Molecular Diagnosis and Cancer Prognosis—A Concise Review
Abstract
1. Introduction
Rapid Point-of-Care Detection
2. Biomarkers
Different Types of Cancer Biomarkers
3. Protein Biomarkers
3.1. Electrochemical Detection
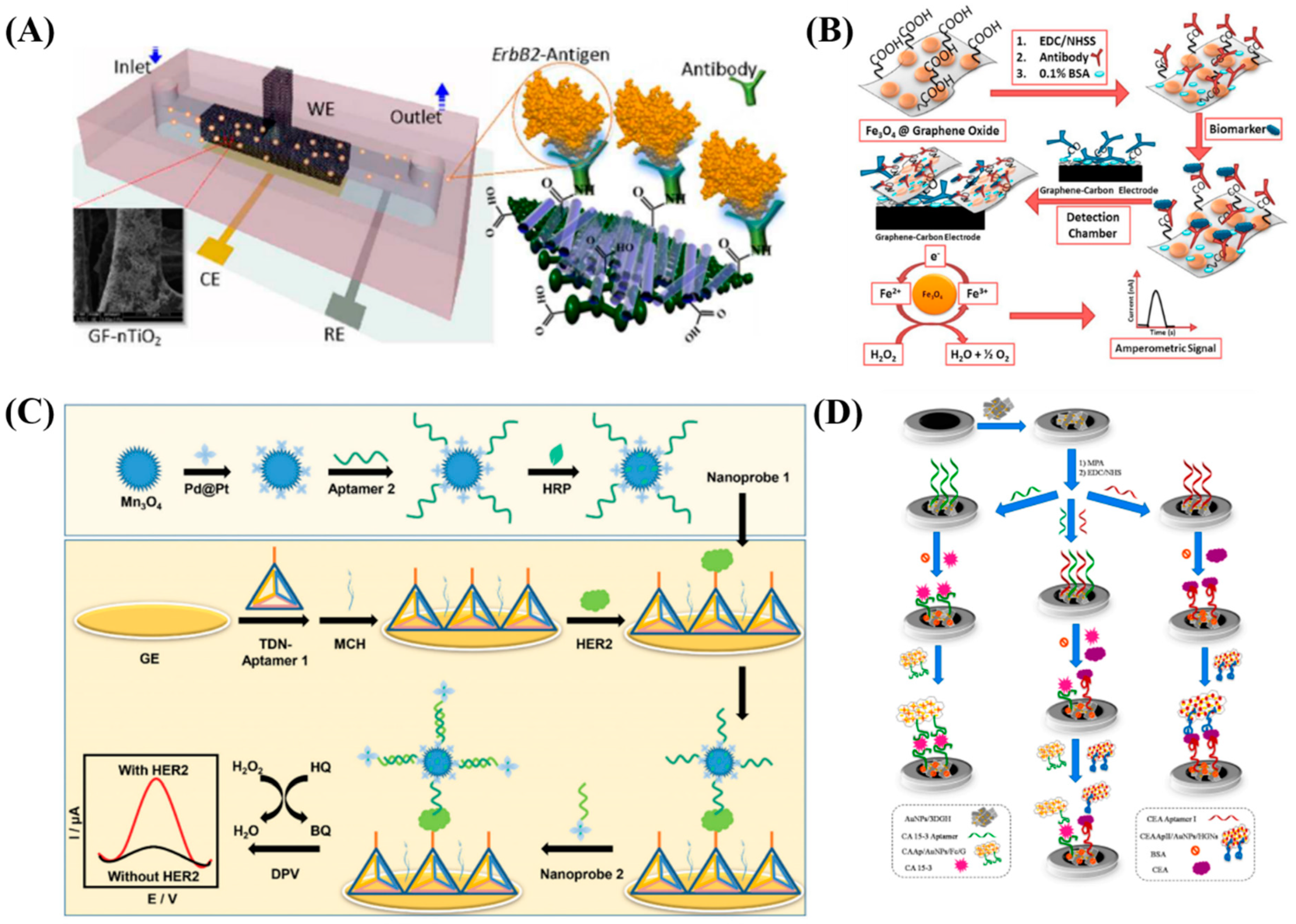
3.2. Optical Based Detection
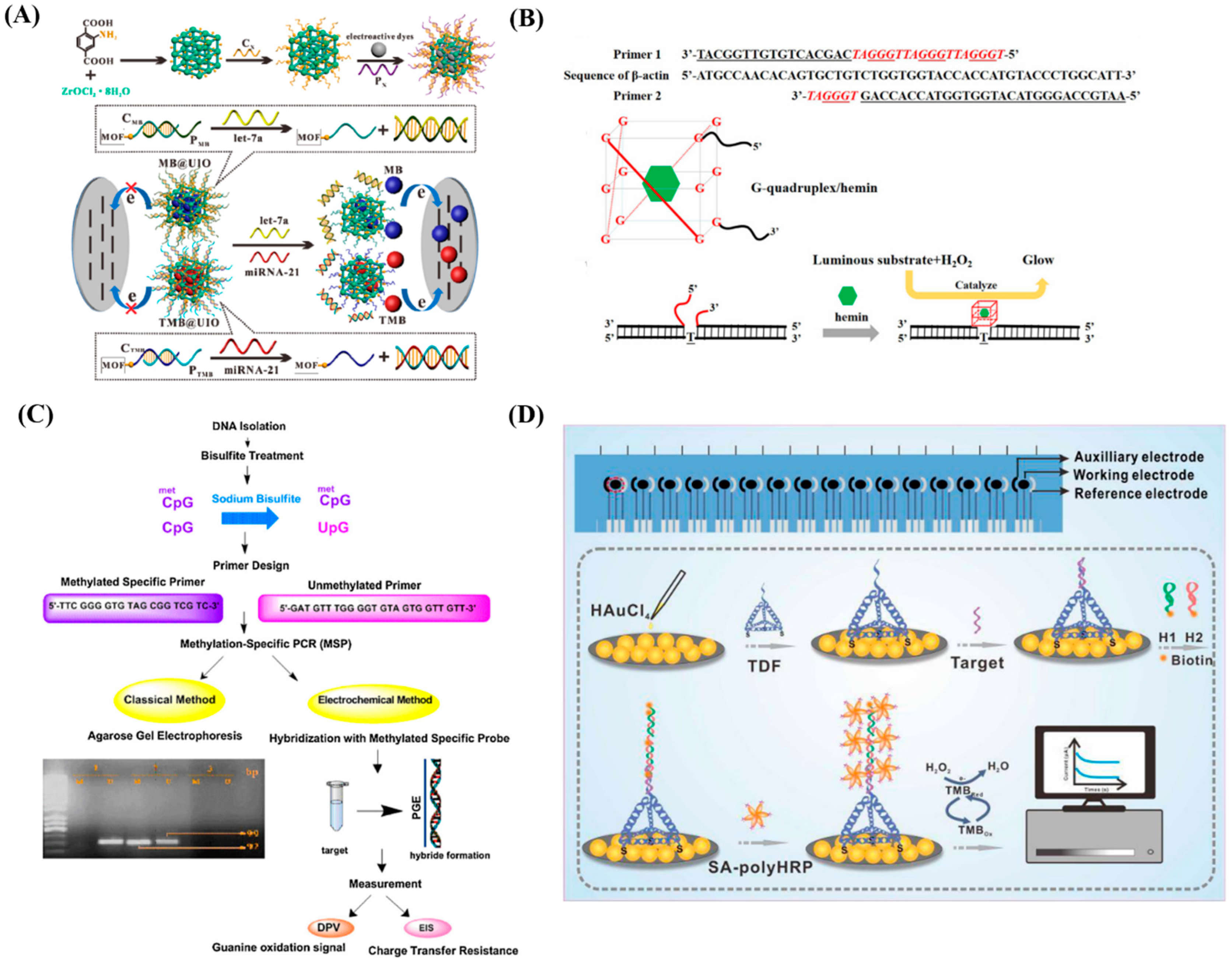
4. Nucleic Acid Based Biomarkers
4.1. DNA Based Nucleic Acid Biomarkers
4.2. RNA Based Nucleic Acid Biomarkers
4.3. CRISPR
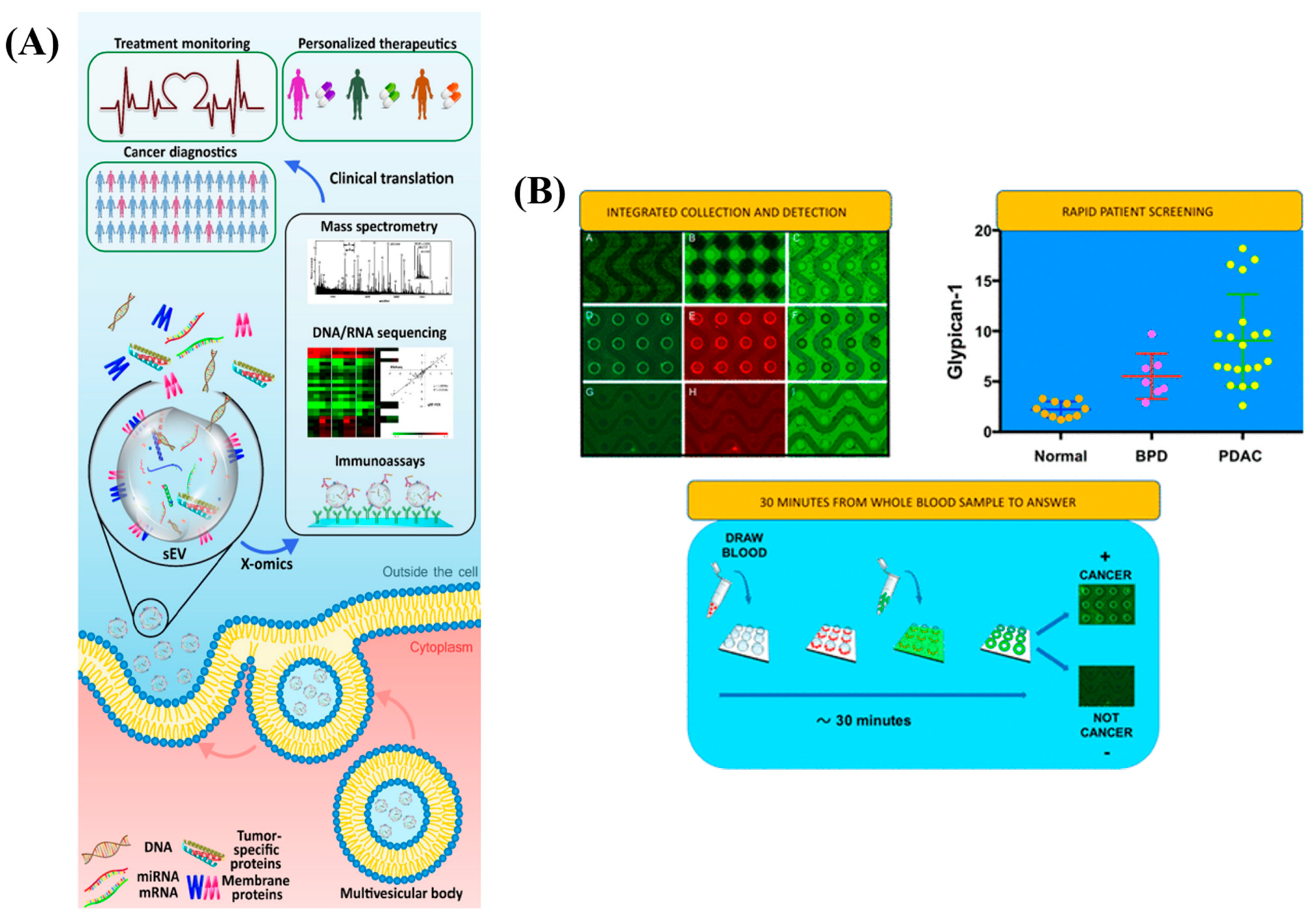
5. Extracellular Vesicles (EV) Based Biomarkers
5.1. Microfluidics, Microarrays and FET Based EVs
5.2. Alternating Current Electrokinetics (ACE)
5.3. Plasmon-Enhanced Fluorescence Detection
5.4. Multiplexed and Other Platforms
5.5. EV in Immune System
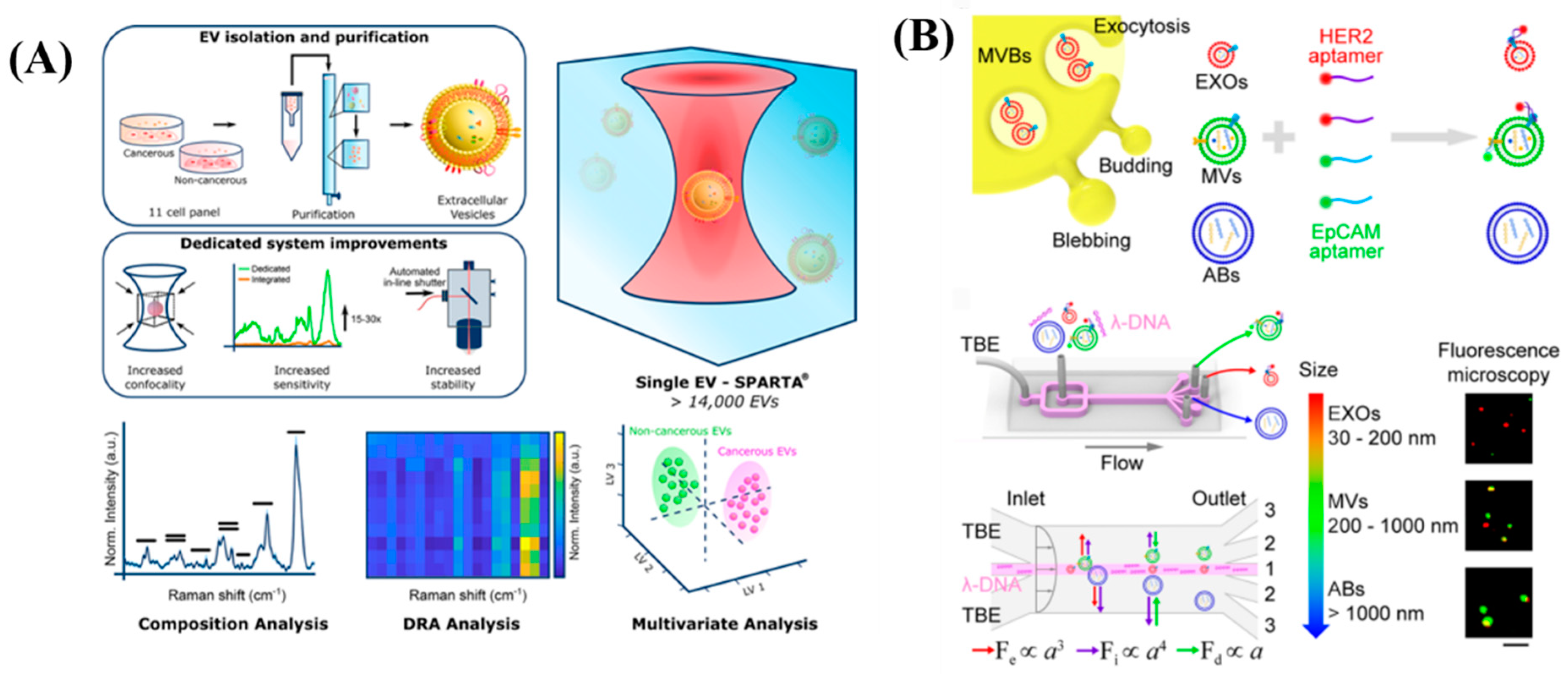
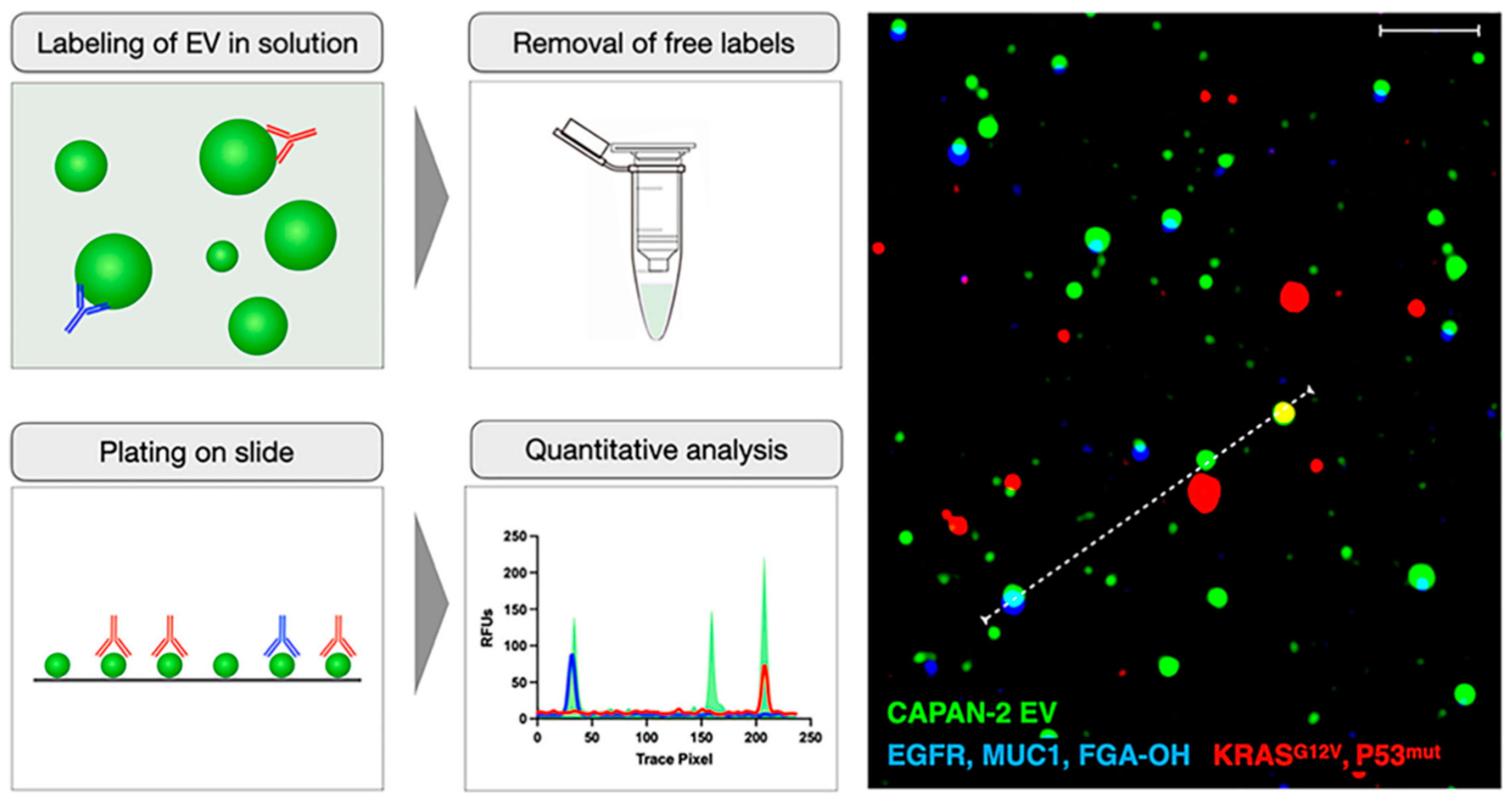
5.6. Single Extracellular Vesicle
6. Conclusions with Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Manuscript Number
References
- Tothill, I.E. Biosensors for Cancer Markers Diagnosis. Semin. Cell Dev. Biol. 2009, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Bohunicky, B.; Mousa, S.A. Biosensors: The New Wave in Cancer Diagnosis. Nanotechnol. Sci. Appl. 2011, 4, 1–10. [Google Scholar] [CrossRef]
- Hasan, M.R.; Ahommed, M.S.; Daizy, M.; Bacchu, M.S.; Ali, M.R.; Al-Mamun, M.R.; Saad Aly, M.A.; Khan, M.Z.H.; Hossain, S.I. Recent Development in Electrochemical Biosensors for Cancer Biomarkers Detection. Biosens. Bioelectron. X 2021, 8, 100075. [Google Scholar] [CrossRef]
- Xu, W.; Wang, D.; Li, D.; Liu, C.C. Recent Developments of Electrochemical and Optical Biosensors for Antibody Detection. Int. J. Mol. Sci. 2020, 21, 134. [Google Scholar] [CrossRef]
- Law, W.C.; Yong, K.T.; Baev, A.; Prasad, P.N. Sensitivity Improved Surface Plasmon Resonance Biosensor for Cancer Biomarker Detection Based on Plasmonic Enhancement. ACS Nano 2011, 5, 4858–4864. [Google Scholar] [CrossRef]
- Garcia-Cordero, J.L.; Maerkl, S.J. Microfluidic Systems for Cancer Diagnostics. Curr. Opin. Biotechnol. 2020, 65, 37–44. [Google Scholar] [CrossRef]
- Prendergast, G.C.; Metz, R.; Muller, A.J. Towards a Genetic Definition of Cancer-Associated Inflammation: Role of the IDO Pathway. Am. J. Pathol. 2010, 176, 2082–2087. [Google Scholar] [CrossRef]
- Henry, N.L.; Hayes, D.F. Cancer Biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef]
- Badr, H.A.; Alsadek, D.M.M.; Darwish, A.A.; Elsayed, A.I.; Bekmanov, B.O.; Khussainova, E.M.; Zhang, X.; Cho, W.C.S.; Djansugurova, L.B.; Li, C.Z. Lectin Approaches for Glycoproteomics in FDA-Approved Cancer Biomarkers. Expert Rev. Proteom. 2014, 11, 227–236. [Google Scholar] [CrossRef]
