Conventional and Deep-Learning-Based Image Reconstructions of Undersampled K-Space Data of the Lumbar Spine Using Compressed Sensing in MRI: A Comparative Study on 20 Subjects
Abstract
1. Introduction
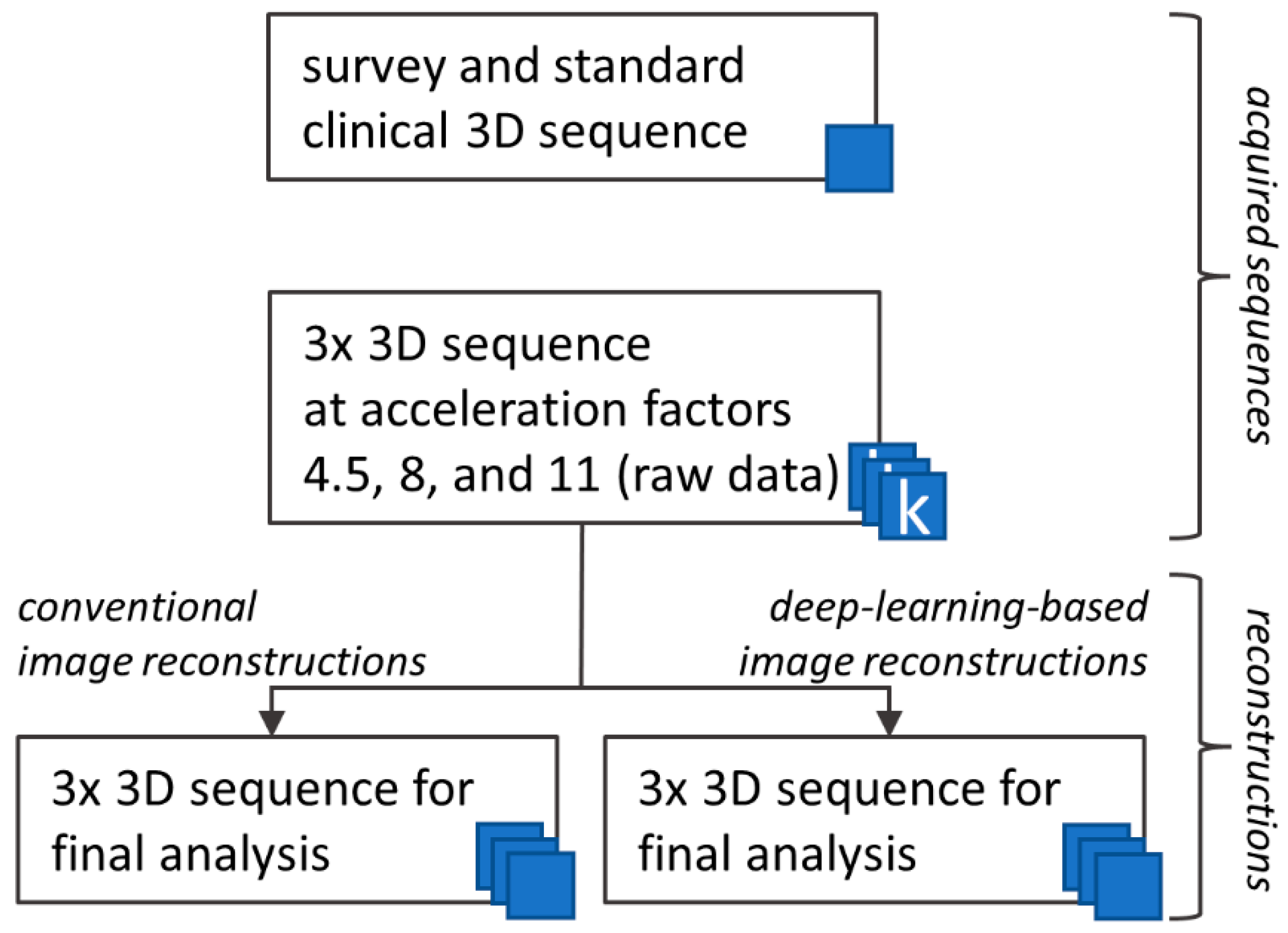
2. Materials and Methods
2.1. Study Population
2.2. MRI Protocol
2.3. Image Analysis
2.4. Objective Image Analysis: ROI-Based
2.5. Objective Image Analysis: Pixel-Based
2.6. Subjective Image Analysis
2.7. Statistical Analysis
3. Results
3.1. Study Population
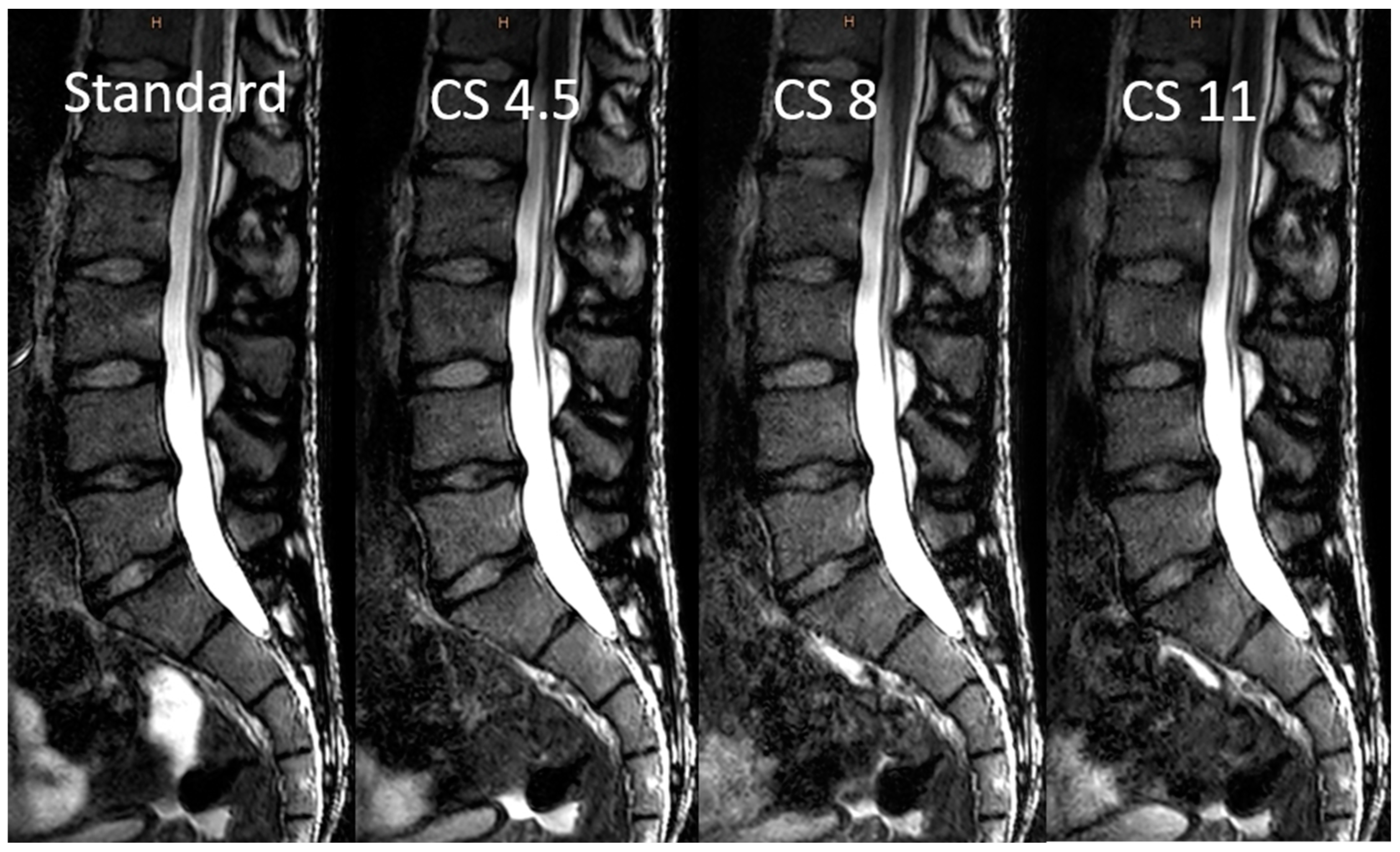
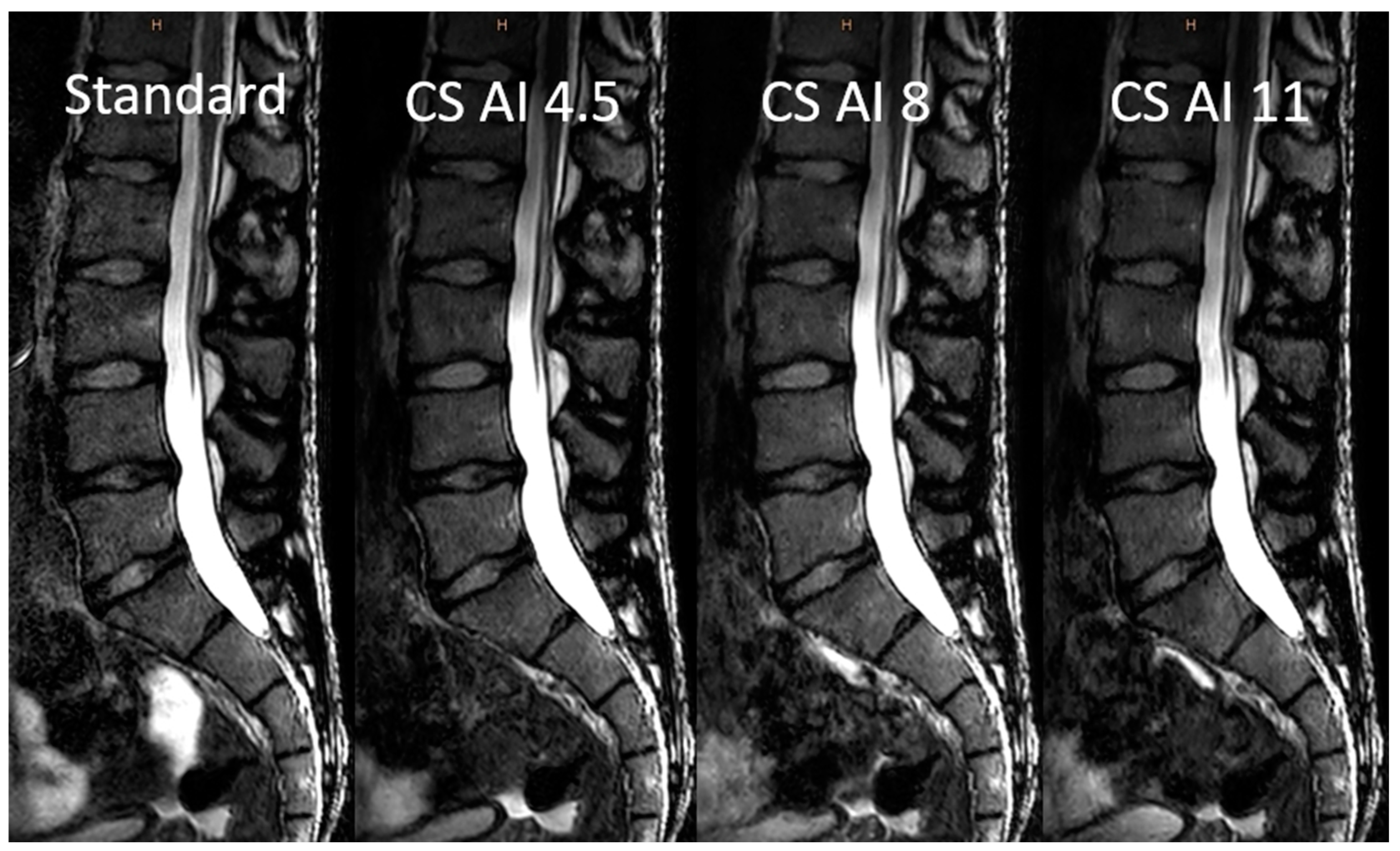
3.2. Image Analysis
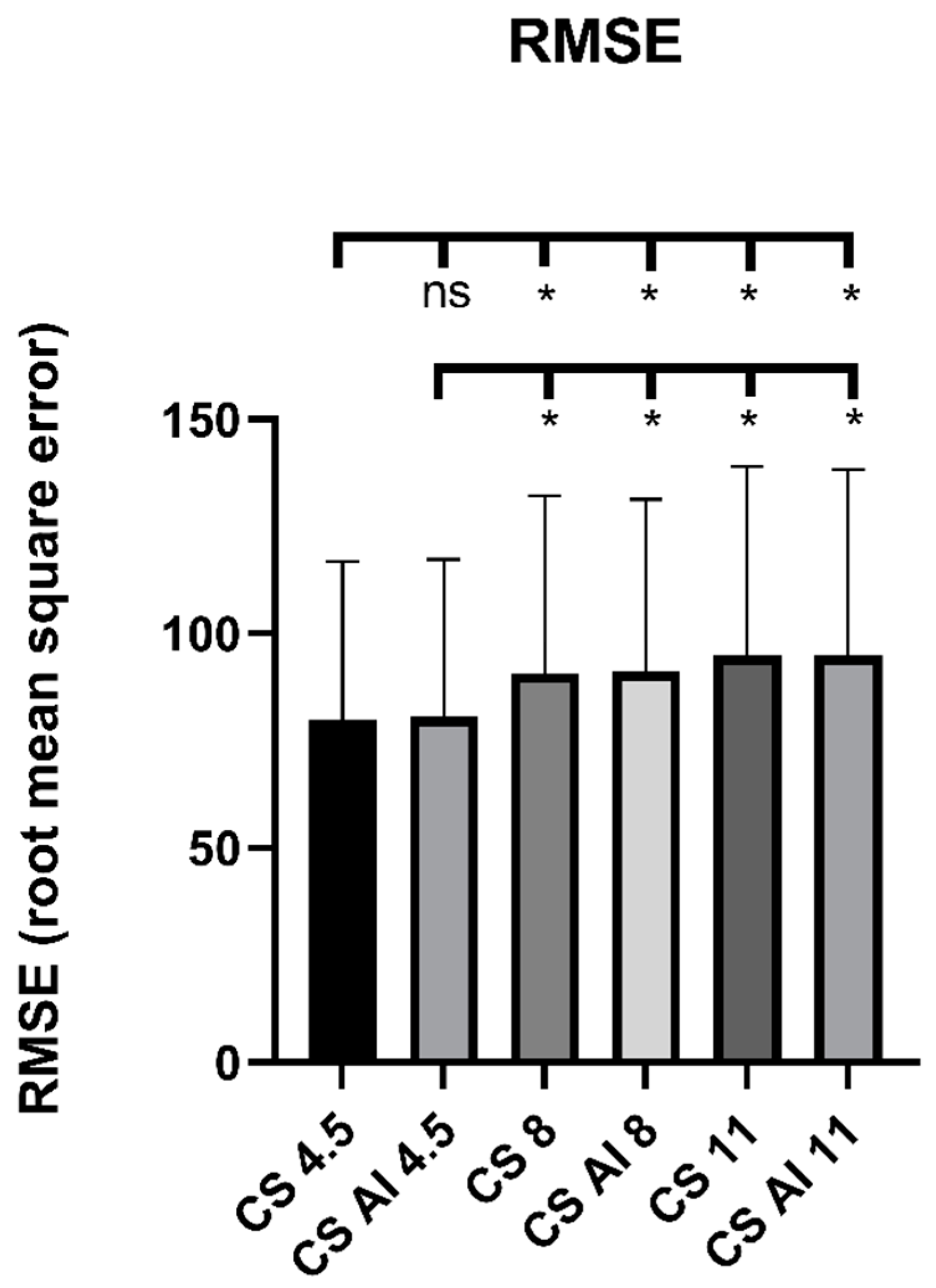
3.3. Objective Image Analysis
3.4. Subjective Image Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Health Care Use—Magnetic Resonance Imaging (MRI) Exams—OECD Data. Available online: https://data.oecd.org/healthcare/magnetic-resonance-imaging-mri-exams.htm (accessed on 2 December 2022).
- Obyn, C.; Cleemput, I. The capital cost and productivity of MRI in a Belgian setting. JBR-BTR 2010, 93, 92. [Google Scholar] [CrossRef] [PubMed]
