Deep-Learning-Based Automatic Segmentation of Parotid Gland on Computed Tomography Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Study Data
2.3. Ground Truth Labeling
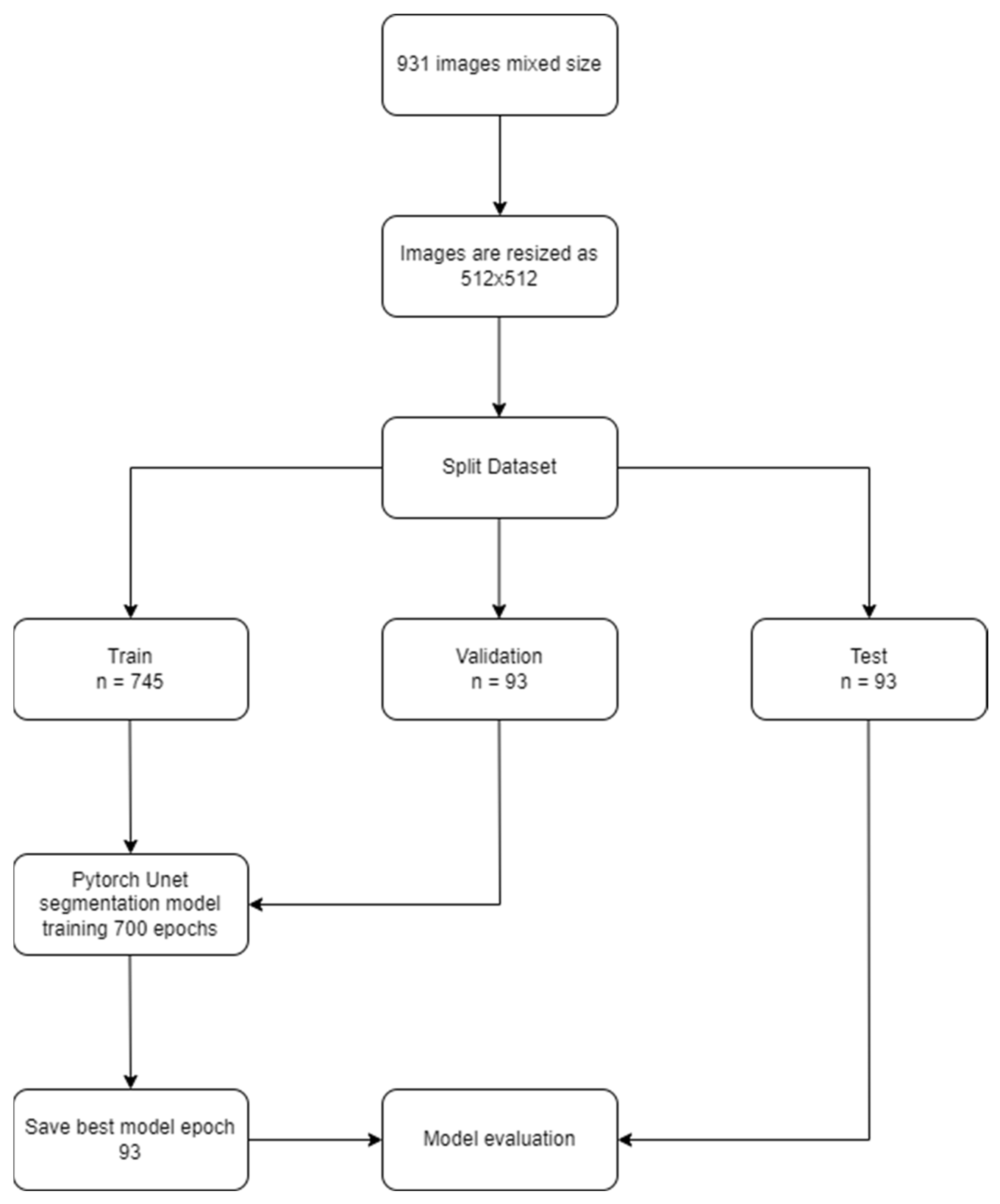
2.4. Data Split
- Training group: 745 (1445 labels);
- Validation group: 93 (178 labels);
- Testing group: 93 (184 labels).