- Thenrajan, T.; Wilson, J. Biosensors for Cancer Theranostics. Biosens. Bioelectron. X 2022, 12, 100232. [Google Scholar] [CrossRef]
- Borrebaeck, C.A.K. Precision Diagnostics: Moving towards Protein Biomarker Signatures of Clinical Utility in Cancer. Nat. Rev. Cancer 2017, 17, 199–204. [Google Scholar] [CrossRef]
- Ranjan, P.; Parihar, A.; Jain, S.; Kumar, N.; Dhand, C.; Murali, S.; Mishra, D.; Sanghi, S.K.; Chaurasia, J.P.; Srivastava, A.K.; et al. Biosensor-Based Diagnostic Approaches for Various Cellular Biomarkers of Breast Cancer: A Comprehensive Review. Anal. Biochem. 2020, 610, 113996. [Google Scholar] [CrossRef] [PubMed]
- Bertok, T.; Lorencova, L.; Chocholova, E.; Jane, E.; Vikartovska, A.; Kasak, P.; Tkac, J. Electrochemical Impedance Spectroscopy Based Biosensors: Mechanistic Principles, Analytical Examples and Challenges towards Commercialization for Assays of Protein Cancer Biomarkers. ChemElectroChem 2019, 6, 989–1003. [Google Scholar] [CrossRef]
- Magar, H.S.; Hassan, R.Y.A.; Mulchandani, A. Electrochemical Impedance Spectroscopy (EIS): Principles, Construction, and Biosensing Applications. Sensors 2021, 21, 6578. [Google Scholar] [CrossRef]
- Park, J.-Y.; Park, S.-M. DNA Hybridization Sensors Based on Electrochemical Impedance Spectroscopy as a Detection Tool. Sensors 2009, 9, 9513–9532. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Mondal, K.; Jiao, Y.; Oren, S.; Xu, Z.; Sharma, A.; Dong, L. Microfluidic Immuno-Biochip for Detection of Breast Cancer Biomarkers Using Hierarchical Composite of Porous Graphene and Titanium Dioxide Nanofibers. ACS Appl. Mater. Interfaces 2016, 8, 20570–20582. [Google Scholar] [CrossRef]
- Sharafeldin, M.; Bishop, G.W.; Bhakta, S.; El-Sawy, A.; Suib, S.L.; Rusling, J.F. Fe3O4 Nanoparticles on Graphene Oxide Sheets for Isolation and Ultrasensitive Amperometric Detection of Cancer Biomarker Proteins. Biosens. Bioelectron. 2017, 91, 359–366. [Google Scholar] [CrossRef]
- Ou, D.; Sun, D.; Lin, X.; Liang, Z.; Zhong, Y.; Chen, Z. A Dual-Aptamer-Based Biosensor for Specific Detection of Breast Cancer Biomarker HER2 via Flower-like Nanozymes and DNA Nanostructures. J. Mater. Chem. B 2019, 7, 3661–3669. [Google Scholar] [CrossRef]
- Abrao Nemeir, I.; Saab, J.; Hleihel, W.; Errachid, A.; Jafferzic-Renault, N.; Zine, N. The Advent of Salivary Breast Cancer Biomarker Detection Using Affinity Sensors. Sensors 2019, 19, 2373. [Google Scholar] [CrossRef]
- Shekari, Z.; Zare, H.R.; Falahati, A. Dual Assaying of Breast Cancer Biomarkers by Using a Sandwich–Type Electrochemical Aptasensor Based on a Gold Nanoparticles–3D Graphene Hydrogel Nanocomposite and Redox Probes Labeled Aptamers. Sens. Actuators B Chem. 2021, 332, 129515. [Google Scholar] [CrossRef]