- Bratke, G.; Rau, R.; Weiss, K.; Kabbasch, C.; Sircar, K.; Morelli, J.N.; Persigehl, T.; Maintz, D.; Giese, D.; Haneder, S. Accelerated MRI of the Lumbar Spine Using Compressed Sensing: Quality and Efficiency. J. Magn. Reson. Imaging 2019, 49, e164–e175. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.; Knapp, K.; Benwell, M. An evaluation of MRI lumbar spine scans within a community-based diagnostic setting. Musculoskeletal Care 2021, 19, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Donoho, D.L. Compressed sensing. IEEE Trans. Inf. Theory 2006, 52, 1289–1306. [Google Scholar] [CrossRef]
- Lustig, M.; Donoho, D.; Pauly, J.M. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 2007, 58, 1182–1195. [Google Scholar] [CrossRef]
- Moratal, D.; Vallés-Luch, A.; Martí-Bonmati, L.; Brummers, M.E. k-Space tutorial: An MRI educational tool for a better understanding of k-space. Biomed. Imaging Interv. J. 2008, 4, e15. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, Y.H.; Song, H.T.; Suh, J.S. Rapid acquisition of magnetic resonance imaging of the shoulder using three-dimensional fast spin echo sequence with compressed sensing. Magn. Reson. Imaging 2017, 42, 152–157. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, Y.H.; Suh, J.S. Accelerating knee MR imaging: Compressed sensing in isotropic three-dimensional fast spin-echo sequence. Magn. Reson. Imaging 2018, 46, 90–97. [Google Scholar] [CrossRef]
