Performance Evaluation of Three Antibody Binding Assays, a Neutralizing Antibody Assay, and an Interferon-Gamma Release Assay for SARS-CoV-2 According to Vaccine Type in Vaccinated Group
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Samples
2.2. SARS-CoV-2 IgG Antibody Binding Assays
2.3. SARS-CoV-2 Neutralizing Antibody Assay
2.4. Interferon-Gamma Release Assay for SARS-CoV-2
2.5. Statistical Analysis
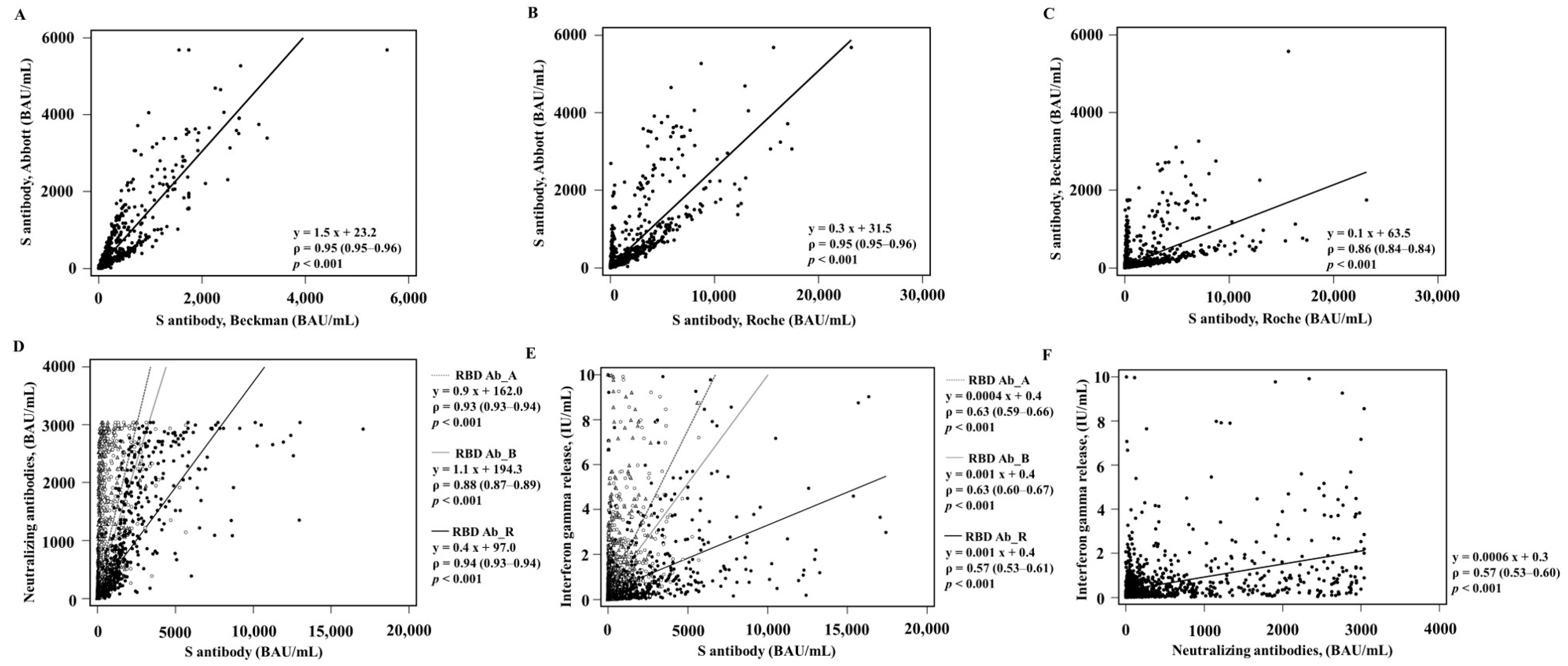
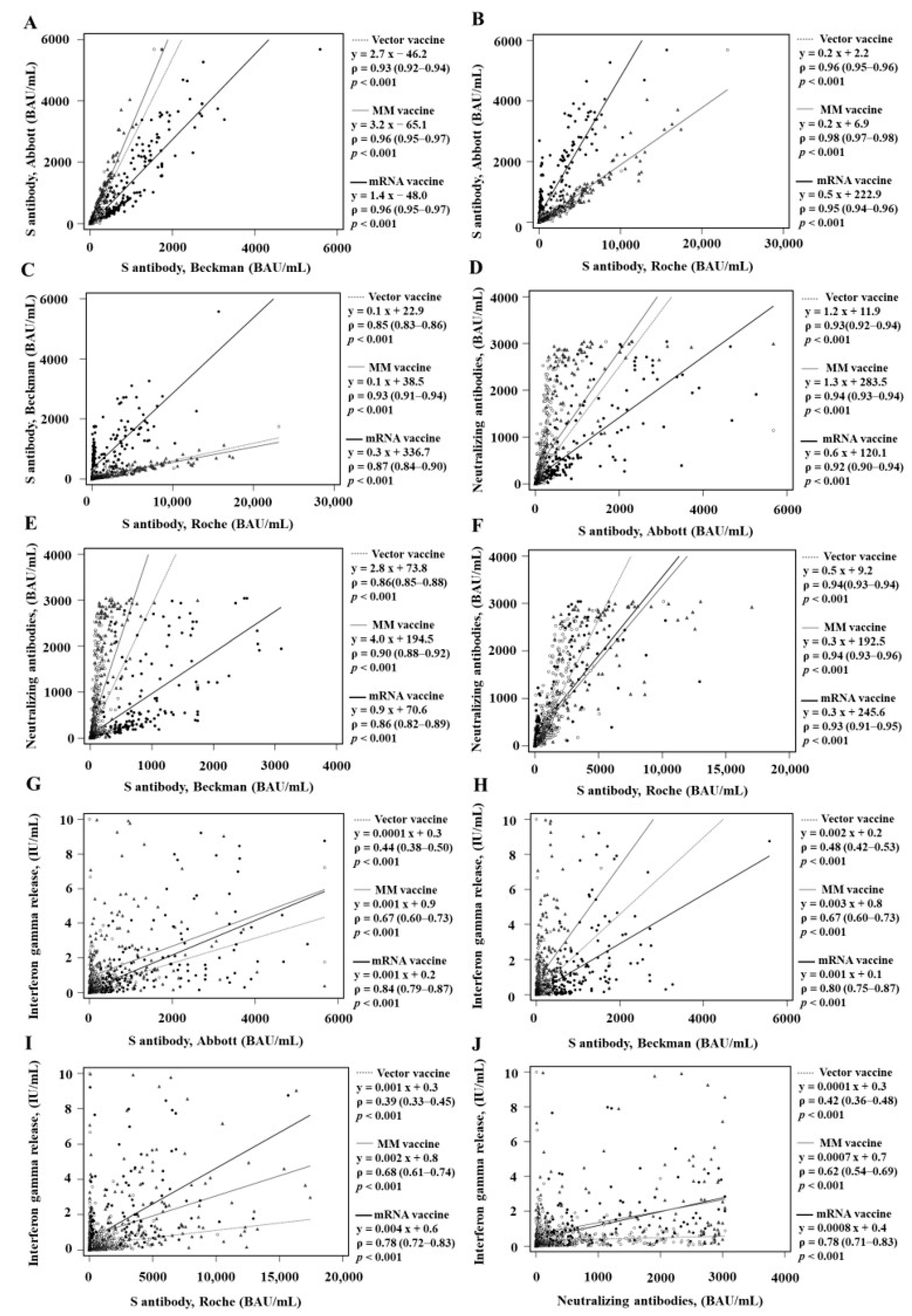
3. Results
3.1. Comparison of Qualitative and Quantitative Values in the Vaccinated Group
3.2. Comparison of Qualitative and Quantitative Values according to the Vaccine Types
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, A.; Tiwari, S.; Deb, M.K.; Marty, J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): A global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054. [Google Scholar] [CrossRef] [PubMed]
- Dopico, X.C.; Ols, S.; Loré, K.; Hedestam, G.B.K. Immunity to SARS-CoV-2 induced by infection or vaccination. J. Intern. Med. 2022, 291, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-Y.; Kim, Y.; Lee, K.-M.; Jang, E.-J.; Woo, C.-H.; Hong, C.-U.; Choi, S.-T.; Xayaheuang, S.; Jang, J.-G.; Ahn, J.-H.; et al. Humoral and cellular responses to COVID-19 vaccines in SARS-CoV-2 infection-naïve and -recovered Korean individuals. Vaccines 2022, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Nam, M.; Yun, S.G.; Kim, S.-W.; Kim, C.G.; Cha, J.H.; Lee, C.; Kang, S.; Park, S.G.; Kim, S.B.; Lee, K.-B.; et al. Humoral and cellular immune responses to vector, mix-and-match, or mRNA vaccines against SARS-CoV-2 and the relationship between the two immune responses. Microbiol. Spectr. 2022, 10, e0249521. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.; Ryu, J.H.; Jang, J.H.; Bae, H.; Yoo, S.-H.; Choi, A.-R.; Jo, S.J.; Lim, J.; Lee, J.; Ryu, H.; et al. Comparison of SARS-CoV-2 antibody responses and seroconversion in COVID-19 patients using twelve commercial immunoassays. Ann. Lab. Med. 2021, 41, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 diagnosis—A review of current methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef] [PubMed]