2.5. Development of the U-Net Based dCNN Model
2.6. Statistics for the Model’s Performance
- True positive (TP): At least 50% of the pixels intersect between the automatic segmentation algorithm and the ground truth;
- False positive (FP): At least 50% of the pixels of the automatic segmentation algorithm do not intersect with the ground truth;
- False negative (FN): At least 50% of the pixels of the ground truth do not intersect with the results of the automatic segmentation algorithm;
- Sensitivity (Recall, True positive rate (TPR)) = TP⁄((TP + FN));
- Precision (Positive predictive value (PPV)) = TP⁄((TP + FP));
- F1-Score = 2TP⁄((2TP + FP + FN)).
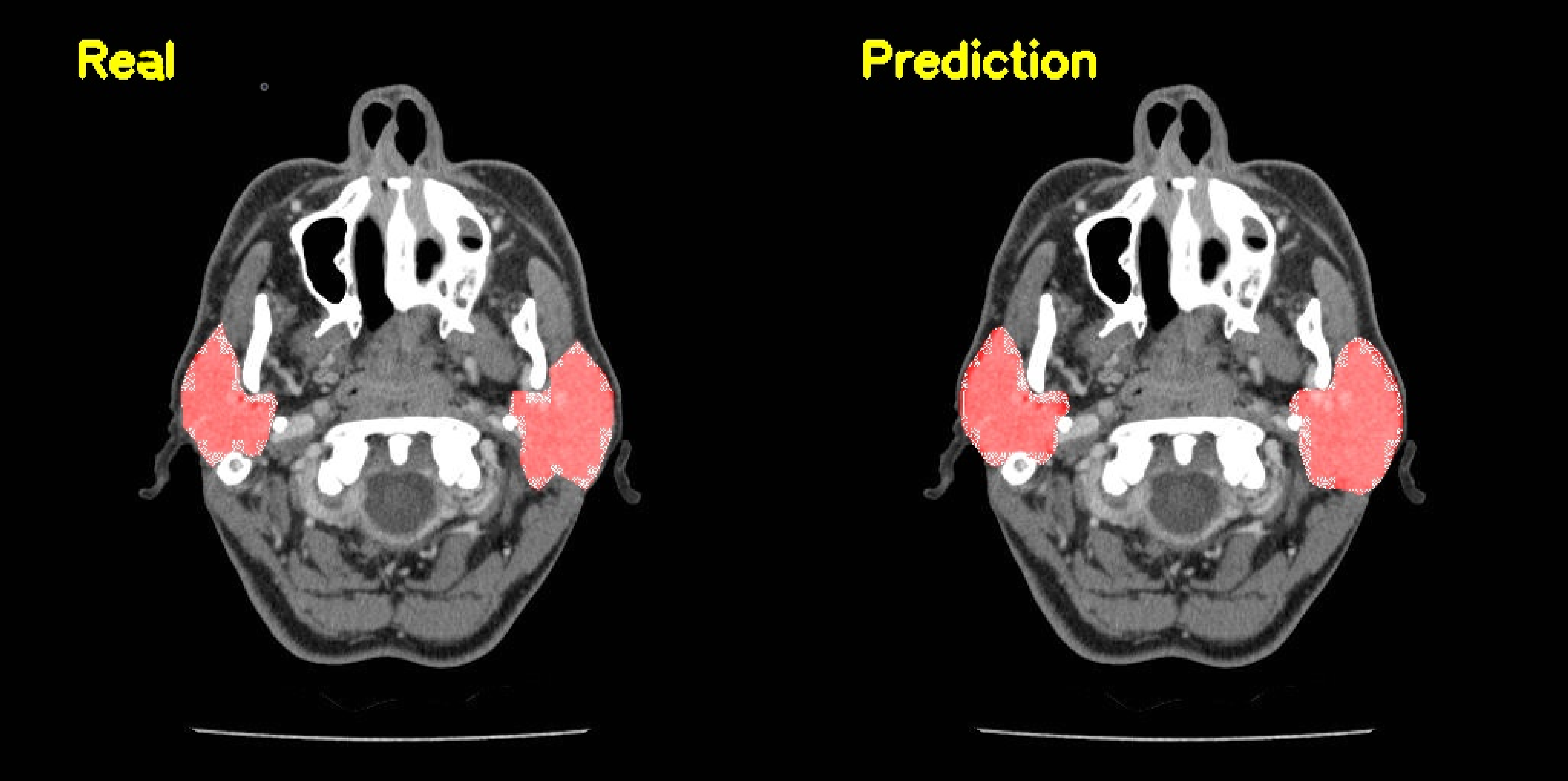
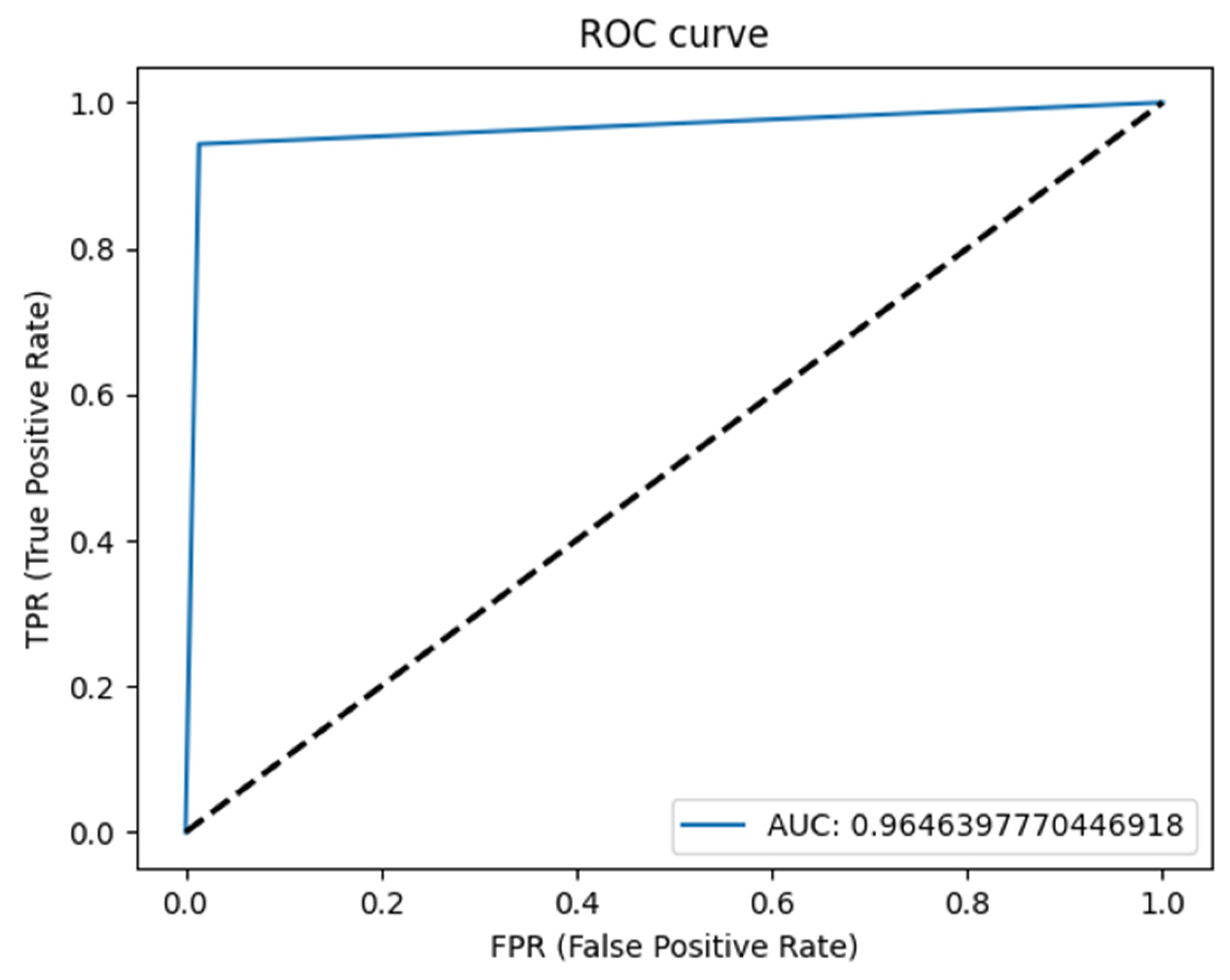
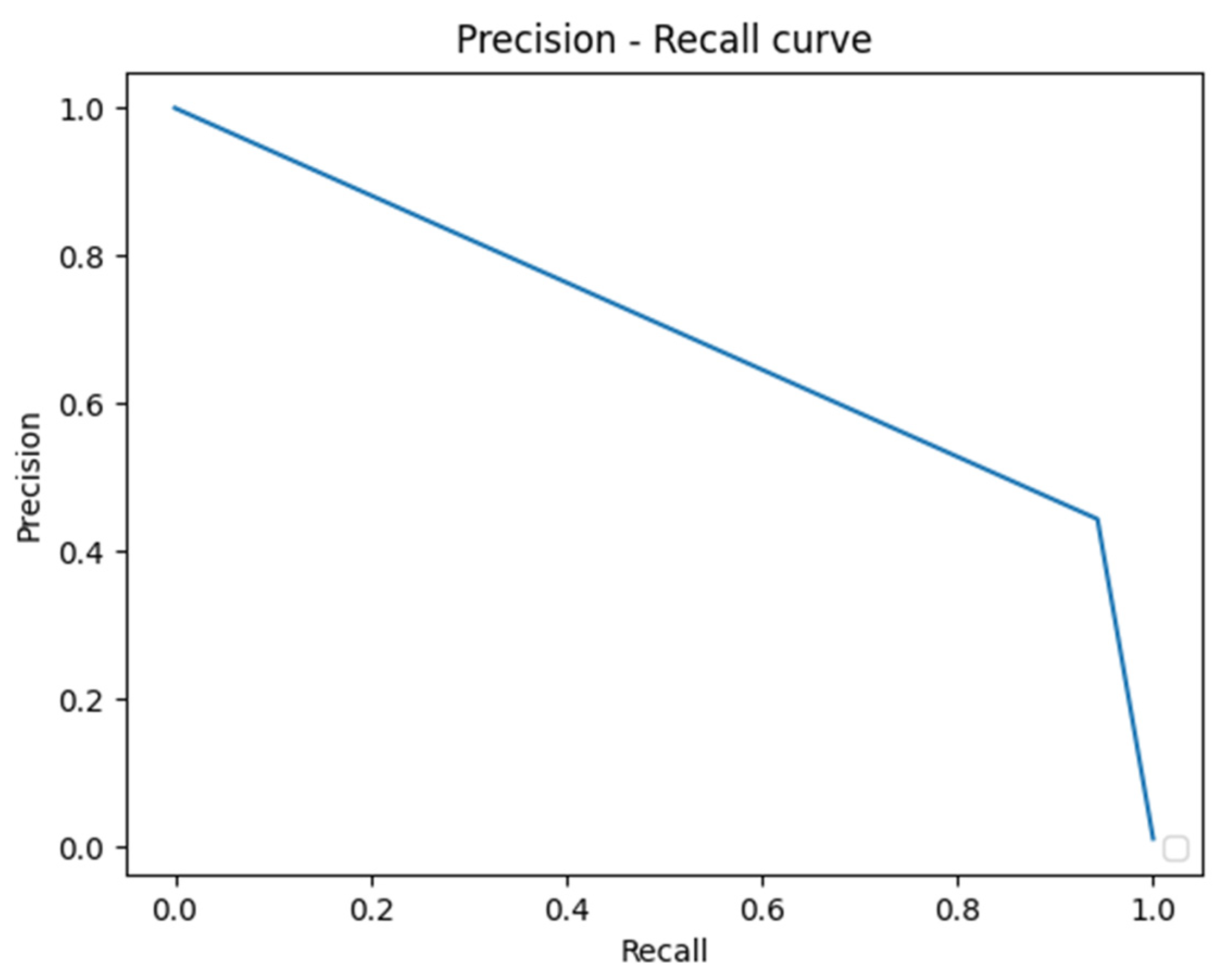
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Stenner, M.; Klussmann, J.P. Current update on established and novel biomarkers in salivary gland carcinoma pathology and the molecular pathways involved. Eur. Arch. Otorhinolaryngol. 2009, 266, 333–341. [Google Scholar] [CrossRef]
- Mortazavi, H.; Baharvand, M.; Movahhedian, A.; Mohammadi, M.; Khodadoustan, A. Xerostomia due to systemic disease: A review of 20 conditions and mechanisms. Ann. Med. Health Sci. Res. 2014, 4, 503–510. [Google Scholar] [CrossRef]
- Dirix, P.; Nuyts, S. Evidence-based organ-sparing radiotherapy in head and neck cancer. Lancet Oncol. 2010, 11, 85–91. [Google Scholar] [CrossRef]
- Lowe, L.H.; Stokes, L.S.; Johnson, J.E.; Heller, R.M.; Royal, S.A.; Wushensky, C.; Hernanz-Schulman, M. Swelling at the angle of the mandible: Imaging of the pediatric parotid gland and periparotid region. Radiographics 2001, 21, 1211–1227. [Google Scholar] [CrossRef]