- Kijowski, R.; Rosas, H.; Samsonov, A.; King, K.; Peters, R.; Liu, F. Knee imaging: Rapid three-dimensional fast spin-echo using compressed sensing. J. Magn. Reson. Imaging 2017, 45, 1712–1722. [Google Scholar] [CrossRef]
- Altahawi, F.F.; Blount, K.J.; Morley, N.P.; Raithel, E.; Omar, I.M. Comparing an accelerated 3D fast spin-echo sequence (CS-SPACE) for knee 3-T magnetic resonance imaging with traditional 3D fast spin-echo (SPACE) and routine 2D sequences. Skeletal Radiol. 2017, 46, 7–15. [Google Scholar] [CrossRef]
- Iuga, A.I.; Abdullayev, N.; Weiss, K.; Haneder, S.; Brüggemann-Bratke, L.; Maintz, D.; Rau, R.; Bratke, G. Accelerated MRI of the knee. Quality and efficiency of compressed sensing. Eur. J. Radiol. 2020, 132, 109273. [Google Scholar] [CrossRef]
- Glockner, J.F.; Hu, H.H.; Stanley, D.W.; Angelos, L.; King, K. Parallel MR Imaging: A User’s Guide1. Radio Graphics 2005, 25, 1279–1297. [Google Scholar] [CrossRef] [PubMed]
- Bratke, G.; Rau, R.; Kabbasch, C.; Zäske, C.; Maintz, D.; Haneder, S.; Große Hokamp, N.; Persigehl, T.; Siedek, F.; Weiss, K. Speeding up the clinical routine: Compressed sensing for 2D imaging of lumbar spine disc herniation. Eur. J. Radiol. 2021, 140, 109738. [Google Scholar] [CrossRef]
