DTLCx: An Improved ResNet Architecture to Classify Normal and Conventional Pneumonia Cases from COVID-19 Instances with Grad-CAM-Based Superimposed Visualization Utilizing Chest X-ray Images
Abstract
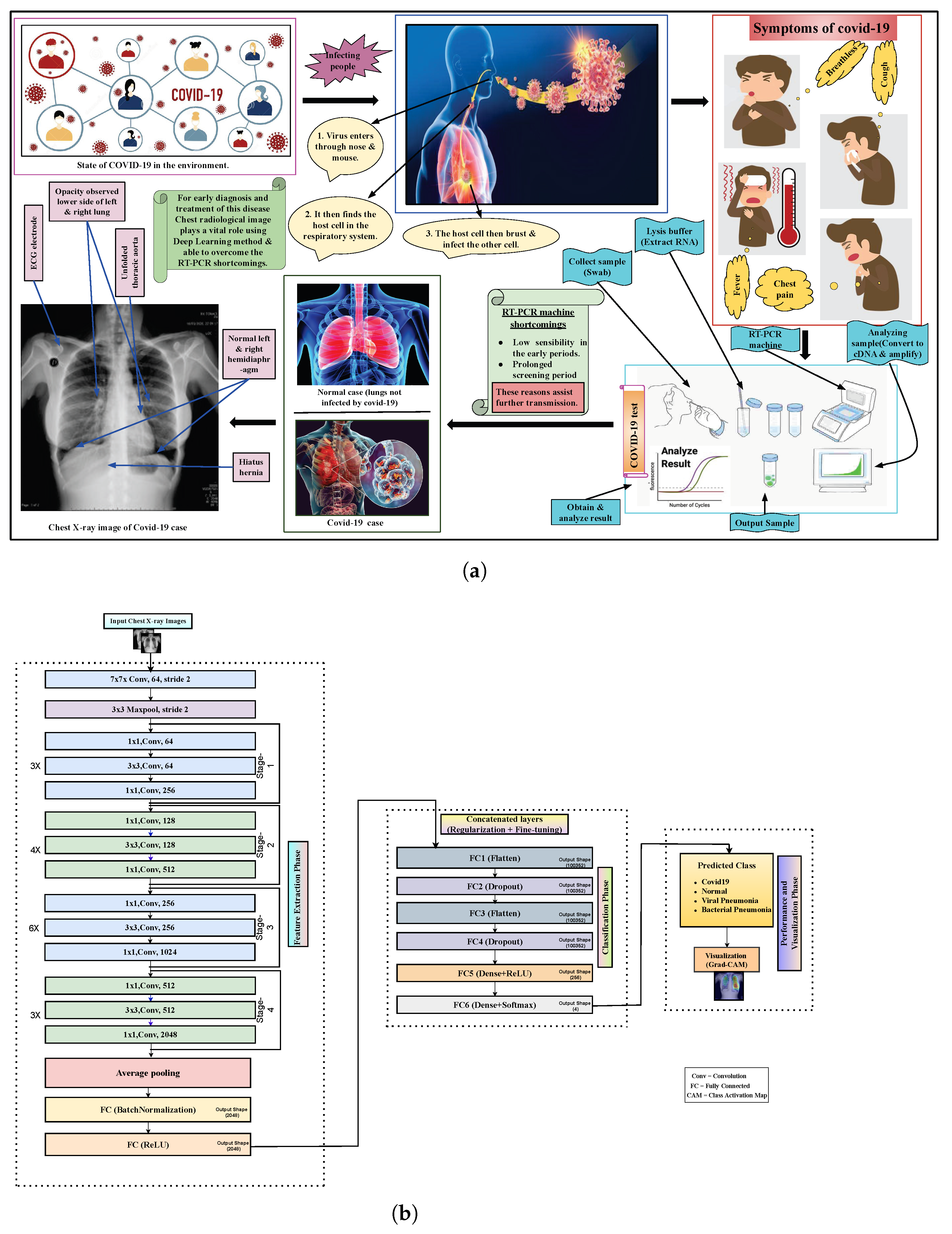
:1. Introduction
- Using a more sophisticated methodology for diagnosis and identifying COVID-19 patients through a deep learning approach, which is based on X-ray images.
- Developing and examining a deep learning model to automatically detect and early diagnose COVID-19-infected patients in an effective manner.
- Performing an experimental analysis of the proposed model while classifying COVID-19 cases by using operable X-ray images. It has a lower implementation cost than other existing models that are trained using CT scan.
- Classifying COVID-19 images as four classes, three classes and binary classes: namely COVID-19, normal, bacterial, and viral pneumonia from normal and regular pneumonia.