- Adelstein, D.J.; Koyfman, S.A.; El-Naggar, A.K.; Hanna, E.Y. Biology and management of salivary gland cancers. Semin. Radiat. Oncol. 2012, 22, 245–253. [Google Scholar] [CrossRef]
- Lewis, A.G.; Tong, T.; Maghami, E. Diagnosis and Management of Malignant Salivary Gland Tumors of the Parotid Gland. Otolaryngol. Clin. N. Am. 2016, 49, 343–380. [Google Scholar] [CrossRef]
- Stenner, M.; Molls, C.; Luers, J.C.; Beutner, D.; Klussmann, J.P.; Huettenbrink, K.B. Occurrence of lymph node metastasis in early-stage parotid gland cancer. Eur. Arch. Otorhinolaryngol. 2012, 269, 643–648. [Google Scholar] [CrossRef]
- Yue, D.; Feng, W.; Ning, C.; Han, L.X.; YaHong, L. Myoepithelial carcinoma of the salivary gland: Pathologic and CT imaging characteristics (report of 10 cases and literature review). Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017, 123, e182–e187. [Google Scholar] [CrossRef]
- Kim, K.H.; Sung, M.-W.; Yun, J.B.; Han, M.H.; Baek, C.-H.; Chu, K.-C.; Kim, J.H.; Lee, K.-S. The significance of CT scan or MRI in the evaluation of salivary gland tumors. Auris Nasus Larynx 1998, 25, 397–402. [Google Scholar] [CrossRef]
- Yousem, D.M.; Kraut, M.A.; Chalian, A.A. Major salivary gland imaging. Radiology 2000, 216, 19–29. [Google Scholar] [CrossRef]
- Dong, Y.; Lei, G.W.; Wang, S.W.; Zheng, S.W.; Ge, Y.; Wei, F.C. Diagnostic value of CT perfusion imaging for parotid neoplasms. Dentomaxillofac. Radiol. 2014, 43, 20130237. [Google Scholar] [CrossRef]
- Ginat, D.T.; Christoforidis, G. High-Resolution MRI Microscopy Coil Assessment of Parotid Masses. Ear Nose Throat J. 2019, 98, 562–565. [Google Scholar] [CrossRef]
- Mikaszewski, B.; Markiet, K.; Smugała, A.; Stodulski, D.; Szurowska, E.; Stankiewicz, C. An algorithm for preoperative differential diagnostics of parotid tumours on the basis of their dynamic and diffusion-weighted magnetic resonance images: A retrospective analysis of 158 cases. Folia Morphol. 2018, 77, 29–35. [Google Scholar] [CrossRef]
