipriani, R.; Marra, E.S.; Barbui, A.M.; Cavallo, R.; Costa, C. Rapid Identification of Microorganisms from Positive Blood Culture by MALDI-TOF MS After Short-Term Incubation on Solid Medium. Curr. Microbiol. 2017, 74, 97–102. [Google Scholar] [CrossRef]
- Lallemand, E.; Coiffier, G.; Arvieux, C.; Brillet, E.; Guggenbuhl, P.; Jolivet-Gougeon, A. MALDI-TOF MS performance compared to direct examination, culture, and 16S rDNA PCR for the rapid diagnosis of bone and joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Kuo, F.C.; Chien, C.C.; Lee, M.S.; Wang, J.W.; Lin, P.C.; Lee, C.H. Rapid diagnosis of periprosthetic joint infection from synovial fluid in blood culture bottles by direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS ONE 2020, 15, e0239290. [Google Scholar] [CrossRef] [PubMed]
- Gomez, E.; Cazanave, C.; Cunningham, S.A.; Greenwood-Quaintance, K.E.; Steckelberg, J.M.; Uhl, J.R.; Hanssen, A.D.; Karau, M.J.; Schmidt, S.M.; Osmon, D.R.; et al. Prosthetic joint infection diagnosis using broad-range PCR of biofilms dislodged from knee and hip arthroplasty surfaces using sonication. J. Clin. Microbiol. 2012, 50, 3501–3508. [Google Scholar] [CrossRef] [PubMed]
- Mullegama, S.V.; Alberti, M.O.; Au, C.; Li, Y.; Toy, T.; Tomasian, V.; Xian, R.R. Nucleic Acid Extraction from Human Biological Samples. Methods Mol. Biol. 2019, 1897, 359–383. [Google Scholar] [CrossRef] [PubMed]
- Aubin, G.G.; Bemer, P.; Guillouzouic, A.; Cremet, L.; Touchais, S.; Fraquet, N.; Boutoille, D.; Reynaud, A.; Lepelletier, D.; Corvec, S. First report of a hip prosthetic and joint infection caused by Lactococcus garvieae in a woman fishmonger. J. Clin. Microbiol. 2011, 49, 2074–2076. [Google Scholar] [CrossRef]
- Bémer, P.; Plouzeau, C.; Tande, D.; Léger, J.; Giraudeau, B.; Valentin, A.S.; Jolivet-Gougeon, A.; Vincent, P.; Corvec, S.; Gibaud, S.; et al. Evaluation of 16S rRNA gene PCR sensitivity and specificity for diagnosis of prosthetic joint infection: A prospective multicenter cross-sectional study. J. Clin. Microbiol. 2014, 52, 3583–3589. [Google Scholar] [CrossRef]
- Marin, M.; Garcia-Lechuz, J.M.; Alonso, P.; Villanueva, M.; Alcala, L.; Gimeno, M.; Cercenado, E.; Sanchez-Somolinos, M.; Radice, C.; Bouza, E. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J. Clin. Microbiol. 2012, 50, 583–589. [Google Scholar] [CrossRef]
- Titécat, M.; Loïez, C.; Demaeght, F.; Leclerc, J.T.; Martin, T.; Dezèque, H.; Migaud, H.; Senneville, E. Challenging Methicillin Resistance Detection in Bone and Joint Infections: Focus on the MRSA/SA SSTI® Strategy. Front. Med. 2021, 8, 553965. [Google Scholar] [CrossRef]
- Cazanave, C.; Greenwood-Quaintance, K.E.; Hanssen, A.D.; Patel, R. Corynebacterium prosthetic joint infection. J. Clin. Microbiol. 2012, 50, 1518–1523. [Google Scholar] [CrossRef]
- Prieto-Borja, L.; Rodriguez-Sevilla, G.; Auñon, A.; Pérez-Jorge, C.; Sandoval, E.; Garcia-Cañete, J.; Gadea, I.; Fernandez-Roblas, R.; Blanco, A.; Esteban, J. Evaluación de una PCR múltiple comercial (Unyvero i60©) diseñada para el diagnóstico de infecciones osteoarticulares utilizando prótesis articulares sonicadas. Enferm. Infecc. Microbiol. Clin. 2017, 35, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.A.; Wolf, M.J.; Strasburg, A.P.; Daniels, M.L.; Starkey, J.C.; Donadio, A.D.; Abdel, M.P.; Greenwood-Quaintance, K.E.; Patel, R. Comparison of the BioFire Joint Infection Panel to 16S Ribosomal RNA Gene-Based Targeted Metagenomic Sequencing for Testing Synovial Fluid from Patients with Knee Arthroplasty Failure. J. Clin. Microbiol. 2022, 60, e0112622. [Google Scholar] [CrossRef]