- Demonstrating comparative performance analysis of the proposed work with the other previous state-of-the-art works and Grad-CAM-based visualization marking the most flawless classification results of the COVID-19 cases using the datasets of chest X-ray images.
2. Related Background
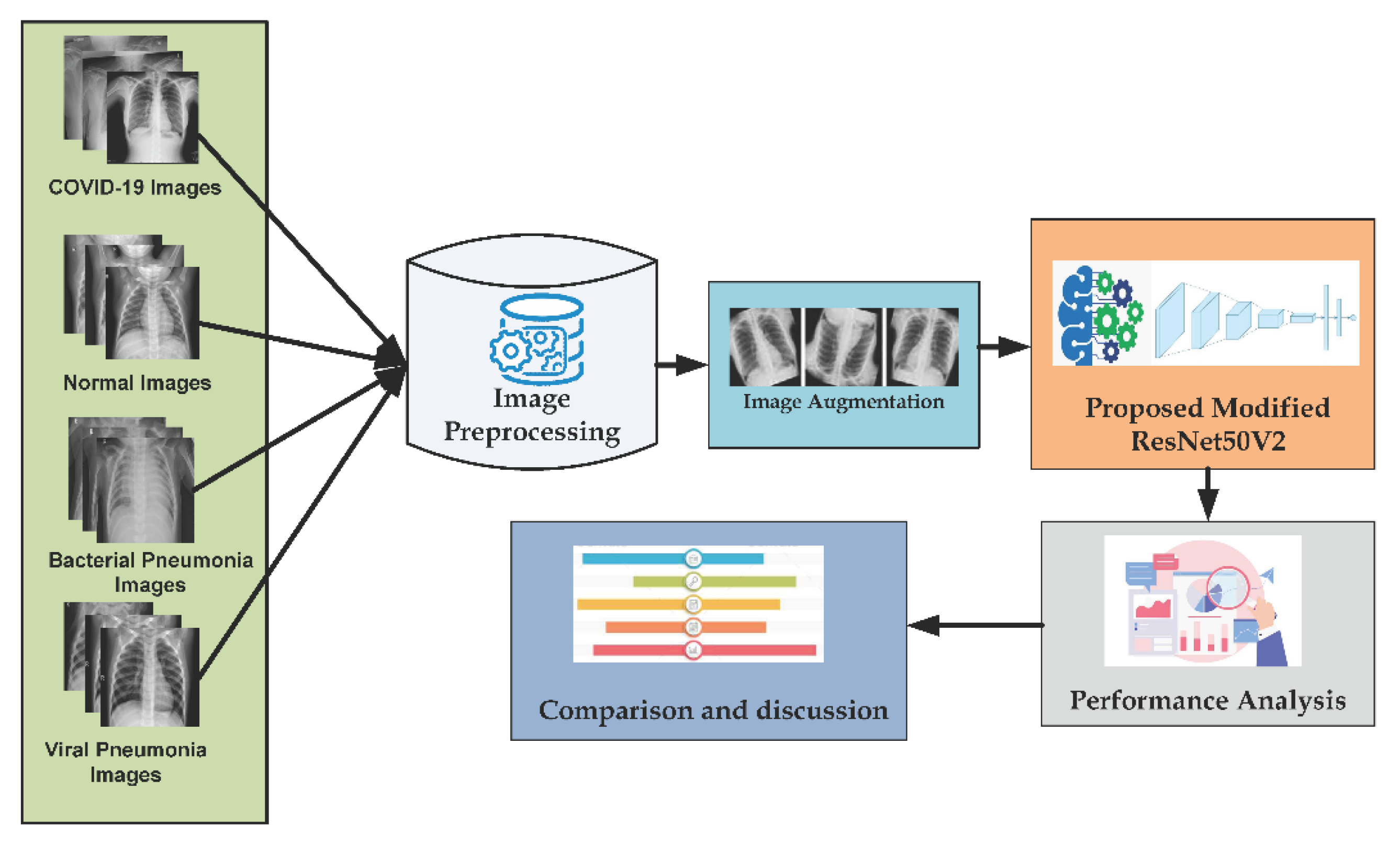
3. Methodology
3.1. Data Preprocessing
3.2. Data Augmentation
3.3. Transfer Learning with Convolutional Neural Networks
3.3.1. Convolutional Layer
3.3.2. Pooling Layer
3.3.3. Fully Connected Layer
3.4. Proposed Architecture with Six Extra Layers
4. Experimental Results
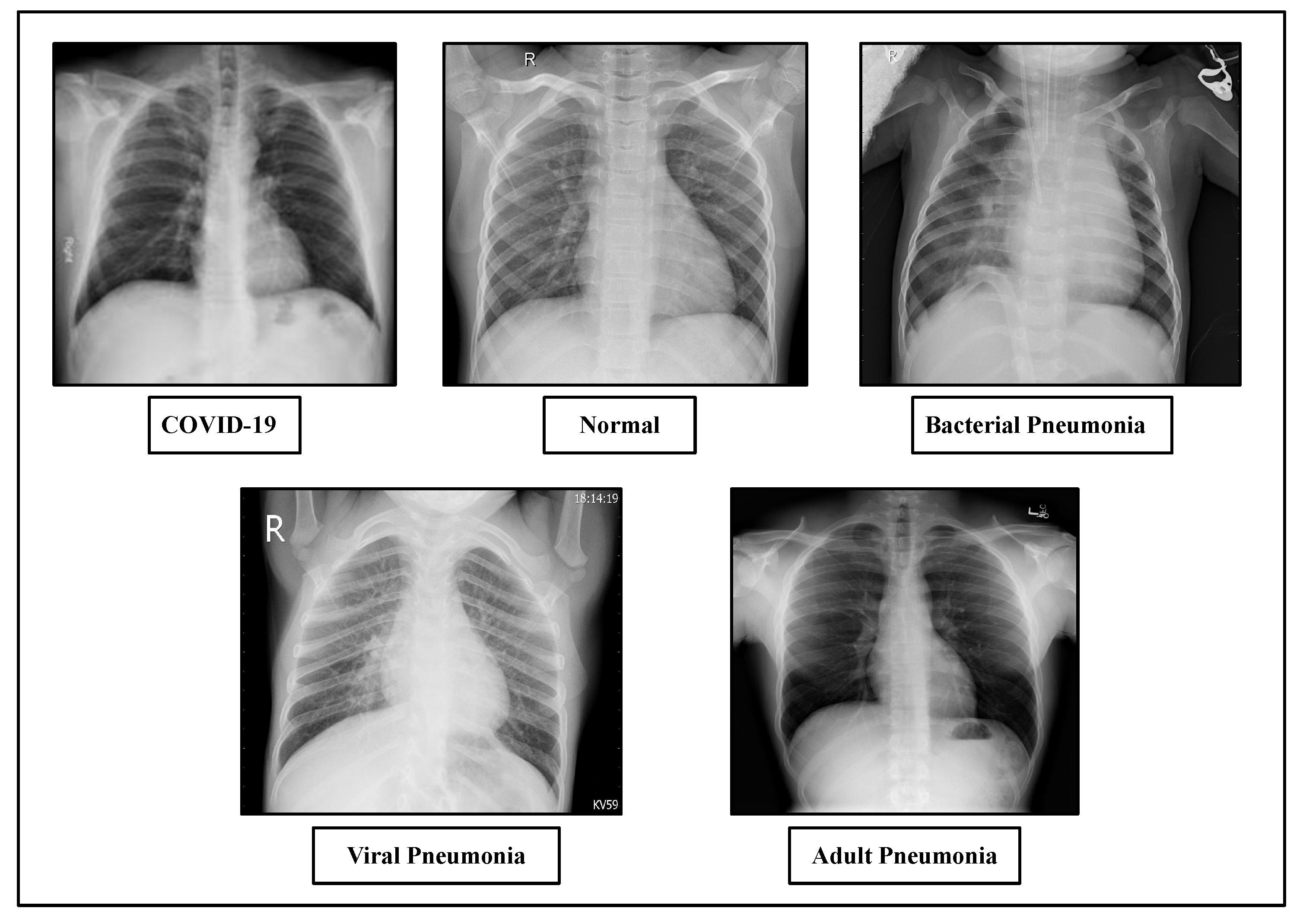
4.1. Dataset Description
4.2. Experimental Configuration and Implementation
4.3. Evaluation Metrics
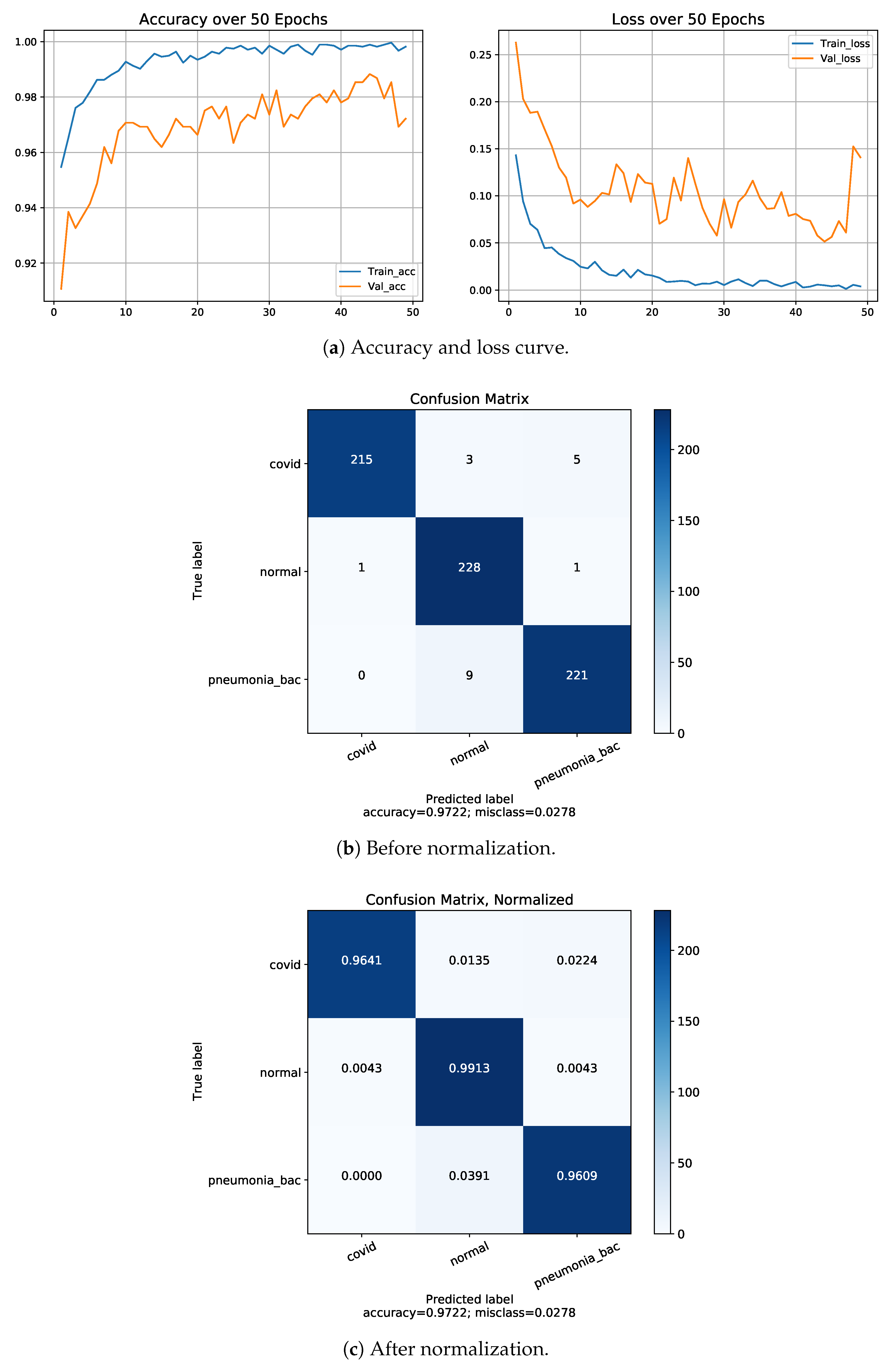
4.4. Normalization of Confusion Matrix
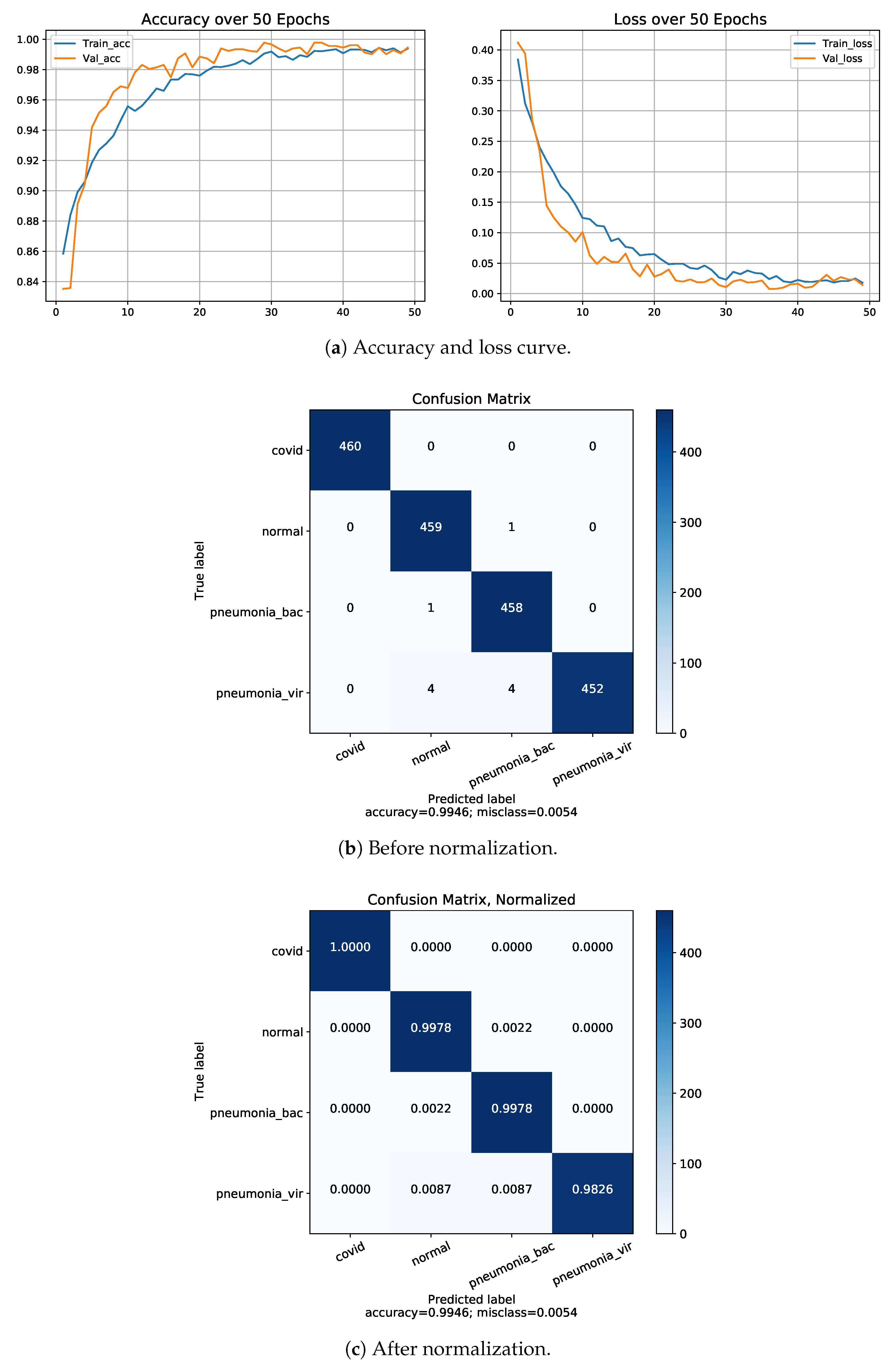
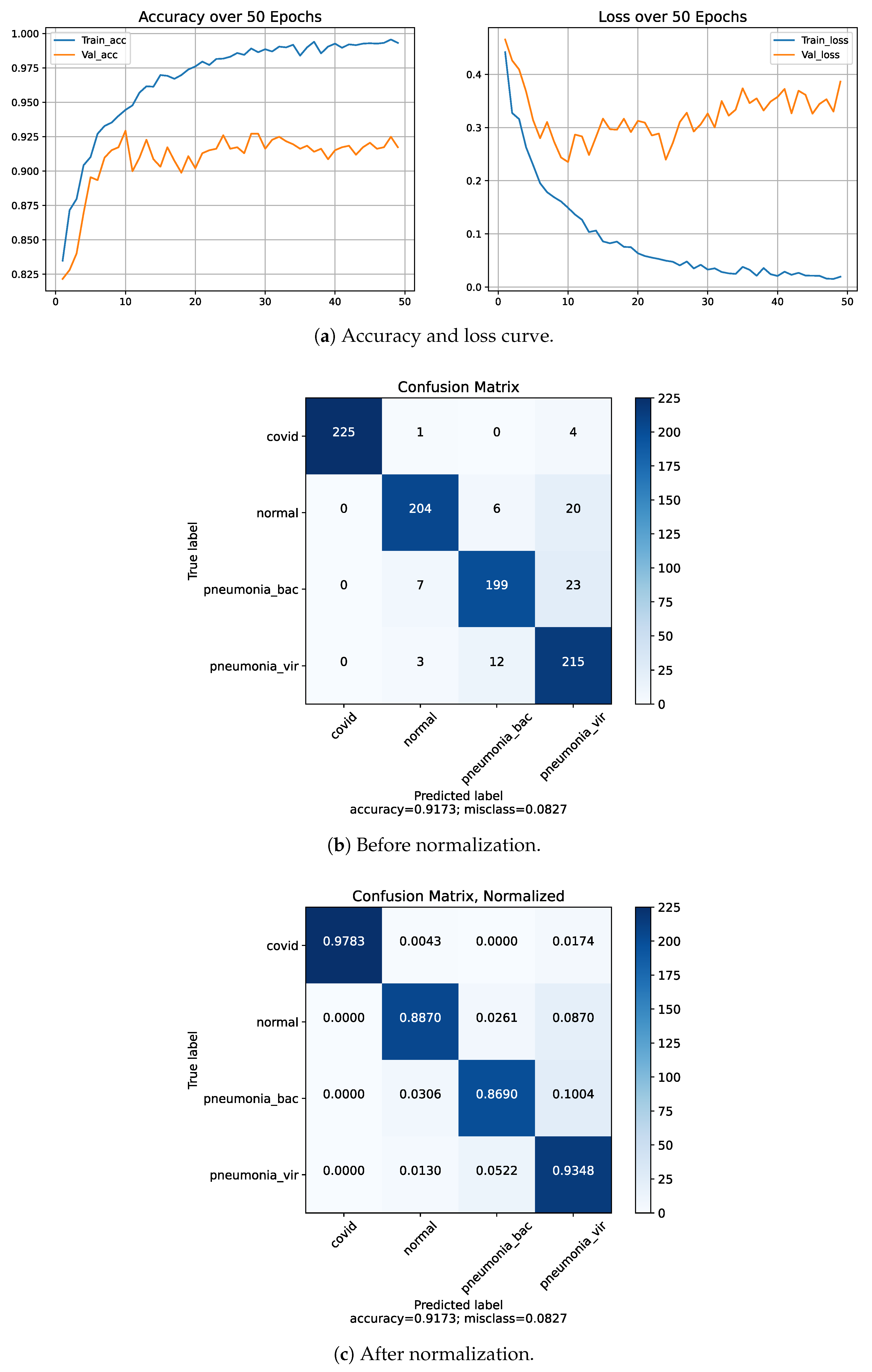
4.5. Classification Performance on Dataset-1
4.6. Classification Performance on Dataset-2
4.7. K-Fold Cross Validation
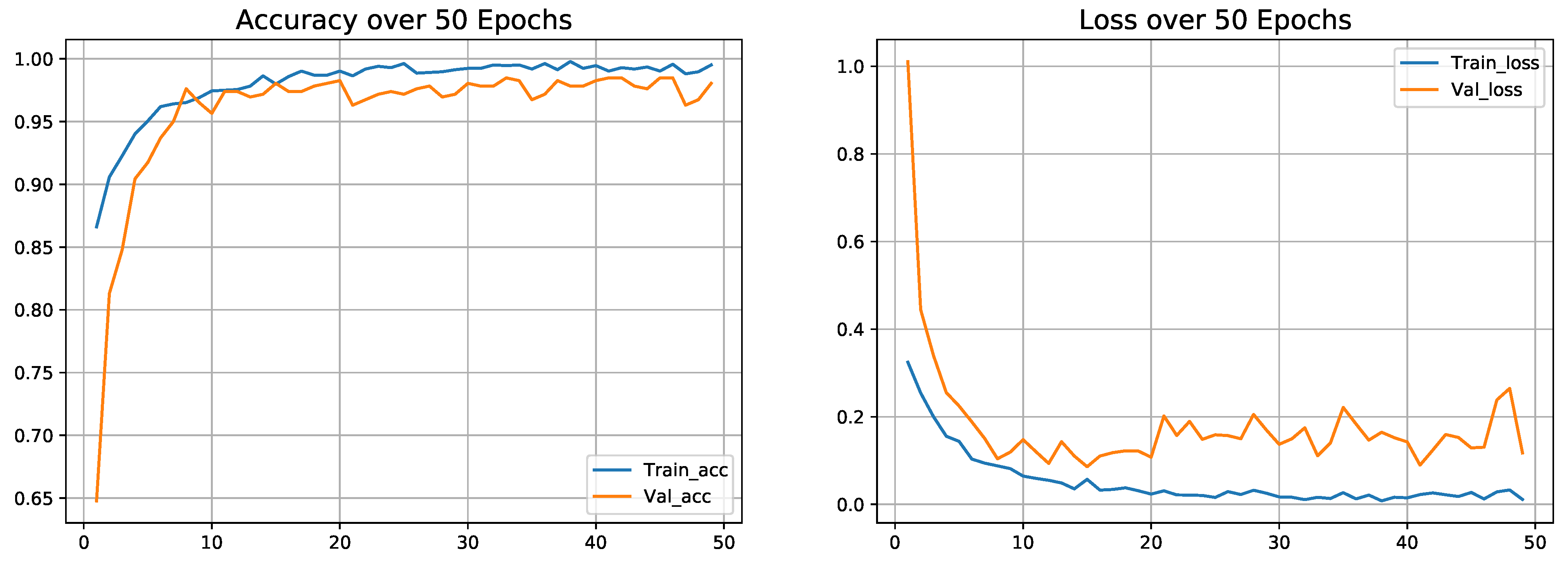
4.8. Classification Performance on Dataset-3
5. Discussion