- Peeters, H.; Chung, H.; Valvano, G.; Van Gemert, J.; De Weerdt, E.; Van De Ven, K.; Smartspeed, W. Philips SmartSpeed Whitepaper No compromise Image Quality and Speed at Your Fingertips. Available online: https://images.philips.com/is/content/PhilipsConsumer/Campaigns/HC20140401_DG/Documents/HC05072022-white_paper_philips_smartspeed.pdf (accessed on 30 November 2022).
- Pezzotti, N.; Yousefi, S.; Elmahdy, M.S.; van Gemert, J.H.F.; Schuelke, C.; Doneva, M.; Nielsen, T.; Kastryulin, S.; Lelieveldt, B.P.F.; van Osch, M.J.P.; et al. An adaptive intelligence algorithm for undersampled knee MRI reconstruction. IEEE Access 2020, 8, 204825–204838. [Google Scholar] [CrossRef]
- Pezzotti, N.; de Weerdt, E.; Yousefi, S.; Elmahdy, M.S.; van Gemert, J.; Schülke, C.; Doneva, M.; Nielsen, T.; Kastryulin, S.; Lelieveldt, B.P.F.; et al. Adaptive-CS-Net: FastMRI with Adaptive Intelligence. arXiv 2019, arXiv:1912.12259. [Google Scholar] [CrossRef]