- Chen, X.; Jia, X.; Han, J.; Ma, J.; Ma, Z. Electrochemical Immunosensor for Simultaneous Detection of Multiplex Cancer Biomarkers Based on Graphene Nanocomposites. Biosens. Bioelectron. 2013, 50, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Kailemia, M.J.; Park, D.; Lebrilla, C.B. Glycans and Glycoproteins as Specific Biomarkers for Cancer. Anal. Bioanal. Chem. 2017, 409, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.W.; Pui, T.-S. Cytokine and Cancer Biomarkers Detection: The Dawn of Electrochemical Paper-Based Biosensor. Sensors 2020, 20, 1854. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Li, S.; Turner, A.P.F. Electrochemical Immunosensor with N-Doped Graphene-Modified Electrode for Label-Free Detection of the Breast Cancer Biomarker CA 15-3. Biosens. Bioelectron. 2013, 43, 25–29. [Google Scholar] [CrossRef]
- Gohring, J.T.; Dale, P.S.; Fan, X. Detection of HER2 Breast Cancer Biomarker Using the Opto-Fluidic Ring Resonator Biosensor. Sens. Actuators B Chem. 2010, 146, 226–230. [Google Scholar] [CrossRef]
- Retolaza, A.; Martinez-Perdiguero, J.; Merino, S.; Morales-Vidal, M.; Boj, P.G.; Quintana, J.A.; Villalvilla, J.M.; Díaz-García, M.A. Organic Distributed Feedback Laser for Label-Free Biosensing of ErbB2 Protein Biomarker. Sens. Actuators B Chem. 2016, 223, 261–265. [Google Scholar] [CrossRef]
- Gao, X.; Niu, S.; Ge, J.; Luan, Q.; Jie, G. 3D DNA Nanosphere-Based Photoelectrochemical Biosensor Combined with Multiple Enzyme-Free Amplification for Ultrasensitive Detection of Cancer Biomarkers. Biosens. Bioelectron. 2020, 147, 111778. [Google Scholar] [CrossRef]
- Cui, M.; Wang, Y.; Wang, H.; Wu, Y.; Luo, X. A Label-Free Electrochemical DNA Biosensor for Breast Cancer Marker BRCA1 Based on Self-Assembled Antifouling Peptide Monolayer. Sens. Actuators B Chem. 2017, 244, 742–749. [Google Scholar] [CrossRef]
- Cheng, N.; Fu, J. An Approach to the Simultaneous Detection of Multiple Biomarkers for the Early Diagnosis of Liver Cancer Using Quantum Dot Nanoprobes. Infect. Microbes Dis. 2022, 4, 34–40. [Google Scholar] [CrossRef]
- Loyez, M.; Lobry, M.; Hassan, E.M.; DeRosa, M.C.; Caucheteur, C.; Wattiez, R. HER2 Breast Cancer Biomarker Detection Using a Sandwich Optical Fiber Assay. Talanta 2021, 221, 121452. [Google Scholar] [CrossRef]
- Mahmudunnabi, R.G.; Farhana, F.Z.; Kashaninejad, N.; Firoz, S.H.; Shim, Y.B.; Shiddiky, M.J.A. Nanozyme-Based Electrochemical Biosensors for Disease Biomarker Detection. Analyst 2020, 145, 4398–4420. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, A.; Wang, R.; Zhang, Q.; Cui, D. A Review on Metal- and Metal Oxide-Based Nanozymes: Properties, Mechanisms, and Applications; Springer: Singapore, 2021; Volume 13, ISBN 0123456789. [Google Scholar]