- Syeda-Mahmood, T. Role of Big Data and Machine Learning in Diagnostic Decision Support in Radiology. J. Am. Coll. Radiol. 2018, 15, 569–576. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Waymel, Q.; Badr, S.; Demondion, X.; Cotten, A.; Jacques, T. Impact of the rise of artificial intelligence in radiology: What do radiologists think? Diagn. Interv. Imaging 2019, 100, 327–336. [Google Scholar] [CrossRef]
- (ESR) ESoR. What the radiologist should know about artificial intelligence—An ESR white paper. Insights Imaging 2019, 10, 44. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Ravi, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.-Z. Deep Learning for Health Informatics. IEEE J. Biomed. Health Inform. 2017, 21, 4–21. [Google Scholar] [CrossRef]
- Du, G.; Cao, X.; Liang, J.; Chen, X.; Zhan, Y. Medical image segmentation based on u-net: A review. J. Imaging Sci. Technol. 2020, 64, 1–12. [Google Scholar] [CrossRef]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-net and its variants for medical image segmentation: A review of theory and applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical image segmentation review: The success of u-net. arXiv 2022, arXiv:221114830. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Fritscher, K.; Raudaschl, P.; Zaffino, P.; Spadea, M.F.; Sharp, G.C.; Schubert, R. Deep neural networks for fast segmentation of 3D medical images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 158–165. [Google Scholar] [CrossRef]
- Ibragimov, B.; Xing, L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med. Phys. 2017, 44, 547–557. [Google Scholar] [CrossRef]