- Knoll, F.; Zbontar, J.; Sriram, A.; Muckley, M.J.; Bruno, M.; Defazio, A.; Parente, M.; Geras, K.J.; Katsnelsn, J.; Chandarana, H.; et al. FastMRI: A publicly available raw k-space and DICOM dataset of knee images for accelerated MR image reconstruction using machine learning. Radiol. Artif. Intell. 2020, 2, e190007. [Google Scholar] [CrossRef]
- FastMRI. Available online: https://fastmri.org/leaderboards/challenge/2019/ (accessed on 30 November 2022).
- Foreman, S.C.; Neumann, J.; Han, J.; Harrasser, N.; Weiss, K.; Peeters, J.M.; Karampinos, D.C.; Makowski, M.R.; Gersing, A.S.; Woertler, K. Deep learning-based acceleration of Compressed Sense MR imaging of the ankle. Eur. Radiol. 2022, 32, 8376–8385. [Google Scholar] [CrossRef]
- Roudsari, B.; Jarvik, J.G. Lumbar spine MRI for low back pain: Indications and yield. Am. J. Roentgenol. 2010, 195, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, N.J. Magnetic resonance imaging for low back pain: Indications and limitations. Postgrad. Med. J. 2010, 86, 7–11. [Google Scholar] [CrossRef]
- Geerts-Ossevoort, L.; De Weerdt, E.; Duijndam, A.; Van Ijperen, G.; Peeters, H.; Doneva, M.; Nijenhuis, M.; Huang, A. Compressed SENSE Speed Done Right. Every time. Available online: https://www.philips.com/c-dam/b2bhc/de/resourcecatalog/landingpages/ingeniaelition/White_Paper_Compressed_SENSE-opt.pdf (accessed on 30 November 2022).