Discriminative Visualization by Proposed Model Using Grad-Cam
- By considering the normal X-rays, there is no opacity in normal X-rays, which distinguishes normal cases from all types of pneumonia cases who have opacities with various types [81,82,83]. For normal X-rays, no substantial region is localized, as shown in Figure 13. Since it is more recognizable, it is easier to distinguish from the other patients.
- When looking at the heatmaps for classical viral pneumonia, it is observed that our proposed model has localized the regions with bilateral multifocal Ground Glass Opacities (GGO) as well as patchy consolidations in some cases. These localized characteristics are also commonly recognized as radiological features of classic viral pneumonia [9,84,85].
- Localized activation heatmaps in the event of bacterial pneumonia generally involve opacities with consolidation on the lower and upper lobes. Furthermore, both unilateral and bilateral, as well as peripheral, participation is seen. These characteristics, according to [81,82], are primarily associated with bacterial pneumonia.
- According to [9,85], COVID-19 and typical viral pneumonia have many similarities, including bilateral GGOs and patchy consolidations. Peripheral and diffuse distributions, vascular thickening, micro-reticular opacity, and typical viral-like Ground Glass Opacities (GGOs) are some of the more likely hallmarks of COVID-19-induced pneumonia [86,87]. By closely inspecting the generated heatmap from several COVID-19 infected X-rays (Figure 13), it is observed that the opacities are distributed in a peripheral and diffuse manner. Furthermore, vascular thickening, as well as other conventional viral features, is localized in some of the cases.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. Author Correction: A new coronavirus associated with human respiratory disease in China. Nature 2020, 580, E7. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 CORONAVIRUS PANDEMIC. 2020. Available online: https://www.worldometers.info/coronavirus/ (accessed on 20 March 2021).