- Kemp, T.J.; Hempel, H.A.; Pan, Y.; Roy, D.; Cherry, J.; Lowy, D.R.; Pinto, L.A. Assay harmonization study to measure immune response to SARS-CoV-2 infection and vaccines: A serology methods study. Microbiol. Spectr. 2023, 11, e05353-22. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, Y.; Greig, M.; Liu, G.; Driedger, M.; Langlois, M.-A. Humoral responses and serological assays in SARS-CoV-2 infections. Front. Immunol. 2020, 11, 610688. [Google Scholar] [CrossRef]
- Kim, S.J.; Yao, Z.; Marsh, M.C.; Eckert, D.M.; Kay, M.S.; Lyakisheva, A.; Pasic, M.; Bansal, A.; Birnboim, C.; Jha, P.; et al. Homogeneous surrogate virus neutralization assay to rapidly assess neutralization activity of anti-SARS-CoV-2 antibodies. Nat. Commun. 2022, 13, 3716. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.-C.; Tiu, C.; Hu, Z.; Chen, V.C.-W.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef]
- Ameratunga, R.; Woon, S.-T.; Jordan, A.; Longhurst, H.; Leung, E.; Steele, R.; Lehnert, K.; Snell, R.; Brooks, A.E.S. Perspective: Diagnostic laboratories should urgently develop T cell assays for SARS-CoV-2 infection. Expert Rev. Clin. Immunol. 2021, 17, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Abate, D.; Saldan, A.; Mengoli, C.; Fiscon, M.; Silvestre, C.; Fallico, L.; Peracchi, M.; Furian, L.; Cusinato, R.; Bonfante, L.; et al. Comparison of cytomegalovirus (CMV) enzyme-linked immunosorbent spot and CMV quantiferon gamma interferon-releasing assays in assessing risk of CMV infection in kidney transplant recipients. J. Clin. Microbiol. 2013, 51, 2501–2507. [Google Scholar] [CrossRef] [PubMed]
- Pai, M.; Denkinger, C.M.; Kik, S.V.; Rangaka, M.X.; Zwerling, A.; Oxlade, O.; Metcalfe, J.Z.; Cattamanchi, A.; Dowdy, D.W.; Dheda, K.; et al. Gamma interferon release assays for detection of mycobacterium tuberculosis infection. Clin. Microbiol. Rev. 2014, 27, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Echeverría, G.; Guevara, Á.; Coloma, J.; Ruiz, A.M.; Vasquez, M.M.; Tejera, E.; de Waard, J.H. Pre-existing T-cell immunity to SARS-CoV-2 in unexposed healthy controls in Ecuador, as detected with a COVID-19 interferon-gamma release assay. Int. J. Infect. Dis. 2021, 105, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-Y.; Lee, Y.-L.; Lin, Y.-C.; Lee, N.-Y.; Liao, C.-H.; Hung, Y.-P.; Lu, M.-C.; Wu, J.-L.; Tseng, W.-P.; Lin, C.-H.; et al. Multicenter evaluation of two chemiluminescence and three lateral flow immunoassays for the diagnosis of COVID-19 and assessment of antibody dynamic responses to SARS-CoV-2 in Taiwan. Emerg. Microbes Infect. 2020, 9, 2157–2168. [Google Scholar] [CrossRef] [PubMed]