- Van Der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. Scikit-image: Image processing in python. PeerJ 2014, 2014, e453. [Google Scholar] [CrossRef]
- RANDOM.ORG—List Randomizer. Available online: https://www.random.org/lists/ (accessed on 28 November 2022).
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef] [PubMed]
- Quantify Interrater Agreement with Kappa. Available online: https://www.graphpad.com/quickcalcs/kappa1/ (accessed on 28 November 2022).
- Sartoretti, E.; Wyss, M.; Alfieri, A.; Binkert, C.A.; Erne, C.; Sartoretti-Schefer, S.; Sartoretti, T. Introduction and reproducibility of an updated practical grading system for lumbar foraminal stenosis based on high-resolution MR imaging. Sci. Rep. 2021, 11, 12000. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, L.; Katemann, C.; Isaak, A.; Mesropyan, N.; Wichtmann, B.; Kravchenko, D.; Endler, C.; Kuetting, D.; Pieper, C.; Ellinger, J.; et al. T2 Turbo Spin Echo with Compressed Sensing and Propeller Acquisition (Sampling k-Space by Utilizing Rotating Blades) for Fast and Motion Robust Prostate MRI: Comparison With Conventional Acquisition. Investig. Radiol. 2022. ahead of print. [Google Scholar] [CrossRef] [PubMed]

| Sequence | Standard 3D | CS 4.5/CS-AI 4.5 | CS 8/CS-AI 8 | CS 11/CS-AI 11 |
|---|---|---|---|---|
| Echo time (ms) | 168 | 168 | 168 | 168 |
| Repetition time (ms) | 3261 | 3261 | 3261 | 3261 |
| Flip angle (deg.) | 90 | 90 | 90 | 90 |
| Field of view (mm) | 180 × 300 × 90 | 180 × 300 × 90 | 180 × 300 × 90 | 180 × 300 × 90 |
| Gap (mm) | 0 | 0 | 0 | 0 |
| Acquisition voxel size (mm) | 1 × 1 × 1 | 1 × 1 × 1 | 1 × 1 × 1 | 1 × 1 × 1 |
| Reconstruction voxel size (mm) | 0.47 × 0.47 × 0.5 | 0.47 × 0.47 × 0.5 | 0.47 × 0.47 × 0.5 | 0.47 × 0.47 × 0.5 |
| Turbo factor/ echo train length | 64 | 64 | 64 | 64 |
| CS factor | SENSE 2.5 | CS 4.5 | CS 8 | CS 11 |
| Scan time (s) | 427 | 261 | 149 | 109 |
| Saved scan time (s) | 0 | 166 | 278 | 228 |
| Scan time reduction (%) | 0 | 38.88 | 65.11 | 74.47 |
| Standard | CS 4.5 | CS AI 4.5 | CS 8 | CS AI 8 | CS 11 | CS AI 11 | |
|---|---|---|---|---|---|---|---|