- Hafeez, A.; Ahmad, S.; Siddqui, S.A.; Ahmad, M.; Mishra, S. A review of COVID-19 (Coronavirus Disease-2019) diagnosis, treatments and prevention. Ejmo 2020, 4, 116–125. [Google Scholar]
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Franquet, T. Imaging of community-acquired pneumonia. J. Thorac. Imaging 2018, 33, 282–294. [Google Scholar] [CrossRef]
- Zu, Z.Y.; Jiang, M.D.; Xu, P.P.; Chen, W.; Ni, Q.Q.; Lu, G.M.; Zhang, L.J. Coronavirus disease 2019 (COVID-19): A perspective from China. Radiology 2020, 296, E15–E25. [Google Scholar] [CrossRef]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology 2020, 296, E65–E72. [Google Scholar] [CrossRef]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef]
- Dhiman, C.; Vishwakarma, D.K. View-invariant deep architecture for human action recognition using two-stream motion and shape temporal dynamics. IEEE Trans. Image Process. 2020, 29, 3835–3844. [Google Scholar]
- Deepak, S.; Ameer, P. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, H.; Bidgoli, F.A.; Zhou, B.; Iribarren, C.; Molloi, S.; Baldi, P. Detecting cardiovascular disease from mammograms with deep learning. IEEE Trans. Med Imaging 2017, 36, 1172–1181. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhong, Z.; Xie, X.; Yu, Q.; Liu, J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: A multicenter study. AJR Am. J. Roentgenol. 2020, 214, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xia, L. Coronavirus disease 2019 (COVID-19): Role of chest CT in diagnosis and management. Ajr Am. J. Roentgenol. 2020, 214, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Yuan, S.; Kok, K.H.; To, K.K.W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.Y.; Poon, R.W.S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Yoon, S.H.; Lee, K.H.; Kim, J.Y.; Lee, Y.K.; Ko, H.; Kim, K.H.; Park, C.M.; Kim, Y.H. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): Analysis of nine patients treated in Korea. Korean J. Radiol. 2020, 21, 494–500. [Google Scholar] [CrossRef]
- Kong, W.; Agarwal, P.P. Chest imaging appearance of COVID-19 infection. Radiol. Cardiothorac. Imaging 2020, 2, e200028. [Google Scholar] [CrossRef]
- Infante, M.; Lutman, R.; Imparato, S.; Di Rocco, M.; Ceresoli, G.; Torri, V.; Morenghi, E.; Minuti, F.; Cavuto, S.; Bottoni, E.; et al. Differential diagnosis and management of focal ground-glass opacities. Eur. Respir. J. 2009, 33, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Kanne, J.P.; Little, B.P.; Chung, J.H.; Elicker, B.M.; Ketai, L.H. Essentials for radiologists on COVID-19: An update—Radiology scientific expert panel. Radiology 2020, 296, E113–E114. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Cao, D.; Zeng, K.; Wang, J.; Sharma, P.K.; Ma, X.; Liu, Y.; Zhou, S. Bert-based deep spatial-temporal network for taxi demand prediction. IEEE Trans. Intell. Transp. Syst. 2021, 23, 9442–9454. [Google Scholar] [CrossRef]
- Ahmed, I.; Ahmad, A.; Jeon, G. An IoT-based deep learning framework for early assessment of COVID-19. IEEE Internet Things J. 2020, 8, 15855–15862. [Google Scholar] [CrossRef] [PubMed]
- Awan, M.J.; Bilal, M.H.; Yasin, A.; Nobanee, H.; Khan, N.S.; Zain, A.M. Detection of COVID-19 in chest X-ray images: A big data enabled deep learning approach. Int. J. Environ. Res. Public Health 2021, 18, 10147. [Google Scholar] [CrossRef] [PubMed]
- Sohail, A.; Yu, Z.; Nutini, A. COVID-19 Variants and Transfer Learning for the Emerging Stringency Indices. Neural Process. Lett. 2022, 1–10. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Talo, M.; Yildirim, O.; Baloglu, U.B.; Aydin, G.; Acharya, U.R. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput. Med Imaging Graph. 2019, 78, 101673. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Ahamed, K.U.; Islam, M.; Uddin, A.; Akhter, A.; Paul, B.K.; Yousuf, M.A.; Uddin, S.; Quinn, J.M.; Moni, M.A. A deep learning approach using effective preprocessing techniques to detect COVID-19 from chest CT-scan and X-ray images. Comput. Biol. Med. 2021, 139, 105014. [Google Scholar] [CrossRef]
- Comelli, A.; Coronnello, C.; Dahiya, N.; Benfante, V.; Palmucci, S.; Basile, A.; Vancheri, C.; Russo, G.; Yezzi, A.; Stefano, A. Lung segmentation on high-resolution computerized tomography images using deep learning: A preliminary step for radiomics studies. J. Imaging 2020, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Talukder, M.A.; Islam, M.M.; Uddin, M.A.; Akhter, A.; Hasan, K.F.; Moni, M.A. Machine learning-based lung and colon cancer detection using deep feature extraction and ensemble learning. Expert Syst. Appl. 2022, 205, 117695. [Google Scholar] [CrossRef]
- Bullock, J.; Luccioni, A.; Pham, K.H.; Lam, C.S.N.; Luengo-Oroz, M. Mapping the landscape of artificial intelligence applications against COVID-19. J. Artif. Intell. Res. 2020, 69, 807–845. [Google Scholar] [CrossRef]
- Modegh, R.G.; Hamidi, M.; Masoudian, S.; Mohseni, A.; Lotfalinezhad, H.; Kazemi, M.A.; Moradi, B.; Ghafoori, M.; Motamedi, O.; Pournik, O.; et al. Accurate and rapid diagnosis of COVID-19 pneumonia with batch effect removal of chest CT-scans and interpretable Artificial intelligence. arXiv 2020, arXiv:2011.11736. [Google Scholar]
- Shalbaf, A.; Vafaeezadeh, M.; Gifani, P. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 115–123. [Google Scholar]
- Zheng, C.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Wang, X. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv 2020, 2020-03. [Google Scholar]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Wang, R.; Zhao, H.; Chong, Y.; et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 2775–2780. [Google Scholar] [CrossRef]
- Purohit, K.; Kesarwani, A.; Ranjan Kisku, D.; Dalui, M. COVID-19 detection on chest x-ray and ct scan images using multi-image augmented deep learning model. In Seventh International Conference on Mathematics and Computing; Springer: Berlin/Heidelberg, Germany, 2022; pp. 395–413. [Google Scholar]
- Dansana, D.; Kumar, R.; Bhattacharjee, A.; Hemanth, D.J.; Gupta, D.; Khanna, A.; Castillo, O. Early diagnosis of COVID-19-affected patients based on X-ray and computed tomography images using deep learning algorithm. Soft Comput. 2020, 1–9. [Google Scholar] [CrossRef]
- Panwar, H.; Gupta, P.; Siddiqui, M.K.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals 2020, 140, 110190. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Gupta, K.D.; Islam, M.M.; Sen, S.; Rahman, M.; Hossain, M.S. Study of different deep learning approach with explainable ai for screening patients with COVID-19 symptoms: Using ct scan and chest x-ray image dataset. arXiv 2020, arXiv:2007.12525. [Google Scholar]
- Jaiswal, A.K.; Tiwari, P.; Rathi, V.K.; Qian, J.; Pandey, H.M.; Albuquerque, V.H.C. Covidpen: A novel covid-19 detection model using chest x-rays and ct scans. Medrxiv 2020, 2020-07. [Google Scholar]
- Maghdid, H.S.; Asaad, A.T.; Ghafoor, K.Z.; Sadiq, A.S.; Mirjalili, S.; Khan, M.K. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. In Proceedings of the Multimodal Image Exploitation and Learning 2021, SPIE, Online, 12–15 April 2021; Volume 11734, pp. 99–110. [Google Scholar]
- Kassania, S.H.; Kassanib, P.H.; Wesolowskic, M.J.; Schneidera, K.A.; Detersa, R. Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: A machine learning based approach. Biocybern. Biomed. Eng. 2021, 41, 867–879. [Google Scholar] [CrossRef]
- Mukherjee, H.; Ghosh, S.; Dhar, A.; Obaidullah, S.M.; Santosh, K.; Roy, K. Deep neural network to detect COVID-19: One architecture for both CT Scans and Chest X-rays. Appl. Intell. 2021, 51, 2777–2789. [Google Scholar] [CrossRef] [PubMed]
- Kamil, M.Y. A deep learning framework to detect COVID-19 disease via chest X-ray and CT scan images. Int. J. Electr. Comput. Eng. 2021, 11, 844–850. [Google Scholar] [CrossRef]
- Krishnamraju, K.; Chandini, T.; Vamsi, S.; Tejaswarrao, S.; Devika, T.; Madhu, C. Detection and Analysis of COVID-19 Using Chest X-ray Images. Int. J. Signal Process. 2022, 7. [Google Scholar]
- Mousavi, Z.; Shahini, N.; Sheykhivand, S.; Mojtahedi, S.; Arshadi, A. COVID-19 detection using chest X-ray images based on a developed deep neural network. SLAS Technol. 2022, 27, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Luz, E.; Silva, P.; Silva, R.; Silva, L.; Guimarães, J.; Miozzo, G.; Moreira, G.; Menotti, D. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res. Biomed. Eng. 2022, 38, 149–162. [Google Scholar] [CrossRef]
- Al-Waisy, A.S.; Al-Fahdawi, S.; Mohammed, M.A.; Abdulkareem, K.H.; Mostafa, S.A.; Maashi, M.S.; Arif, M.; Garcia-Zapirain, B. COVID-CheXNet: Hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020, 1–16. [Google Scholar] [CrossRef]
- Chen, A.; Jaegerman, J.; Matic, D.; Inayatali, H.; Charoenkitkarn, N.; Chan, J. Detecting COVID-19 in chest X-rays using transfer learning with VGG16. In Proceedings of the CSBio’20: Eleventh International Conference on Computational Systems-Biology and Bioinformatics, Bangkok, Thailand, 19–21 November 2020; pp. 93–96. [Google Scholar]
- Wang, L.; Lin, Z.Q.; Wong, A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Gupta, A.; Shreyansh, G.; Katarya, R. InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl. Soft Comput. 2021, 99, 106859. [Google Scholar] [CrossRef]
- Chalifah, A.; Purbojo, S.; Umitaibatin, R.; Rudiyanto, D. Comparative Study of Convolutional Neural Network Feature Extractors Used for COVID-19 Detection from Chest X-Ray Images. 2020. Available online: https://www.researchgate.net/publication/344656730_Comparative_Study_of_Convolutional_Neural_Network_Feature_Extractors_Used_for_COVID-19_Detection_From_Chest_X-Ray_Images (accessed on 30 January 2023).