- Yang, X.; Wu, N.; Cheng, G.; Zhou, Z.; Yu, D.S.; Beitler, J.J.; Curran, W.J.; Liu, T. Automated segmentation of the parotid gland based on atlas registration and machine learning: A longitudinal MRI study in head-and-neck radiation therapy. Int. J. Radiat. Oncol. 2014, 90, 1225–1233. [Google Scholar] [CrossRef]
- Močnik, D.; Ibragimov, B.; Xing, L.; Strojan, P.; Likar, B.; Pernuš, F.; Vrtovec, T. Segmentation of parotid glands from registered CT and MR images. Phys. Med. 2018, 52, 33–41. [Google Scholar] [CrossRef]
- Hänsch, A.; Schwier, M.; Gass, T.; Morgas, T.; Haas, B.; Dicken, V.; Meine, H.; Klein, J.; Hahn, H.K. Evaluation of deep learning methods for parotid gland segmentation from CT images. J. Med. Imaging 2019, 6, 011005. [Google Scholar] [CrossRef]
- Shan, T.; Tay, F.R.; Gu, L. Application of Artificial Intelligence in Dentistry. J. Dent. Res. 2021, 100, 232–244. [Google Scholar] [CrossRef]
- Carrillo-Perez, F.; Pecho, O.E.; Msc, J.C.M.; Paravina, R.D.; Della Bona, A.; Ghinea, R.; Pulgar, R.; Pérez, M.D.M.; Herrera, L.J.; Msc, F.C.-P.; et al. Applications of artificial intelligence in dentistry: A comprehensive review. J. Esthet. Restor. Dent. 2022, 34, 259–280. [Google Scholar] [CrossRef]
- Raudaschl, P.F.; Zaffino, P.; Sharp, G.C.; Spadea, M.F.; Chen, A.; Dawant, B.M.; Albrecht, T.; Gass, T.; Langguth, C.; Lüthi, M.; et al. Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015. Med. Phys. 2017, 44, 2020–2036. [Google Scholar] [CrossRef]
- Kim, H.; Jung, J.; Kim, J.; Cho, B.; Kwak, J.; Jang, J.Y.; Lee, S.-W.; Lee, J.-G.; Yoon, S.M. Abdominal multi-organ auto-segmentation using 3D-patch-based deep convolutional neural network. Sci. Rep. 2020, 10, 6204. [Google Scholar] [CrossRef]
- Hu, P.; Wu, F.; Peng, J.; Liang, P.; Kong, D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys. Med. Biol. 2016, 61, 8676–8698. [Google Scholar] [CrossRef]