- Plouzeau, C.; Bémer, P.; Valentin, A.S.; Héry-Arnaud, G.; Tandé, D.; Jolivet-Gougeon, A.; Vincent, P.; Kempf, M.; Lemarié, C.; Guinard, J.; et al. First experience of a multicenter external quality assessment of molecular 16S rRNA gene detection in bone and joint infections. J. Clin. Microbiol. 2015, 53, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.X.; Huang, Z.; Fang, X.; Li, W.; Yang, B.; Zhang, W. Comparison of broad-range polymerase chain reaction and metagenomic next-generation sequencing for the diagnosis of prosthetic joint infection. Int. J. Infect. Dis. 2020, 95, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Stylianakis, A.; Schinas, G.; Thomaidis, P.C.; Papaparaskevas, J.; Ziogas, D.C.; Gamaletsou, M.N.; Daikos, G.L.; Pneumaticos, S.; Sipsas, N.V. Combination of conventional culture, vial culture, and broad-range PCR of sonication fluid for the diagnosis of prosthetic joint infection. Diagn. Microbiol. Infect. Dis. 2018, 92, 13–18. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.P.; Petti, C.A. Detection and Identification of Microorganisms by Gene Amplification and Sequencing. Clin. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar] [CrossRef]
- Fenollar, F.; Roux, V.; Stein, A.; Drancourt, M.; Raoult, D. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J. Clin. Microbiol. 2006, 44, 1018–1028. [Google Scholar] [CrossRef]
- Jun, Y.; Jianghua, L. Diagnosis of Periprosthetic Joint Infection Using Polymerase Chain Reaction: An Updated Systematic Review and Meta-Analysis. Surg. Infect. 2018, 19, 555–565. [Google Scholar] [CrossRef]
- Kommedal, Ø.; Kvello, K.; Skjåstad, R.; Langeland, N.; Wiker, H.G. Direct 16S rRNA gene sequencing from clinical specimens, with special focus on polybacterial samples and interpretation of mixed DNA chromatograms. J. Clin. Microbiol. 2009, 47, 3562–3568. [Google Scholar] [CrossRef]
- Yang, F.; Choe, H.; Kobayashi, N.; Tezuka, T.; Oba, M.; Miyamae, Y.; Morita, A.; Abe, K.; Inaba, Y. An automated real-time PCR assay for synovial fluid improves the preoperative etiological diagnosis of periprosthetic joint infection and septic arthritis. J. Orthop. Res. 2021, 39, 348–355. [Google Scholar] [CrossRef]
- Hartley, J.C.; Harris, K.A. Molecular Techniques for Diagnosing Prosthetic Joint Infections-Search Results-PubMed. J. Antimicrob. Chemother. 2014, 69, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Fu, J.; Yu, B.; Sun, W.; Chen, J.; Hao, L. Meta-analysis of sonication prosthetic fluid PCR for diagnosing periprosthetic joint infection. PLoS ONE 2018, 13, e0196418. [Google Scholar] [CrossRef] [PubMed]
- Saeed, K.; Ahmad-Saeed, N.; Annett, R.; Barlow, G.; Barrett, L.; Boyd, S.E.; Boran, N.; Davies, P.; Hughes, H.; Jones, G.; et al. A multicentre evaluation and expert recommendations of use of the newly developed BioFire Joint Infection polymerase chain reaction panel. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 1, 29–37. [Google Scholar] [CrossRef]
- Malandain, D.; Bémer, P.; Leroy, A.G.; Léger, J.; Plouzeau, C.; Valentin, A.S.; Jolivet-Gougeon, A.; Tandé, D.; Héry-Arnaud, G.; Lemarié, C.; et al. Assessment of the automated multiplex-PCR Unyvero i60 ITI® cartridge system to diagnose prosthetic joint infection: A multicentre study. Clin. Microbiol. Infect. 2018, 24, 83.e1–83.e6. [Google Scholar] [CrossRef] [PubMed]