- Hosseinkazemi, H.; Samani, S.; O’Neill, A.; Soezi, M.; Moghoofei, M.; Azhdari, M.H.; Aavani, F.; Nazbar, A.; Keshel, S.H.; Doroudian, M. Applications of Iron Oxide Nanoparticles against Breast Cancer. J. Nanomater. 2022, 2022, 1–12. [Google Scholar] [CrossRef]
- Kermani, F.; Vojdani-Saghir, A.; Mollazadeh Beidokhti, S.; Nazarnezhad, S.; Mollaei, Z.; Hamzehlou, S.; El-Fiqi, A.; Baino, F.; Kargozar, S. Iron (Fe)-Doped Mesoporous 45S5 Bioactive Glasses: Implications for Cancer Therapy. Transl. Oncol. 2022, 20, 101397. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Truong, P.L.; Lee, D.; Ko, S.H. Metal-Oxide Nanomaterials Synthesis and Applications in Flexible and Wearable Sensors. ACS Nanosci. Au 2022, 2, 64–92. [Google Scholar] [CrossRef]
- Leung, F.; Kulasingam, V.; Diamandis, E.P.; Hoon, D.S.B.; Kinzler, K.; Pantel, K.; Alix-Panabières, C. Circulating Tumor DNA as a Cancer Biomarker: Fact or Fiction? Clin. Chem. 2016, 62, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H. Circulating Nucleic Acids as Biomarkers in Breast Cancer. Breast Cancer Res. 2013, 15, 211. [Google Scholar] [CrossRef]
- Chang, J.; Wang, X.; Wang, J.; Li, H.; Li, F. Nucleic Acid-Functionalized Metal-Organic Framework-Based Homogeneous Electrochemical Biosensor for Simultaneous Detection of Multiple Tumor Biomarkers. Anal. Chem. 2019, 91, 3604–3610. [Google Scholar] [CrossRef]
- Hao, T.B.; Shi, W.; Shen, X.J.; Qi, J.; Wu, X.H.; Wu, Y.; Tang, Y.Y.; Ju, S.Q. Circulating Cell-Free DNA in Serum as a Biomarker for Diagnosis and Prognostic Prediction of Colorectal Cancer. Br. J. Cancer 2014, 111, 1482–1489. [Google Scholar] [CrossRef]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and Characterization of Lung Cancer Using Cell-Free DNA Fragmentomes. Nat. Commun. 2021, 12, 5060. [Google Scholar] [CrossRef]
- Ying, H.; Fengying, S.; Feng, H.; Yanhong, W.; Xianru, X.; Xiaolei, T. Diagnostic Value of Quantification of Circulating Free DNA for Gall Bladder Cancer Using a Chemiluminescence DNA Biosensor System Based on DNA G-Quadruplex/Hemin Enzyme. Transl. Oncol. 2021, 14, 100928. [Google Scholar] [CrossRef]
- Ponti, G.; Maccaferri, M.; Mandrioli, M.; Manfredini, M.; Micali, S.; Cotugno, M.; Bianchi, G.; Ozben, T.; Pellacani, G.; Del Prete, C.; et al. Seminal Cell-Free DNA Assessment as a Novel Prostate Cancer Biomarker. Pathol. Oncol. Res. 2018, 24, 941–945. [Google Scholar] [CrossRef]
- Ruiyi, L.; Ling, L.; Hongxia, B.; Zaijun, L. Nitrogen-Doped Multiple Graphene Aerogel/Gold Nanostar as the Electrochemical Sensing Platform for Ultrasensitive Detection of Circulating Free DNA in Human Serum. Biosens. Bioelectron. 2016, 79, 457–466. [Google Scholar] [CrossRef]