- Demonbreun, A.R.; Sancilio, A.; Velez, M.P.; Ryan, D.T.; Saber, R.; Vaught, L.A.; Reiser, N.L.; Hsieh, R.R.; D’Aquila, R.T.; Mustanski, B.; et al. Comparison of IgG and neutralizing antibody responses after one or two doses of COVID-19 mRNA vaccine in previously infected and uninfected individuals. EClinicalMedicine 2021, 38, 101018. [Google Scholar] [CrossRef] [PubMed]
- Lamikanra, A.; Nguyen, D.; Simmonds, P.; Williams, S.; Bentley, E.M.; Rowe, C.; Otter, A.D.; Brooks, T.; Gilmour, K.; Mai, A.; et al. Comparability of six different immunoassays measuring SARS-CoV-2 antibodies with neutralizing antibody levels in convalescent plasma: From utility to prediction. Transfusion 2021, 61, 2837–2843. [Google Scholar] [CrossRef]
- Nandakumar, V.; Profaizer, T.; Lozier, B.K.; Elgort, M.G.; Larragoite, E.T.; Williams, E.S.C.P.; Solis-Leal, A.; Lopez, B.J.B.; Berges, B.K.; Planelles, V.; et al. Evaluation of a surrogate enzyme-linked immunosorbent assay-based severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cPass neutralization antibody detection assay and correlation with immunoglobulin G commercial serology assays. Arch. Pathol. Lab. Med. 2021, 145, 1212–1220. [Google Scholar] [CrossRef]
- Jeong, S.; Lee, N.; Lee, S.K.; Cho, E.-J.; Hyun, J.; Park, M.-J.; Song, W.; Jung, E.J.; Woo, H.; Bin Seo, Y.; et al. Comparison of the results of five SARS-CoV-2 antibody assays before and after the first and second ChAdOx1 nCoV-19 vaccinations among health care workers: A prospective multicenter study. J. Clin. Microbiol. 2021, 59, e0178821. [Google Scholar] [CrossRef]
- Krüttgen, A.; Klingel, H.; Haase, G.; Haefner, H.; Imöhl, M.; Kleines, M. Evaluation of the QuantiFERON SARS-CoV-2 interferon-ɣ release assay in mRNA-1273 vaccinated health care workers. J. Virol. Methods 2021, 298, 114295. [Google Scholar] [CrossRef]
- Kalimuddin, S.; Tham, C.Y.L.; Qui, M.; de Alwis, R.; Sim, J.X.Y.; Lim, J.M.E.; Tan, H.-C.; Syenina, A.; Zhang, S.L.; Le Bert, N.; et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med 2021, 2, 682–688.e4. [Google Scholar] [CrossRef] [PubMed]