- Vasoo, S.; Cunningham, S.A.; Greenwood-Quaintance, K.E.; Mandrekar, J.N.; Hanssen, A.D.; Abdel, M.P.; Osmon, D.R.; Berbari, E.F.; Patel, R. Evaluation of the FilmArray blood culture ID panel on biofilms dislodged from explanted arthroplasties for prosthetic joint infection diagnosis. J. Clin. Microbiol. 2015, 53, 2790–2792. [Google Scholar] [CrossRef]
- Dekker, J.P.; Dekkera, J.P. Metagenomics for Clinical Infectious Disease Diagnostics Steps Closer to Reality. J. Clin. Microbiol. 2018, 56, 9. [Google Scholar] [CrossRef]
- Street, T.L.; Sanderson, N.D.; Atkins, B.L.; Brent, A.J.; Cole, K.; Foster, D.; McNally, M.A.; Oakley, S.; Peto, L.; Taylor, A.; et al. Molecular diagnosis of orthopedic-device-related infection directly from sonication fluid by metagenomic sequencing. J. Clin. Microbiol. 2017, 55, 2334–2347. [Google Scholar] [CrossRef]
- Thoendel, M.J.; Jeraldo, P.R.; Greenwood-Quaintance, K.E.; Yao, J.Z.; Chia, N.; Hanssen, A.D.; Abdel, M.P.; Patel, R.P. Identification of Prosthetic Joint Infection Pathogens Using a Shotgun Metagenomics Approach-PubMed. Clin. Infect. Dis. 2018, 67, 1333–1338. [Google Scholar] [CrossRef]
- Yohe, S.; Thyagarajan, B. Review of Clinical Next-Generation Sequencing. Arch. Pathol. Lab. Med. 2017, 141, 1544–1557. [Google Scholar] [CrossRef]
- Zhong, Y.; Xu, F.; Wu, J.; Schubert, J.; Li, M.M. Application of Next Generation Sequencing in Laboratory Medicine. Ann. Lab. Med. 2021, 41, 25–43. [Google Scholar] [CrossRef]
- Church, D.L.; Cerutti, L.; Gürtler, A.; Griener, T.; Zelazny, A.; Emler, S. Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory. Clin. Microbiol. Rev. 2020, 33, 1–74. [Google Scholar] [CrossRef] [PubMed]
- Deurenberg, R.H.; Bathoorn, E.; Chlebowicz, M.A.; Couto, N.; Ferdous, M.; García-Cobos, S.; Kooistra-Smid, A.M.D.; Raangs, E.C.; Rosema, S.; Veloo, A.C.M.; et al. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 2017, 243, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.L.; Flurin, L.; Thoendel, M.J.; Wolf, M.J.; Abdel, M.P.; Greenwood-Quaintance, K.E.; Patel, R. Targeted versus Shotgun Metagenomic Sequencing-Based Detection of Microorganisms in Sonicate Fluid for Periprosthetic Joint Infection Diagnosis. Clin. Infect. Dis. 2022, 76, e1456–e1462. [Google Scholar] [CrossRef] [PubMed]
- Hantouly, A.T.; Alzobi, O.; Toubasi, A.A.; Zikria, B.; Al Dosari, M.A.A.; Ahmed, G. Higher sensitivity and accuracy of synovial next-generation sequencing in comparison to culture in diagnosing periprosthetic joint infection: A systematic review and meta-analysis. Knee Surg. Sport. Traumatol. Arthrosc. 2022. [Google Scholar] [CrossRef]
- Indelli, P.F.; Ghirardelli, S.; Violante, B.; Amanatullah, D.F. Next generation sequencing for pathogen detection in periprosthetic joint infections. EFORT Open Rev. 2021, 6, 236–244. [Google Scholar] [CrossRef]
- Eyre, D.W.; Golubchik, T.; Gordon, N.C.; Bowden, R.; Piazza, P.; Batty, E.M.; Ip, C.L.C.; Wilson, D.J.; Didelot, X.; O’Connor, L.; et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2012, 2, e001124. [Google Scholar] [CrossRef]
- Reuter, S.; Ellington, M.J.; Cartwright, E.J.P.; Köser, C.U.; Török, M.E.; Gouliouris, T.; Harris, S.R.; Brown, N.M.; Holden, M.T.G.; Quail, M.; et al. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern. Med. 2013, 173, 1397–1404. [Google Scholar] [CrossRef]
- Wildeman, P.; Tevell, S.; Eriksson, C.; Lagos, A.C.; Söderquist, B.; Stenmark, B. Genomic characterization and outcome of prosthetic joint infections caused by Staphylococcus aureus. Sci. Rep. 2020, 10, 5938. [Google Scholar] [CrossRef]