- Topkaya, S.N.; Ozkan-Ariksoysal, D.; Kosova, B.; Ozel, R.; Ozsoz, M. Electrochemical DNA Biosensor for Detecting Cancer Biomarker Related to Glutathione S-Transferase P1 (GSTP1) Hypermethylation in Real Samples. Biosens. Bioelectron. 2012, 31, 516–522. [Google Scholar] [CrossRef]
- Wang, C.; Wang, W.; Xu, Y.; Zhao, X.; Li, S.; Qian, Q.; Mi, X. Tetrahedral DNA Framework-Programmed Electrochemical Biosenors with Gold Nanoparticles for Ultrasensitive Cell-Free DNA Detection. Nanomaterials 2022, 12, 666. [Google Scholar] [CrossRef]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef]
- Xi, X.; Li, T.; Huang, Y.; Sun, J.; Zhu, Y.; Yang, Y.; Lu, Z.J. RNA Biomarkers: Frontier of Precision Medicine for Cancer. Non-Coding RNA 2017, 3, 9. [Google Scholar] [CrossRef]
- Chen, Y.; Nakamoto, K.; Niwa, O.; Corn, R.M. On-Chip Synthesis of RNA Aptamer Microarrays for Multiplexed Protein Biosensing with SPR Imaging Measurements. Langmuir 2012, 28, 8281–8285. [Google Scholar] [CrossRef]
- Chen, M.; Wu, D.; Tu, S.; Yang, C.; Chen, D.J.; Xu, Y. A Novel Biosensor for the Ultrasensitive Detection of the LncRNA Biomarker MALAT1 in Non-Small Cell Lung Cancer. Sci. Rep. 2021, 11, 3666. [Google Scholar] [CrossRef]
- Huertas, C.S.; Fariña, D.; Lechuga, L.M. Direct and Label-Free Quantification of Micro-RNA-181a at Attomolar Level in Complex Media Using a Nanophotonic Biosensor. ACS Sens. 2016, 1, 748–756. [Google Scholar] [CrossRef]
- Nabok, A.; Abu-Ali, H.; Takita, S.; Smith, D.P. Electrochemical Detection of Prostate Cancer Biomarker Pca3 Using Specific Rna-Based Aptamer Labelled with Ferrocene. Chemosensors 2021, 9, 59. [Google Scholar] [CrossRef]
- Mello, H.J.N.P.D.; Bachour Junior, B.; Mulato, M. Polyaniline-Based Field Effect Transistor for DNA/RNA Biomarker Sensing: Comparison to Electrochemical Impedance and Inorganic Layer. Sens. Actuators A Phys. 2021, 318, 112481. [Google Scholar] [CrossRef]
- Gutiérrez-Gálvez, L.; García-Mendiola, T.; Gutiérrez-Sánchez, C.; Guerrero-Esteban, T.; García-Diego, C.; Buendía, I.; García-Bermejo, M.L.; Pariente, F.; Lorenzo, E. Carbon Nanodot–Based Electrogenerated Chemiluminescence Biosensor for MiRNA-21 Detection. Microchim. Acta 2021, 188, 398. [Google Scholar] [CrossRef]
- Aziz, N.B.; Mahmudunnabi, R.G.; Umer, M.; Sharma, S.; Rashid, M.A.; Alhamhoom, Y.; Shim, Y.-B.; Salomon, C.; Shiddiky, M.J.A. MicroRNAs in Ovarian Cancer and Recent Advances in the Development of MicroRNA-Based Biosensors. Analyst 2020, 145, 2038–2057. [Google Scholar] [CrossRef]
- D’Agata, R.; Giuffrida, M.; Spoto, G. Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis. Molecules 2017, 22, 1951. [Google Scholar] [CrossRef]