| RMSE | - | 79.80 ± 37.03 | 80.53 ± 36.77 | 90.66 ± 41.37 | 91.08 ± 40.26 | 94.87 ± 44.07 | 94.79 ± 43.49 |
| SSIM | - | 0.86 ± 0.04 | 0.86 ± 0.04 | 0.84 ± 0.04 | 0.84 ± 0.04 | 0.83 ± 0.04 | 0.83 ± 0.04 |
| aSNR bone | 6.00 ± 1.69 | 6.51 ± 1.79 | 6.60 ± 1.97 | 5.60 ± 1.38 | 6.18 ± 1.64 | 5.05 ± 1.21 | 5.72 ± 1.70 |
| aSNR spinal cord | 11.36 ± 4.18 | 10.39 ± 3.65 | 10.19 ± 3.79 | 10.56 ± 3.71 | 10.64 ± 3.61 | 10.40 ± 3.36 | 10.22 ± 3.59 |
| aSNR CSF | 26.26 ± 14.05 | 28.17 ± 13.53 | 28.58 ± 13.58 | 26.89 ± 13.17 | 27.01 ± 13.06 | 26.42 ± 12.91 | 27.69 ± 13.79 |
| aCNR bone/CSF | 17.41 ± 7.88 | 19.70 ± 7.70 | 19.79 ± 7.72 | 18.46 ± 7.24 | 18.83 ± 7.25 | 17.63 ± 6.48 | 18.57 ± 6.60 |
| aCNR spinal cord/CSF | 21.72 ± 11.39 | 23.18 ± 9.90 | 23.36 ± 9.89 | 22.26 ± 9.79 | 22.27 ± 9.79 | 21.92 ± 9.58 | 22.63 ± 9.43 |
| Standard | CS 4.5 | CS AI 4.5 | CS 8 | CS AI 8 | CS 11 | CS AI 11 | |
|---|---|---|---|---|---|---|---|
| Bone marrow | 0.568 | 0.627 | 0.743 | 0.914 | 0.691 | 0.733 | 0.915 |
| Intervertebral discs | 0.559 | 0.84 | 0.916 | 0.731 | 0.69 | 0.821 | 0.839 |
| Spinal cord | 0.836 | 0.655 | 0.769 | 0.701 | 0.846 | 0.844 | 0.73 |
| CSF | 0.914 | 1.000 | 0.856 | 0.774 | 0.733 | 0.685 | 0.688 |
| Nerve roots | 0.713 | 0.841 | 0.918 | 0.669 | 0.71 | 0.491 | 0.827 |
| Neuroforamina | 0.931 | 0.853 | 0.854 | 0.712 | 0.844 | 0.767 | 0.838 |
| Overall image impression | 0.914 | 0.747 | 0.923 | 0.73 | 0.843 | 0.754 | 0.766 |
| Standard | CS 4.5 | CS AI 4.5 | CS 8 | CS AI 8 | CS 11 | CS AI 11 | |
|---|---|---|---|---|---|---|---|
| Bone marrow | 3.10 ± 0.81 | 3.68 ± 0.80 | 4.03 ± 0.80 | 2.98 ± 0.66 | 3.68 ± 0.76 | 2.45 ± 0.71 | 3.23 ± 0.73 |
| Intervertebral discs | 3.00 ± 0.64 | 3.28 ± 0.68 | 3.90 ± 0.74 | 2.78 ± 0.58 | 3.63 ± 0.81 | 2.48 ± 0.60 | 3.10 ± 0.74 |
| Spinal cord | 3.08 ± 0.73 | 3.48 ± 0.72 | 3.85 ± 0.77 | 2.88 ± 0.82 | 3.70 ± 0.79 | 2.83 ± 0.75 | 3.00 ± 0.68 |
| CSF | 3.43 ± 0.75 | 3.70 ± 0.65 | 3.83 ± 0.87 | 3.20 ± 0.79 | 3.78 ± 0.62 | 3.05 ± 0.75 | 3.45 ± 0.75 |
| Nerve roots | 3.72 ± 0.88 | 3.58 ± 0.75 | 3.75 ± 0.87 | 3.38 ± 0.70 | 3.70 ± 0.85 | 2.98 ± 0.77 | 3.28 ± 0.75 |