- Parry, H.; Bruton, R.; Tut, G.; Ali, M.; Stephens, C.; Greenwood, D.; Faustini, S.; Hughes, S.; Huissoon, A.; Meade, R.; et al. Immunogenicity of single vaccination with BNT162b2 or ChAdOx1 nCoV-19 at 5–6 weeks post vaccine in participants aged 80 years or older: An exploratory analysis. Lancet Healthy Longev. 2021, 2, e554–e560. [Google Scholar] [CrossRef] [PubMed]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Akoglu, H. User’s guide to correlation coefficients. Turk. J. Emerg. Med. 2018, 18, 91–93. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Kontou, P.I.; Braliou, G.G.; Dimou, N.L.; Nikolopoulos, G.; Bagos, P.G. Antibody tests in detecting SARS-CoV-2 infection: A meta-analysis. Diagnostics 2020, 10, 319. [Google Scholar] [CrossRef]
- Pinto, D.; Park, Y.-J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Almendro-Vázquez, P.; Laguna-Goya, R.; Paz-Artal, E. Defending against SARS-CoV-2: The T cell perspective. Front. Immunol. 2023, 14, 1107803. [Google Scholar] [CrossRef]
- Valcourt, E.J.; Manguiat, K.; Robinson, A.; Chen, J.C.-Y.; Dimitrova, K.; Philipson, C.; Lamoureux, L.; McLachlan, E.; Schiffman, Z.; Drebot, M.A.; et al. Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Diagn. Microbiol. Infect. Dis. 2021, 99, 115294. [Google Scholar] [CrossRef]
- Kristiansen, P.A.; Page, M.; Bernasconi, V.; Mattiuzzo, G.; Dull, P.; Makar, K.; Plotkin, S.; Knezevic, I. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet 2021, 397, 1347–1348. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, S.; Karapanagiotidis, T.; Khvorov, A.; Douros, C.; Mordant, F.; Bond, K.; Druce, J.; Williamson, D.A.; Purcell, D.; Lewin, S.R.; et al. Evaluation of 6 commercial SARS-CoV-2 serology assays detecting different antibodies for clinical testing and serosurveillance. Open Forum Infect. Dis. 2021, 8, ofab239. [Google Scholar] [CrossRef] [PubMed]
- Groß, R.; Zanoni, M.; Seidel, A.; Conzelmann, C.; Gilg, A.; Krnavek, D.; Erdemci-Evin, S.; Mayer, B.; Hoffmann, M.; Pöhlmann, S.; et al. Heterologous ChAdOx1 nCoV-19 and BNT162b2 prime-boost vaccination elicits potent neutralizing antibody responses and T cell reactivity against prevalent SARS-CoV-2 variants. EBioMedicine 2022, 75, 103761. [Google Scholar] [CrossRef] [PubMed]