- Amri, C.; Shukla, A.K.; Lee, J.-H. Recent Advancements in Nanoparticle-Based Optical Biosensors for Circulating Cancer Biomarkers. Materials 2021, 14, 1339. [Google Scholar] [CrossRef]
- Huang, C.H.; Lee, K.C.; Doudna, J.A. Applications of CRISPR-Cas Enzymes in Cancer Therapeutics and Detection. Trends Cancer 2018, 4, 499–512. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Mubthasima, P.P.; Pande, K.; Prakash, R.; Kannan, A. CRISPR Cas/Exosome Based Diagnostics: Future of Early Cancer Detection. In Rural Health; IntechOpen: London, UK, 2022; pp. 225–240. [Google Scholar]
- Chen, K.; Shen, Z.; Wang, G.; Gu, W.; Zhao, S.; Lin, Z.; Liu, W.; Cai, Y.; Mushtaq, G.; Jia, J.; et al. Research Progress of CRISPR-Based Biosensors and Bioassays for Molecular Diagnosis. Front. Bioeng. Biotechnol. 2022, 10, 986233. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Wang, X.; Wang, W. Recent Advances in CRISPR/Cas-Based Biosensors for Protein Detection. Bioengineering 2022, 9, 512. [Google Scholar] [CrossRef]
- Zhang, H.; Qin, C.; An, C.; Zheng, X.; Wen, S.; Chen, W.; Liu, X.; Lv, Z.; Yang, P.; Xu, W.; et al. Application of the CRISPR/Cas9-Based Gene Editing Technique in Basic Research, Diagnosis, and Therapy of Cancer. Mol. Cancer 2021, 20, 126. [Google Scholar] [CrossRef]
- Choi, J.H.; Lim, J.; Shin, M.; Paek, S.H.; Choi, J.W. CRISPR-Cas12a-Based Nucleic Acid Amplification-Free DNA Biosensor via Au Nanoparticle-Assisted Metal-Enhanced Fluorescence and Colorimetric Analysis. Nano Lett. 2021, 21, 693–699. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, X.; Chen, X.; Qiu, X.; Qing, G.; Zhang, H.; Zhang, L.; Hu, X.; He, Z.; Zhong, D.; et al. Rolling Circular Amplification (RCA)-Assisted CRISPR/Cas9 Cleavage (RACE) for Highly Specific Detection of Multiple Extracellular Vesicle MicroRNAs. Anal. Chem. 2020, 92, 2176–2185. [Google Scholar] [CrossRef]
- Chen, M.; Wu, D.; Tu, S.; Yang, C.; Chen, D.J.; Xu, Y. CRISPR/Cas9 Cleavage Triggered ESDR for Circulating Tumor DNA Detection Based on a 3D Graphene/AuPtPd Nanoflower Biosensor. Biosens. Bioelectron. 2021, 173, 112821. [Google Scholar] [CrossRef]
- Ganbaatar, U.; Liu, C. NEXT CRISPR: An Enhanced CRISPR-Based Nucleic Acid Biosensing Platform Using Extended CrRNA. Sens. Actuators B Chem. 2022, 369, 132296. [Google Scholar] [CrossRef]
- Hu, T.; Wolfram, J.; Srivastava, S. Extracellular Vesicles in Cancer Detection: Hopes and Hypes. Trends Cancer 2021, 7, 122–133. [Google Scholar] [CrossRef]
- Buzas, E.I. The Roles of Extracellular Vesicles in the Immune System. Nat. Rev. Immunol. 2022, 12. [Google Scholar] [CrossRef]
- Xie, H.; Di, K.; Huang, R.; Khan, A.; Xia, Y.; Xu, H.; Liu, C.; Tan, T.; Tian, X.; Shen, H.; et al. Extracellular Vesicles Based Electrochemical Biosensors for Detection of Cancer Cells: A Review. Chin. Chem. Lett. 2020, 31, 1737–1745. [Google Scholar] [CrossRef]