- Jain, R.; Gupta, M.; Taneja, S.; Hemanth, D.J. Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl. Intell. 2021, 51, 1690–1700. [Google Scholar] [CrossRef]
- Mohammadi, R.; Salehi, M.; Ghaffari, H.; Rohani, A.; Reiazi, R. Transfer learning-based automatic detection of coronavirus disease 2019 (COVID-19) from chest X-ray images. J. Biomed. Phys. Eng. 2020, 10, 559. [Google Scholar] [CrossRef]
- Turkoglu, M. COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl. Intell. 2021, 51, 1213–1226. [Google Scholar] [CrossRef]
- Ouchicha, C.; Ammor, O.; Meknassi, M. CVDNet: A novel deep learning architecture for detection of coronavirus (COVID-19) from chest x-ray images. Chaos Solitons Fractals 2020, 140, 110245. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef] [PubMed]
- Sethy, P.K.; Behera, S.K. Detection of Coronavirus Disease (COVID-19) Based on Deep Features. Preprints 2020. [Google Scholar]
- Aslan, M.F.; Unlersen, M.F.; Sabanci, K.; Durdu, A. CNN-based transfer learning–BiLSTM network: A novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021, 98, 106912. [Google Scholar] [CrossRef] [PubMed]
- Arsenovic, M.; Sladojevic, S.; Orcic, S.; Anderla, A.; Sladojevic, M. Detection of COVID-19 Cases by Utilizing Deep Learning Algorithms on X-ray Images. 2020, pp. 1–8. Available online: https://www.researchgate.net/publication/344649501_Detection_of_COVID-19_cases_by_utilizing_deep_learning_algorithms_on_X-ray_images (accessed on 30 January 2023).
- Chowdhury, N.K.; Rahman, M.; Kabir, M.A. PDCOVIDNet: A parallel-dilated convolutional neural network architecture for detecting COVID-19 from chest X-ray images. Health Inf. Sci. Syst. 2020, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Makris, A.; Kontopoulos, I.; Tserpes, K. COVID-19 detection from chest X-Ray images using Deep Learning and Convolutional Neural Networks. In Proceedings of the 11th Hellenic Conference on Artificial Intelligence, Athens, Greece, 2 September 2020; pp. 60–66. [Google Scholar]
- Civit-Masot, J.; Luna-Perejón, F.; Domínguez Morales, M.; Civit, A. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl. Sci. 2020, 10, 4640. [Google Scholar] [CrossRef]
- Mahmud, T.; Rahman, M.A.; Fattah, S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020, 122, 103869. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 630–645. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Demir, A.; Yilmaz, F.; Kose, O. Early detection of skin cancer using deep learning architectures: Resnet-101 and inception-v3. In Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO), IEEE-2019, Izmir, Turkey, 3–5 October 2019; pp. 1–4. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Ketkar, N. Introduction to keras. In Deep Learning with Python; Springer: Berlin/Heidelberg, Germany, 2017; pp. 97–111. [Google Scholar]
- Gulli, A.; Pal, S. Deep Learning with Keras; Packt Publishing Ltd: Birmingham, UK, 2017. [Google Scholar]
- Lih, O.S.; Jahmunah, V.; San, T.R.; Ciaccio, E.J.; Yamakawa, T.; Tanabe, M.; Kobayashi, M.; Faust, O.; Acharya, U.R. Comprehensive electrocardiographic diagnosis based on deep learning. Artif. Intell. Med. 2020, 103, 101789. [Google Scholar] [CrossRef]
- Dekhtiar, J.; Durupt, A.; Bricogne, M.; Eynard, B.; Rowson, H.; Kiritsis, D. Deep learning for big data applications in CAD and PLM–Research review, opportunities and case study. Comput. Ind. 2018, 100, 227–243. [Google Scholar] [CrossRef]
- Rahimzadeh, M.; Attar, A. Detecting and counting pistachios based on deep learning. Iran J. Comput. Sci. 2022, 5, 69–81. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Sharma, S.; Sharma, S.; Athaiya, A. Activation functions in neural networks. Towards Data Sci. 2017, 6, 310–316. [Google Scholar] [CrossRef]
- COVID-19 Radiography Database. 2020. Available online: https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database (accessed on 20 March 2022).
- Chest X-ray Images (Pneumonia). 2020. Available online: https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia (accessed on 20 March 2022).
- RSNA Pneumonia Detection Challenge. 2018. Available online: https://www.kaggle.com/competitions/rsna-pneumonia-detection-challenge/data (accessed on 20 March 2022).