| Neuroforamina | 3.45 ± 0.96 | 3.65 ± 0.86 | 3.73 ± 0.88 | 3.05 ± 0.90 | 3.68 ± 0.76 | 2.80 ± 0.79 | 3.28 ± 0.78 |
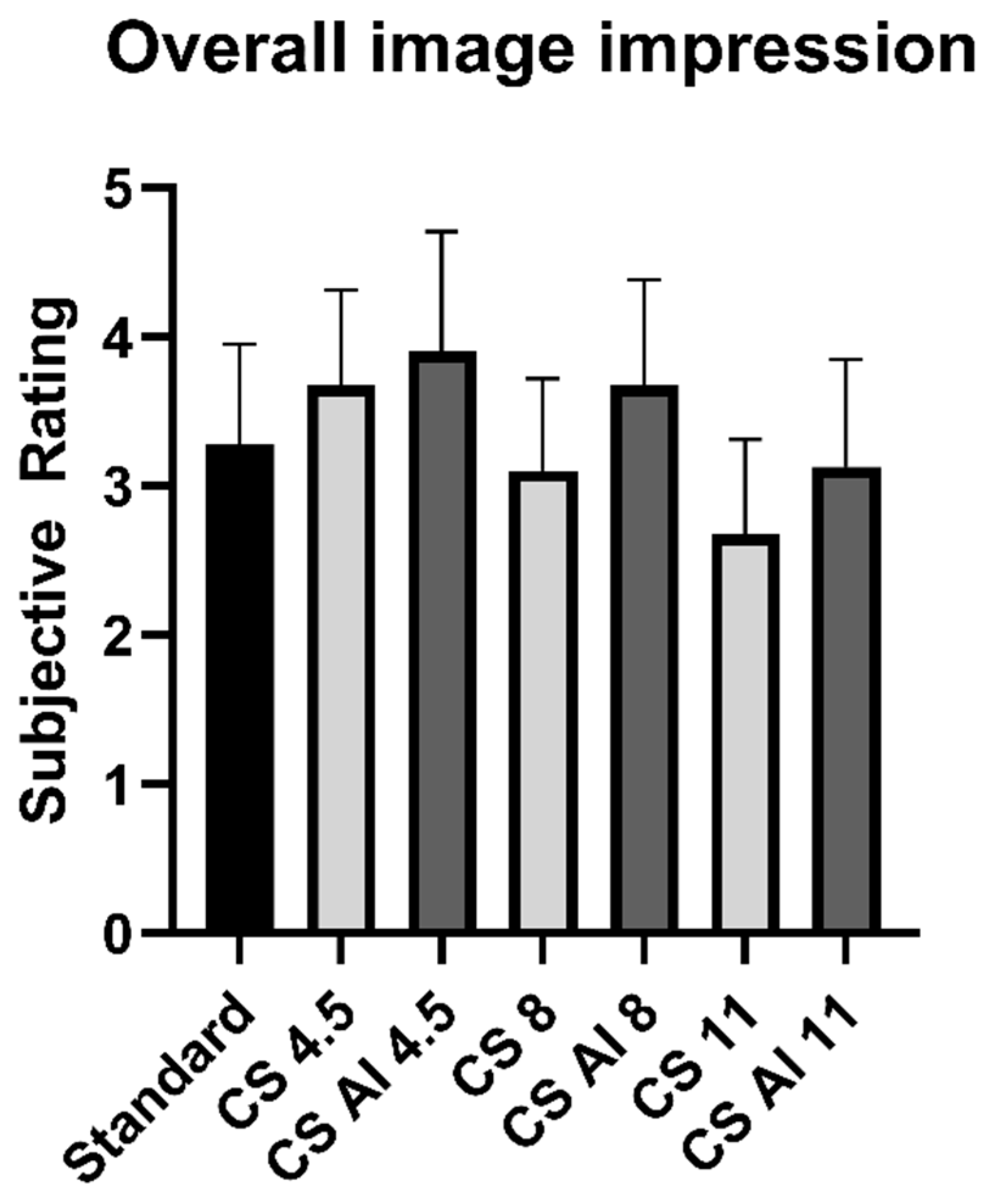
| Overall impression | 3.28 ± 0.68 | 3.68 ± 0.66 | 3.90 ± 0.81 | 3.10 ± 0.67 | 3.68 ± 0.76 | 2.68 ± 0.69 | 3.13 ± 0.76 |
| Standard | CS 4.5 | CS AI 4.5 | CS 8 | CS AI 8 | CS 11 | CS AI 11 | |
|---|---|---|---|---|---|---|---|
| Acceptable for use in a clinical context | 92.50% | 97.50% | 97.50% | 75.00% | 95.00% | 32.50% | 70.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fervers, P.; Zaeske, C.; Rauen, P.; Iuga, A.-I.; Kottlors, J.; Persigehl, T.; Sonnabend, K.; Weiss, K.; Bratke, G. Conventional and Deep-Learning-Based Image Reconstructions of Undersampled K-Space Data of the Lumbar Spine Using Compressed Sensing in MRI: A Comparative Study on 20 Subjects. Diagnostics 2023, 13, 418. https://doi.org/10.3390/diagnostics13030418
Fervers P, Zaeske C, Rauen P, Iuga A-I, Kottlors J, Persigehl T, Sonnabend K, Weiss K, Bratke G. Conventional and Deep-Learning-Based Image Reconstructions of Undersampled K-Space Data of the Lumbar Spine Using Compressed Sensing in MRI: A Comparative Study on 20 Subjects. Diagnostics. 2023; 13(3):418. https://doi.org/10.3390/diagnostics13030418
Chicago/Turabian StyleFervers, Philipp, Charlotte Zaeske, Philip Rauen, Andra-Iza Iuga, Jonathan Kottlors, Thorsten Persigehl, Kristina Sonnabend, Kilian Weiss, and Grischa Bratke. 2023. "Conventional and Deep-Learning-Based Image Reconstructions of Undersampled K-Space Data of the Lumbar Spine Using Compressed Sensing in MRI: A Comparative Study on 20 Subjects" Diagnostics 13, no. 3: 418. https://doi.org/10.3390/diagnostics13030418
APA StyleFervers, P., Zaeske, C., Rauen, P., Iuga, A.-I., Kottlors, J., Persigehl, T., Sonnabend, K., Weiss, K., & Bratke, G. (2023). Conventional and Deep-Learning-Based Image Reconstructions of Undersampled K-Space Data of the Lumbar Spine Using Compressed Sensing in MRI: A Comparative Study on 20 Subjects. Diagnostics, 13(3), 418. https://doi.org/10.3390/diagnostics13030418