- Didelot, X.; Eyre, D.W.; Cule, M.; Ip, C.L.C.; Ansari, M.A.; Griffiths, D.; Vaughan, A.; O’Connor, L.; Golubchik, T.; Batty, E.M.; et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol. 2012, 13, R118. [Google Scholar] [CrossRef]
- Eyre, D.W.; Fawley, W.N.; Best, E.L.; Griffiths, D.; Stoesser, N.E.; Crook, D.W.; Peto, T.E.A.; Walker, A.S.; Wilcox, M.H. Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J. Clin. Microbiol. 2013, 51, 4141–4149. [Google Scholar] [CrossRef]
- Walker, T.M.; Lalor, M.K.; Broda, A.; Ortega, L.S.; Morgan, M.; Parker, L.; Churchill, S.; Bennett, K.; Golubchik, T.; Giess, A.P.; et al. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007–2012, with whole pathogen genome sequences: An observational study. Lancet. Respir. Med. 2014, 2, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Stoesser, N.; Batty, E.M.; Eyre, D.W.; Morgan, M.; Wyllie, D.H.; Del Ojo Elias, C.; Johnson, J.R.; Walker, A.S.; Peto, T.E.A.; Crook, D.W. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J. Antimicrob. Chemother. 2013, 68, 2234–2244. [Google Scholar] [CrossRef] [PubMed]
- Gordon, N.C.; Price, J.R.; Cole, K.; Everitt, R.; Morgan, M.; Finney, J.; Kearns, A.M.; Pichon, B.; Young, B.; Wilson, D.J.; et al. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J. Clin. Microbiol. 2014, 52, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.M.; Kohl, T.A.; Omar, S.V.; Hedge, J.; Del Ojo Elias, C.; Bradley, P.; Iqbal, Z.; Feuerriegel, S.; Niehaus, K.E.; Wilson, D.J.; et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: A retrospective cohort study. Lancet. Infect. Dis. 2015, 15, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Metcalf, B.J.; Chochua, S.; Li, Z.; Gertz, R.E.; Walker, H.; Hawkins, P.A.; Tran, T.; Whitney, C.G.; McGee, L.; et al. Penicillin-Binding Protein Transpeptidase Signatures for Tracking and Predicting β-Lactam Resistance Levels in Streptococcus pneumoniae. MBio 2016, 7, e00756-16. [Google Scholar] [CrossRef]
- Lüftinger, L.; Ferreira, I.; Frank, B.J.H.; Beisken, S.; Weinberger, J.; von Haeseler, A.; Rattei, T.; Hofstaetter, J.G.; Posch, A.E.; Materna, A. Predictive Antibiotic Susceptibility Testing by Next-Generation Sequencing for Periprosthetic Joint Infections: Potential and Limitations. Biomedicines 2021, 9, 910. [Google Scholar] [CrossRef]
- Street, T.L.; Sanderson, N.D.; Kolenda, C.; Kavanagh, J.; Pickford, H.; Hoosdally, S.; Cregan, J.; Taunt, C.; Jones, E.; Oakley, S.; et al. Clinical Metagenomic Sequencing for Species Identification and Antimicrobial Resistance Prediction in Orthopedic Device Infection. J. Clin. Microbiol. 2022, 60, e0215621. [Google Scholar] [CrossRef]
- Kildow, B.J.; Ryan, S.P.; Danilkowicz, R.; Lazarides, A.L.; Penrose, C.; Bolognesi, M.P.; Jiranek, W.; Seyler, T.M. Next-generation sequencing not superior to culture in periprosthetic joint infection diagnosis. Bone Joint J. 2021, 103-B, 26–31. [Google Scholar] [CrossRef]
- Flurin, L.; Wolf, M.J.; Greenwood-Quaintance, K.E.; Sanchez-Sotelo, J.; Patel, R. Targeted next generation sequencing for elbow periprosthetic joint infection diagnosis. Diagn. Microbiol. Infect. Dis. 2021, 101, 115448. [Google Scholar] [CrossRef]
- Cai, Y.; Fang, X.; Chen, Y.; Huang, Z.; Zhang, C.; Li, W.; Yang, B.; Zhang, W. Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int. J. Infect. Dis. 2020, 96, 573–578. [Google Scholar] [CrossRef]