- Raj, A.N.J.; Zhu, H.; Khan, A.; Zhuang, Z.; Yang, Z.; Mahesh, V.G.V.; Karthik, G. ADID-UNET-a segmentation model for COVID-19 infection from lung CT scans. PeerJ Comput. Sci. 2021, 7, e349. [Google Scholar] [CrossRef]
- Yuan, Y.; Chao, M.; Lo, Y. Automatic Skin Lesion Segmentation Using Deep Fully Convolutional Networks with Jaccard Distance. IEEE Trans. Med. Imaging 2017, 36, 1876–1886. [Google Scholar] [CrossRef]
- Elharrouss, O.; Subramanian, N.; Al-Maadeed, S. An Encoder-Decoder-Based Method for Segmentation of COVID-19 Lung Infection in CT Images. SN Comput. Sci. 2022, 3, 13. [Google Scholar] [CrossRef]
- Schultheiss, M.; Schmette, P.; Bodden, J.; Aichele, J.; Müller-Leisse, C.; Gassert, F.G.; Gassert, F.T.; Gawlitza, J.F.; Hofmann, F.C.; Sasse, D.; et al. Lung nodule detection in chest X-rays using synthetic ground-truth data comparing CNN-based diagnosis to human performance. Sci. Rep. 2021, 11, 15857. [Google Scholar] [CrossRef]
- Dot, G.; Schouman, T.; Dubois, G.; Rouch, P.; Gajny, L. Fully automatic segmentation of craniomaxillofacial CT scans for computer-assisted orthognathic surgery planning using the nnU-Net framework. Eur. Radiol. 2022, 32, 3639–3648. [Google Scholar] [CrossRef]
- Park, J.; Hwang, J.; Ryu, J.; Nam, I.; Kim, S.-A.; Cho, B.-H.; Shin, S.-H.; Lee, J.-Y. Deep learning based airway segmentation using key point prediction. Appl. Sci. 2021, 11, 3501. [Google Scholar] [CrossRef]
- Verhelst, P.-J.; Smolders, A.; Beznik, T.; Meewis, J.; Vandemeulebroucke, A.; Shaheen, E.; Van Gerven, A.; Willems, H.; Politis, C.; Jacobs, R. Layered deep learning for automatic mandibular segmentation in cone-beam computed tomography. J. Dent. 2021, 114, 103786. [Google Scholar] [CrossRef]