- Lu, J.; Pang, J.; Chen, Y.; Dong, Q.; Sheng, J.; Luo, Y.; Lu, Y.; Lin, B.; Liu, T. Application of Microfluidic Chips in Separation and Analysis of Extracellular Vesicles in Liquid Biopsy for Cancer. Micromachines 2019, 10, 390. [Google Scholar] [CrossRef]
- Xie, F.; Zhou, X.; Fang, M.; Li, H.; Su, P.; Tu, Y.; Zhang, L.; Zhou, F. Extracellular Vesicles in Cancer Immune Microenvironment and Cancer Immunotherapy. Adv. Sci. 2019, 6, 1901779. [Google Scholar] [CrossRef]
- Lambrecht, J.; Poortmans, P.J.; Verhulst, S.; Reynaert, H.; Mannaerts, I.; van Grunsven, L.A. Circulating ECV-Associated MiRNAs as Potential Clinical Biomarkers in Early Stage HBV and HCV Induced Liver Fibrosis. Front. Pharmacol. 2017, 8, 56. [Google Scholar] [CrossRef]
- Penders, J.; Nagelkerke, A.; Cunnane, E.M.; Pedersen, S.V.; Pence, I.J.; Coombes, R.C.; Stevens, M.M. Single Particle Automated Raman Trapping Analysis of Breast Cancer Cell-Derived Extracellular Vesicles as Cancer Biomarkers. ACS Nano 2021, 15, 18192–18205. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, J.; Tian, F.; Chang, J.; Zhang, W.; Sun, J. I-DNA- A Nd Aptamer-Mediated Sorting and Analysis of Extracellular Vesicles. J. Am. Chem. Soc. 2019, 141, 3817–3821. [Google Scholar] [CrossRef]
- Huang, C.C.; Kuo, Y.H.; Chen, Y.S.; Huang, P.C.; Lee, G. Bin A Miniaturized, DNA-FET Biosensor-Based Microfluidic System for Quantification of Two Breast Cancer Biomarkers. Microfluid. Nanofluid. 2021, 25, 33. [Google Scholar] [CrossRef]
- Zhang, P.; He, M.; Zeng, Y. Ultrasensitive Microfluidic Analysis of Circulating Exosomes Using a Nanostructured Graphene Oxide/Polydopamine Coating. Lab. Chip. 2016, 16, 3033–3042. [Google Scholar] [CrossRef]
- Verel-Yilmaz, Y.; Fernández, J.P.; Schäfer, A.; Nevermann, S.; Cook, L.; Gercke, N.; Helmprobst, F.; Jaworek, C.; Pogge von Strandmann, E.; Pagenstecher, A.; et al. Extracellular Vesicle-Based Detection of Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9, 697939. [Google Scholar] [CrossRef]
- Hinestrosa, J.P.; Kurzrock, R.; Lewis, J.M.; Schork, N.J.; Schroeder, G.; Kamat, A.M.; Lowy, A.M.; Eskander, R.N.; Perrera, O.; Searson, D.; et al. Early-Stage Multi-Cancer Detection Using an Extracellular Vesicle Protein-Based Blood Test. Commun. Med. 2022, 2, 29. [Google Scholar] [CrossRef]
- Lewis, J.M.; Vyas, A.D.; Qiu, Y.; Messer, K.S.; White, R.; Heller, M.J. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano 2018, 12, 3311–3320. [Google Scholar] [CrossRef]
- Gaillard, M.; Thuaire, A.; Nonglaton, G.; Agache, V.; Roupioz, Y.; Raillon, C. Biosensing Extracellular Vesicles: Contribution of Biomolecules in Affinity-Based Methods for Detection and Isolation. Analyst 2020, 145, 1997–2013. [Google Scholar] [CrossRef]