- Yin, H.; Xu, D.; Wang, D. Diagnostic value of next-generation sequencing to detect periprosthetic joint infection. BMC Musculoskelet. Disord. 2021, 22. [Google Scholar] [CrossRef]
- Wang, C.; Huang, Z.; Li, W.; Fang, X.; Zhang, W. Can metagenomic next-generation sequencing identify the pathogens responsible for culture-negative prosthetic joint infection? BMC Infect. Dis. 2020, 20, 253. [Google Scholar] [CrossRef]
- Ivy, M.I.; Thoendel, M.J.; Jeraldo, P.R.; Greenwood-Quaintance, K.E.; Hanssen, A.D.; Abdel, M.P.; Chia, N.; Yao, J.Z.; Tande, A.J.; Mandrekar, J.N.; et al. Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J. Clin. Microbiol. 2018, 56, e00402-18. [Google Scholar] [CrossRef]
- Tarabichi, M.; Shohat, N.; Goswami, K.; Alvand, A.; Silibovsky, R.; Belden, K.; Parvizi, J. Diagnosis of periprosthetic joint infection: The potential of next-generation sequencing. J. Bone Jt. Surg.-Am. Vol. 2018, 100, 147–154. [Google Scholar] [CrossRef]
- Torchia, M.T.; Austin, D.C.; Kunkel, S.T.; Dwyer, K.W.; Moschetti, W.E. Next-Generation Sequencing vs Culture-Based Methods for Diagnosing Periprosthetic Joint Infection After Total Knee Arthroplasty: A Cost-Effectiveness Analysis. J. Arthroplast. 2019, 34, 1333–1341. [Google Scholar] [CrossRef]
- Adelantado Lacasa, M.; Portillo, M.E.; Lobo Palanco, J.; Chamorro, J.; Ezpeleta Baquedano, C. Molecular Epidemiology of Multidrug-Resistant Pseudomonas aeruginosa Acquired in a Spanish Intensive Care Unit: Using Diverse Typing Methods to Identify Clonal Types. Microorganisms 2022, 10, 1791. [Google Scholar] [CrossRef]
- Cazanave, C.; Greenwood-Quaintance, K.E.; Hanssen, A.D.; Karau, M.J.; Schmidt, S.M.; Gomez Urena, E.O.; Mandrekar, J.N.; Osmon, D.R.; Lough, L.E.; Pritt, B.S.; et al. Rapid molecular microbiologic diagnosis of prosthetic joint infection. J. Clin. Microbiol. 2013, 51, 2280–2287. [Google Scholar] [CrossRef] [PubMed]
- Jacovides, C.L.; Kreft, R.; Adeli, B.; Hozack, B.; Ehrlich, G.D.; Parvizi, J. Successful identification of pathogens by polymerase chain reaction (PCR)-based electron spray ionization time-of-flight mass spectrometry (ESI-TOF-MS) in culture-negative periprosthetic joint infection. J. Bone Jt. Surg. 2012, 94, 2247–2254. [Google Scholar] [CrossRef]
- Natoli, R.M.; Marinos, D.P.; Montalvo, R.N.; Degani, Y.; Ochenjele, G.; Griffith, C.; Ding, A.; Gitajn, I.L.; Manson, T.T.; Johnson, A.J.; et al. Poor Agreement Between Next-Generation DNA Sequencing and Bacterial Cultures in Orthopaedic Trauma Procedures. J. Bone Joint Surg. Am. 2022, 104, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Namdari, S.; Nicholson, T.; Abboud, J.; Lazarus, M.; Ramsey, M.L.; Williams, G.; Parvizi, J. Comparative study of cultures and next-generation sequencing in the diagnosis of shoulder prosthetic joint infections. J. Shoulder Elb. Surg. 2019, 28, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Goswami, K.; Clarkson, S.; Phillips, C.D.; Dennis, D.A.; Klatt, B.A.; O’Malley, M.J.; Smith, E.L.; Gililland, J.M.; Pelt, C.E.; Peters, C.L.; et al. An Enhanced Understanding of Culture-Negative Periprosthetic Joint Infection with Next-Generation Sequencing: A Multicenter Study. J. Bone Joint Surg. Am. 2022, 104, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Kullar, R.; Chisari, E.; Snyder, J.; Cooper, C.; Parvizi, J.; Sniffen, J. Next-Generation Sequencing Supports Targeted Antibiotic Treatment for Culture Negative Orthopedic Infections. Clin. Infect. Dis. 2022, 76, 359–364. [Google Scholar] [CrossRef]