- Ushinsky, A.; Bardis, M.; Glavis-Bloom, J.; Uchio, E.; Chantaduly, C.; Nguyentat, M.; Chow, D.; Chang, P.; Houshyar, R. A 3D-2D Hybrid U-Net Convolutional Neural Network Approach to Prostate Organ Segmentation of Multiparametric MRI. AJR Am. J. Roentgenol. 2021, 216, 111–116. [Google Scholar] [CrossRef] [PubMed]
| Number | TP | FP | FN | Sensitivity | Precision | F1-Score |
|---|---|---|---|---|---|---|
| Sample | 93 | 0 | 0 | 1.0 | 1.0 | 1.0 |
| Label | 184 | 0 | 0 | 1.0 | 1.0 | 1.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Önder, M.; Evli, C.; Türk, E.; Kazan, O.; Bayrakdar, İ.Ş.; Çelik, Ö.; Costa, A.L.F.; Gomes, J.P.P.; Ogawa, C.M.; Jagtap, R.; et al. Deep-Learning-Based Automatic Segmentation of Parotid Gland on Computed Tomography Images. Diagnostics 2023, 13, 581. https://doi.org/10.3390/diagnostics13040581
Önder M, Evli C, Türk E, Kazan O, Bayrakdar İŞ, Çelik Ö, Costa ALF, Gomes JPP, Ogawa CM, Jagtap R, et al. Deep-Learning-Based Automatic Segmentation of Parotid Gland on Computed Tomography Images. Diagnostics. 2023; 13(4):581. https://doi.org/10.3390/diagnostics13040581
Chicago/Turabian StyleÖnder, Merve, Cengiz Evli, Ezgi Türk, Orhan Kazan, İbrahim Şevki Bayrakdar, Özer Çelik, Andre Luiz Ferreira Costa, João Pedro Perez Gomes, Celso Massahiro Ogawa, Rohan Jagtap, and et al. 2023. "Deep-Learning-Based Automatic Segmentation of Parotid Gland on Computed Tomography Images" Diagnostics 13, no. 4: 581. https://doi.org/10.3390/diagnostics13040581
APA StyleÖnder, M., Evli, C., Türk, E., Kazan, O., Bayrakdar, İ. Ş., Çelik, Ö., Costa, A. L. F., Gomes, J. P. P., Ogawa, C. M., Jagtap, R., & Orhan, K. (2023). Deep-Learning-Based Automatic Segmentation of Parotid Gland on Computed Tomography Images. Diagnostics, 13(4), 581. https://doi.org/10.3390/diagnostics13040581