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Franquet, T. Imaging of pneumonia: Trends and algorithms. Eur. Respir. J. 2001, 18, 196–208. [Google Scholar] [CrossRef]
- Vilar, J.; Domingo, M.L.; Soto, C.; Cogollos, J. Radiology of bacterial pneumonia. Eur. J. Radiol. 2004, 51, 102–113. [Google Scholar] [CrossRef]
- Koo, H.J.; Lim, S.; Choe, J.; Choi, S.H.; Sung, H.; Do, K.H. Radiographic and CT features of viral pneumonia. Radiographics 2018, 38, 719–739. [Google Scholar] [CrossRef]
- Franquet, T. Imaging of pulmonary viral pneumonia. Radiology 2011, 260, 18–39. [Google Scholar] [CrossRef]
- Bai, H.X.; Hsieh, B.; Xiong, Z.; Halsey, K.; Choi, J.W.; Tran, T.M.L.; Pan, I.; Shi, L.B.; Wang, D.C.; Mei, J.; et al. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology 2020, 296, E46–E54. [Google Scholar] [CrossRef]
- Shi, H.; Han, X.; Jiang, N.; Cao, Y.; Alwalid, O.; Gu, J.; Fan, Y.; Zheng, C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020, 20, 425–434. [Google Scholar] [CrossRef]
- Ng, M.Y.; Lee, E.Y.; Yang, J.; Yang, F.; Li, X.; Wang, H.; Lui, M.M.S.; Lo, C.S.Y.; Leung, B.; Khong, P.L.; et al. Imaging profile of the COVID-19 infection: Radiologic findings and literature review. Radiol. Cardiothorac. Imaging 2020, 2, e200034. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.A.; Stranieri, A.; Gondal, I.; Balasubramanian, V. Blockchain leveraged decentralized IoT eHealth framework. Internet Things 2020, 9, 100159. [Google Scholar] [CrossRef]






| Author (Month, Year) | Number of Cases and Image | Training Model | VisualizeUsing Gard CAM/Other | Accuracy (%) |
|---|---|---|---|---|
| Krishnamraju K [45] | 1000 COVID-19 and 1000 normal | VGG16+ MobileNet | No | 97 |
| Mousavi Z [46] | 939 healthy cases, 800 COVID-19 and 942 viral pneumonia | Developed LSTM network | No | 90 |
| Luz [47] | 1000 COVID-19, 1000 normal and 1000 pneumonia | Efficient deep learning model | Yes | 93.9 |
| Al-Waisy [48] | 400 COVID-19 and 400 normal | COVID-CheXNet | Yes | 99.99 |
| Aslan [59] | 1341 normal, 219 COVID-19 and 1345 viral pneumonia | mAlexNet +BiLSTM | No | 98.14, for first architecture and 98.70 |
| Chen [49] | 3 datasets total 513 COVID-19 and1984 non-COVID-19 | VGG16 | Yes | 98 |
| Wang [50] | 266 COVID-19, 8,066 normal and 5,538 pneumonia | COVID-Net | Yes | 93.30 |
| Gupta [51] | 361 COVID-19, 1345 pneumonia and 1341 normal | InstaCovNet-19 | Yes | 99.08 for 3 class and 99.53 for 2 class |
| Arsenovic [60] | 434 COVID-19, 1100 normal and 1100 bacterial pneumonia | ResNetCOVID-19 | No | 94.10 |
| Ammar [52] | 150 COVID-19, 150 normal, and 150 pneumonia | MobileNetV2, ResNet50V2, ResNet152V2, Xception, VGG16 and DenseNet12 | No | Highest accuracy 91.28 for MobileNetV2 |
| Jain [53] | 490 COVID-19, 1345 normal and 3632 pneumonia | Xception net, Inception net V3 and ResNeXt, | No | Highest accuracy 97.97 for Xception |
| Mohammadi R [54] | 181 COVID-19 and 364 normal | pre-trained VGG16, InceptionResNetV2, MobileNet and VGG19 | No | Highest accuracy 99.1 MobileNet |
| Chowdhury [61] | 1341 normal, 219 COVID-19 and 1345 viral pneumonia | PDCOVIDNet | Yes | 96.58 |
| Turkoglu [55] | 219 COVID-19, 1583 normal and 4290 pneumonia | COVIDetectioNet | No | 99.18 |
| Makris [62] | 112 COVID-19, 112 normal and 112 pneumonia | 9 well-known pre-trained CNN model | No | 95 for the best two model (Vgg16 and Vgg19) |
| Ouchicha [56] | 1341 normal, 219 COVID-19 and 1345 viral pneumonia | CVDNet | No | 96.69 |
| Civit-Masot [63] | 132 COVID-19, 132 healthy and 132 pneumonia | VGG16 | No | 86.00 |
| Mahmud [64] | 1583 normal, 305 COVID-19, 1493 viral pneumonia, 2780 bacterial pneumonia | CovXNet | Yes | 90.2 accuracy for four class |
| Khan [57] | 1203 normal, 290 COVID-19 931 viral pneumonia, 660 bacterial pneumonia | CoroNet | No | Overall accuracy of 89.6 |
| Ozturk [20] | 125 COVID-19 cases, 500 no findings, 500 pneumonia cases | DarkCovid-Net | Yes | 98.08 for two class and 87.02 |
| Notations | Definition |
|---|---|
| I | denotes as input matrix (e.g., image) |
| K | represents 2D filter by considering dimension m × n |
| F | output of 2D characteristic map |
| I*K | operation of convolution |
| x | represents as input vector |
| Z | represents as output vector |
| Train_acc | represents as training accuracy |
| Val_acc | represents as validation accuracy |
| pneumonia_bac | represents bacterial pneumonia cases |
| pneumonia_vir | represents viral pneumonia cases |
| Input Layer | Output Shape | Number of Trainable |
|---|---|---|
| (Depth*Height*Weight) | Parameters | |
| Input image | (3*224*224) | 0 |
| Conv1 | (64*112*112) | 9472 |
| Maxpool1 | (64*56*56) | 0 |
| Conv2 | (256*28*28) | 16,640 |
| Maxpool2 | (256*28*28) | 0 |
| Conv3 | (512*14*14) | 66,048 |
| Maxpool3 | (512*14*14) | 0 |
| Conv4 | (1024*7*7) | 263,168 |
| Maxpool4 | (1024*7*7) | 0 |
| Conv5 | (2048*7*7) | 1,050,624 |
| FC (Batch_normalization) | (2048*7*7) | 8192 |
| FC (ReLu) | (2048*7*7) | 0 |
| “Concatanated layers” | - | - |
| FC1 (Flatten) | (100,352) | 0 |
| FC2 (Dropout) | (100,352) | 0 |
| FC3 (Flatten) | (100,352) | 0 |
| FC4 (Dropout) | (100,352) | 0 |
| FC5 (ReLu) | (256) | 25,690,368 |
| FC6 (SoftMax) | (4) | 1028 |
| Subset | COVID-19 | Normal | Viral Pneumonia | Bacterial Pneumonia |
|---|---|---|---|---|
| Training Data | 686 | 690 | 690 | 690 |
| Test Data | 457 | 460 | 460 | 460 |
| Total | 1143 | 1150 | 1150 | 1150 |
| Subset | COVID-19 | Normal | Viral Pneumonia | Bacterial Pneumonia |
|---|---|---|---|---|
| Training Data | 1143 | 1150 | 1150 | 1150 |
| Test Data | 457 | 460 | 460 | 460 |
| Total | 1600 | 1610 | 1610 | 1610 |
| Class | Task | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|---|
| Four | COVID-19 | 99.0 | 94.0 | 97.0 | 88.79 |
| Normal | 94.0 | 91.0 | 92.0 | ||
| Bacterial Pneumonia | 80.0 | 94.0 | 87.0 | ||