- Pham, T.T.; Lazarevic, V.; Gaia, N.; Girard, M.; Cherkaoui, A.; Suva, D.; Schrenzel, J. Second Periprosthetic Joint Infection Caused by Streptococcus dysgalactiae: How Genomic Sequencing Can Help Defining the Best Therapeutic Strategy. Front. Med. 2020, 7, 53. [Google Scholar] [CrossRef] [PubMed]
| CFU/mL * | Enrichment Broth | Clinical Significance |
|---|---|---|
| ≥50 | Positive | Yes |
| <50 | Positive | No, except for anaerobes, patients receiving antimicrobials or acute PJI |
| 0 | Positive | No, except for anaerobes or patients receiving antimicrobials |
| PCR | General Description | References |
|---|---|---|
| Broad-range PCR | Detection of genes universally present in microorganisms (requires a subsequent sequencing step for identification) | [68,69,70] |
| Targeted PCR | Specific detection of a particular microorganism and/or resistance mechanism | [71,72] |
| Multiplex PCR | Simultaneous detection of several microorganisms and/or resistance mechanisms by adding primers of interest | [61,73,74] |
| Technology | Definitions |
|---|---|
| Next Generation Sequencing (NGS) | High-throughput, massively parallel sequencing of DNA fragments performed independently and simultaneously. |
| Whole Genome Sequencing (WGS) | Method for analyzing the entire microbial genome. |
| Targeted NGS directly from specimen | Focuses only on specific regions of interest in the genome. It requires a pre-sequencing DNA preparation step called target enrichment, where target DNA sequences are either amplified or captured and then sequenced. |
| Shotgun metagenomic NGS directly from specimen | All nucleic acids detected directly from patient specimens are sequenced. The method enables evaluation of bacterial diversity and detection of the abundance of microorganisms. |
| Reference | Method | Nr of PJIs | Type of Sample | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Kildow et al, 2021 [110] | tNGS | 116 | Synovial fluid and/or swabs | 60.9% | 89.9% |
| Flurin et al, 2021 [111] | tNGS | 47 | Sonication fluid | 85% | 98% |
| Cai et al, 2020 [112] | sNGS | 44 | Periprosthetic tissues | 95.45% | 90.91% |
| Yin et al, 2020 [113] | sNGS | 15 | Synovial fluid | 93.3% | 90% |
| Huang et al, 2020 [76] | sNGS | 49 | Synovial fluid | 95.9% | 95.2% |
| Wang et al, 2019 [114] | sNGS | 97 | Sonication fluid | 94% | 95% |
| Ivy et al, 2018 [115] | sNGS | 107 | Synovial fluid | 84% | 100% |
| Tarabichi et al, 2018 [116] | tNGS | 28 | Periprosthetic tissues | 89.3% | 73% |
| Thoendel et al, 2018 [90] | sNGS | 213 | Sonication fluid | 74.2% | 93% |
| Steet et al, 2017 [89] | sNGS | 97 | Sonication fluid | 88% | 88% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Portillo, M.E.; Sancho, I. Advances in the Microbiological Diagnosis of Prosthetic Joint Infections. Diagnostics 2023, 13, 809. https://doi.org/10.3390/diagnostics13040809
Portillo ME, Sancho I. Advances in the Microbiological Diagnosis of Prosthetic Joint Infections. Diagnostics. 2023; 13(4):809. https://doi.org/10.3390/diagnostics13040809
Chicago/Turabian StylePortillo, Maria Eugenia, and Ignacio Sancho. 2023. "Advances in the Microbiological Diagnosis of Prosthetic Joint Infections" Diagnostics 13, no. 4: 809. https://doi.org/10.3390/diagnostics13040809
APA StylePortillo, M. E., & Sancho, I. (2023). Advances in the Microbiological Diagnosis of Prosthetic Joint Infections. Diagnostics, 13(4), 809. https://doi.org/10.3390/diagnostics13040809