- Min, J.; Son, T.; Hong, J.; Cheah, P.S.; Wegemann, A.; Murlidharan, K.; Weissleder, R.; Lee, H.; Im, H. Plasmon-Enhanced Biosensing for Multiplexed Profiling of Extracellular Vesicles. Adv. Biosyst. 2020, 4, 2000003. [Google Scholar] [CrossRef]
- Jiang, C.; Fu, Y.; Liu, G.; Shu, B.; Davis, J.; Tofaris, G.K. Multiplexed Profiling of Extracellular Vesicles for Biomarker Development; Springer: Singapore, 2022; Volume 14, ISBN 0123456789. [Google Scholar]
- He, N.; Thippabhotla, S.; Zhong, C.; Greenberg, Z.; Xu, L.; Pessetto, Z.; Godwin, A.K.; Zeng, Y.; He, M. Nano Pom-Poms Prepared Exosomes Enable Highly Specific Cancer Biomarker Detection. Commun. Biol. 2022, 5, 660. [Google Scholar] [CrossRef]
- Ji, Y.; Qi, D.; Li, L.; Su, H.; Li, X.; Luo, Y.; Sun, B.; Zhang, F.; Lin, B.; Liu, T.; et al. Multiplexed Profiling of Single-Cell Extracellular Vesicles Secretion. Proc. Natl. Acad. Sci. USA. 2019, 116, 5979–5984. [Google Scholar] [CrossRef]
- Ferguson, S.; Yang, K.S.; Weissleder, R. Single Extracellular Vesicle Analysis for Early Cancer Detection. Trends Mol. Med. 2022, 28, 681–692. [Google Scholar] [CrossRef]





| S. No | Name | Cancer Type |
|---|---|---|
| 1. | AFP | Testicular cancer |
| 2. | ß-hGC | Testicular cancer |
| 3. | CA 19-9 | Pancreatic cancer |
| 4. | CA 125 | Ovarian cancer |
| 5. | CA 15.3 | Breast cancer |
| 6. | CA 27.9 | Breast cancer |
| 7. | CEA | Colorectal cancer |
| 8. | FDP | Bladder cancer |
| 9. | HE4 | Ovarian cancer |
| 10. | PSA | Prostate cancer |
| 11. | TG | Thyroid cancer |
| 12. | EGFR | Colorectal cancer |
| 13. | KIT | Gastrointestinal cancer |
| 14. | ER | Breast cancer |
| 15. | PR | Breast cancer |
| 16. | HER2-neu | Breast cancer |
| 17. | NMP/22 | Bladder cancer |
| 18. | BTA | Bladder cancer |
| 19. | Mw CEA | Bladder cancer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thenrajan, T.; Alwarappan, S.; Wilson, J. Molecular Diagnosis and Cancer Prognosis—A Concise Review. Diagnostics 2023, 13, 766. https://doi.org/10.3390/diagnostics13040766
Thenrajan T, Alwarappan S, Wilson J. Molecular Diagnosis and Cancer Prognosis—A Concise Review. Diagnostics. 2023; 13(4):766. https://doi.org/10.3390/diagnostics13040766
Chicago/Turabian StyleThenrajan, Thatchanamoorthy, Subbiah Alwarappan, and Jeyaraj Wilson. 2023. "Molecular Diagnosis and Cancer Prognosis—A Concise Review" Diagnostics 13, no. 4: 766. https://doi.org/10.3390/diagnostics13040766
APA StyleThenrajan, T., Alwarappan, S., & Wilson, J. (2023). Molecular Diagnosis and Cancer Prognosis—A Concise Review. Diagnostics, 13(4), 766. https://doi.org/10.3390/diagnostics13040766