| Viral Pneumonia | 83.0 | 77.0 | 80.0 | ||
| COVID-19 | 1.00 | 96.0 | 98.0 | ||
| Three | Normal | 95.0 | 99.0 | 97.0 | 97.22 |
| Bacterial Pneumonia | 97.0 | 96.0 | 97.0 | ||
| COVID-19 | 1.00 | 97.0 | 98.0 | ||
| Three | Normal | 86.0 | 98.0 | 91.0 | 93.11 |
| Viral Pneumonia | 95.0 | 85.0 | 90.0 | ||
| COVID-19 | 99.0 | 95.0 | 97.0 | ||
| Three | Viral Pneumonia | 87.0 | 97.0 | 91.0 | 92.10 |
| Bacterial Pneumonia | 95.0 | 85.0 | 90.0 | ||
| Two | COVID-19 | 99.0 | 99.0 | 99.0 | 99.13 |
| Normal | 99.0 | 99.0 | 99.0 | ||
| Two | COVID-19 | 1.00 | 97.0 | 98.0 | 98.47 |
| Bacterial Pneumonia | 97.0 | 1.00 | 98.0 | ||
| Two | COVID-19 | 1.00 | 98.0 | 99.0 | 98.91 |
| Viral Pneumonia | 98.0 | 1.00 | 99.0 |
| Dataset | Class | Task | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| Dataset-1 | Four | COVID-19 | 99.0 | 94.0 | 97.0 | 88.79 |
| Normal | 94.0 | 91.0 | 92.0 | |||
| B. Pneumonia | 80.0 | 94.0 | 87.0 | |||
| V. Pneumonia | 83.0 | 77.0 | 80.0 | |||
| Dataset-2 | Four | COVID-19 | 1.00 | 1.00 | 1.00 | 99.46 |
| Normal | 1.00 | 1.00 | 1.00 | |||
| B. Pneumonia | 1.00 | 98.0 | 99.0 | |||
| V. Pneumonia | 98.0 | 1.00 | 99.0 |
| Class | Dataset-01 | |||
|---|---|---|---|---|
| Train_acc | Val_acc | Train_loss | Val_loss | |
| Three (COVID vs. pneu_vir vs. normal) | 0.9912 | 0.9311 | 0.0088 | 0.0689 |
| Two (COVID vs. pneu_vir) | 0.9935 | 0.9891 | 0.0109 | 0.0065 |
| Two (COVID vs. pneu_bac) | 0.9974 | 0.9847 | 0.0026 | 0.0153 |
| Folds | Precision(%) | Recall(%) | F1-Score%) | Accuracy%) |
|---|---|---|---|---|
| Fold-1 | 89.25 | 88.75 | 88.5 | 88.80 |
| Fold-2 | 91.5 | 91.75 | 91.75 | 91.64 |
| Fold-3 | 88.75 | 88.75 | 88.5 | 88.59 |
| Fold-4 | 92.25 | 91.75 | 91.75 | 91.73 |
| Fold-5 | 88.5 | 88.25 | 87.75 | 88.06 |
| Average | 90.05 | 89.85 | 89.65 | 89.76 |
| Folds | Precision | Recall | F1-Score | Accuracy(%) |
|---|---|---|---|---|
| Fold-1 | 0.985 | 0.985 | 0.985 | 98.48 |
| Fold-2 | 0.99 | 0.99 | 0.99 | 98.91 |
| Fold-3 | 0.975 | 0.975 | 0.98 | 97.61 |
| Fold-4 | 0.985 | 0.985 | 0.98 | 98.04 |
| Fold-5 | 0.985 | 0.985 | 0.98 | 98.26 |
| Average | 0.984 | 0.984 | 0.983 | 98.26 |
| Model | Class | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|---|
| VGG19 | 4 | 90 | 88.5 | 89 | 88.98 |
| ResNet50 | 4 | 88 | 87.25 | 87.5 | 87.5 |
| InceptionV3 | 4 | 87.5 | 87.25 | 87.5 | 87.39 |
| VGG19 | 3 | 93.25 | 94 | 93 | 94.35 |
| ResNet50 | 3 | 95 | 93.5 | 94 | 94.88 |
| InceptionV3 | 3 | 93 | 92.25 | 92.75 | 93.27 |
| VGG19 | 2 | 99 | 98.5 | 98 | 98.5 |
| ResNet50 | 2 | 99 | 98 | 98 | 98.33 |
| InceptionV3 | 2 | 0.98 | 97 | 98 | 97.67 |
| Proposed model (on Dataset-1) | 4 | 90.05 | 89.85 | 89.65 | 89.76 |
| 3 | 0.97 | 89.85 | 0.9633 | 97.22 | |
| 2 | 0.99 | 89.85 | 0.99 | 99.13 | |
| Proposed model (on Dataset-2) | 4 | 99.5 | 99.4 | 99.5 | 99.46 |
| Proposed model (on Dataset-3) | 2 | 98.4 | 98.4 | 98.3 | 98.26 |
| Author | Architecture | Number of Images | Class | Accuracy (%) |
|---|---|---|---|---|
| Khan et al. [57] | CoroNet | 297 COVID-19, 330 bacterial pneumonia, 310 normal, 327 viral pneumonia images. | 4 | 89.6 |
| 3 | 95 | |||
| 2 | 99 | |||
| Mahmud et al. [64] | CovXNet | 305 COVID-19, 305 bacterial pneumonia, 305 normal, 305 viral pneumonia images. | 4 | 90.3 |
| 3 | 89.6 | |||
| 2 | 94.7 | |||
| Ammar et al. [52] | 6 pre-trained models | 150 COVID-19, 150 normal, 150 pneumonia images. | 3 | 91.28 |
| Mousavi Z et al. [46] | Developed LSTM network | 800 COVID-19, 942 viral pneumonia, 939 healthy cases images. | 3 | 90.0 |
| Arsenovic et al. [60] | ResNetCOVID-19 | 434 COVID-19, 1100 normal, 1100 bacterial pneumonia. | 3 | 94.1 |
| Hemdan et al. [79] | COVIDXNet | 25 COVID-19 and 25 normal images. | 2 | 90 |
| Sethy et al. [58] | ResNet50 plus SVM | 25 COVID-19 and 25 non-COVID-19. | 2 | 95.38 |
| Proposed model (Dataset-1) | Tuned ResNet50V2 | 1143 COVID-19, 1150 viral pneumonia, 1150 bacterial pneumonia, 1150 normal images. | 4 | 89.76 |
| 3 | 97.22 | |||
| 2 | 99.13 | |||
| Proposed model (Dataset-2) | Tuned ResNet50V2 | 1143 COVID-19, 1150 viral pneumonia, 1150 bacterial pneumonia, 1150 normal images. | 4 | 99.46 |
| Proposed model (Dataset-3) | Tuned ResNet50V2 | 1143 COVID-19, 1150 adult pneumonia. | 2 | 98.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahamed, M.K.U.; Islam, M.M.; Uddin, M.A.; Akhter, A.; Acharjee, U.K.; Paul, B.K.; Moni, M.A. DTLCx: An Improved ResNet Architecture to Classify Normal and Conventional Pneumonia Cases from COVID-19 Instances with Grad-CAM-Based Superimposed Visualization Utilizing Chest X-ray Images. Diagnostics 2023, 13, 551. https://doi.org/10.3390/diagnostics13030551
Ahamed MKU, Islam MM, Uddin MA, Akhter A, Acharjee UK, Paul BK, Moni MA. DTLCx: An Improved ResNet Architecture to Classify Normal and Conventional Pneumonia Cases from COVID-19 Instances with Grad-CAM-Based Superimposed Visualization Utilizing Chest X-ray Images. Diagnostics. 2023; 13(3):551. https://doi.org/10.3390/diagnostics13030551
Chicago/Turabian StyleAhamed, Md. Khabir Uddin, Md Manowarul Islam, Md. Ashraf Uddin, Arnisha Akhter, Uzzal Kumar Acharjee, Bikash Kumar Paul, and Mohammad Ali Moni. 2023. "DTLCx: An Improved ResNet Architecture to Classify Normal and Conventional Pneumonia Cases from COVID-19 Instances with Grad-CAM-Based Superimposed Visualization Utilizing Chest X-ray Images" Diagnostics 13, no. 3: 551. https://doi.org/10.3390/diagnostics13030551
APA StyleAhamed, M. K. U., Islam, M. M., Uddin, M. A., Akhter, A., Acharjee, U. K., Paul, B. K., & Moni, M. A. (2023). DTLCx: An Improved ResNet Architecture to Classify Normal and Conventional Pneumonia Cases from COVID-19 Instances with Grad-CAM-Based Superimposed Visualization Utilizing Chest X-ray Images. Diagnostics, 13(3), 551. https://doi.org/10.3390/diagnostics13030551