- Hillus, D.; Schwarz, T.; Tober-Lau, P.; Vanshylla, K.; Hastor, H.; Thibeault, C.; Jentzsch, S.; Helbig, E.T.; Lippert, L.J.; Tscheak, P.; et al. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1 nCoV-19 and BNT162b2: A prospective cohort study. Lancet Respir. Med. 2021, 9, 1255–1265. [Google Scholar] [CrossRef]
- Liu, X.; Shaw, R.H.; Stuart, A.S.V.; Greenland, M.; Aley, P.K.; Andrews, N.J.; Cameron, J.C.; Charlton, S.; Clutterbuck, E.A.; Collins, A.M.; et al. Safety and immunogenicity of heterologous versus homologous prime-boost schedules with an adenoviral vectored and mRNA COVID-19 vaccine (Com-COV): A single-blind, randomised, non-inferiority trial. Lancet 2021, 398, 856–869. [Google Scholar] [CrossRef]
| Total (n = 1851) | Vector Vaccine (n = 1264) | MM Vaccine (n = 338) | mRNA Vaccine (n = 249) | p | |
|---|---|---|---|---|---|
| RBD Ab_A (BAU/mL) | |||||
| Total | 25.0 (0.0–117.9) | 22.4 (0.0–83.1) | 24.9 (0.0–286.5) | 133.6 (0.0–864.8) | <0.001 |
| T0 | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.915 |
| T1 | 36.2 (9.4–83.2) | 33.8 (11.2–61.5) | 25.5 (0.0–54.8) | 394.1 (163.2–629.4) | <0.001 |
| T2 | 149.5 (63.5–468.4) | 101.1 (48.1–197.4) | 654.7 (273.6–1164) | 1506 (74.8–2796) | <0.001 |
| RBD Ab_B (BAU/mL) | |||||
| Total | 36.8 (1.8–93.6) | 29.8 (1.8–65.4) | 60.0 (2.3–198.0) | 472.8 (1.5–1056) | <0.001 |
| T0 | 1.0 (0.6–1.7) | 1.0 (0.6–1.7) | 0.9 (0.6–1.6) | 0.9 (0.6–1.6) | 0.432 |
| T1– | 56.1 (29.4–110.6) | 43.2 (25.5–74.5) | 56.3 (29.6–93.9) | 618.9 (462.5–860.9) | <0.001 |
| T2 | 75.4 (42.7–198.0) | 56.0 (32.8–90.4) | 214.6 (95.5–394.9) | 1318 (654.7–1738) | <0.001 |
| RBD Ab_R (BAU/mL) | |||||
| Total | 31.1 (0.0–411.9) | 33.5 (0.0–318.1) | 20.5 (0.0–1766) | 64.7 (0.0–460.5) | 0.044 |
| T0 | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.132 |
| T1 | 44.3 (10.0–93.2) | 43.6 (14.3–81.3) | 21.1 (0.0–51.1) | 152.1 (60.8–219.2) | <0.001 |
| T2 | 982.7 (391.2–2516) | 678.5 (237.9–1307) | 3765 (1766–6099) | 2869 (174–4420) | <0.001 |
| nAb (IU/mL) | |||||
| Total | 39.5 (5.0–238. | 33.6 (4.6–134.8) | 47.6 (9.0–1096) | 208.3 (0.9–619.5) | <0.001 |
| T0 | 1.2 (0.0–6.1) | 1.4 (0.0–5.8) | 5.3 (0.0–9.6) | 0.0 (0.0–2.0) | <0.001 |
| T1 | 47.4 (22.5–108.9) | 39.5 (19.5–81.6) | 37.5 (19.5–81.6) | 276.4 (188.5–481.5) | <0.001 |
| T2 | 416.1 (126.0–1473) | 235.8 (90.0–653.1) | 1861 (706.5–2713) | 1791 (1091–2506) | <0.001 |
| IGRA, class I (IU/mL) | |||||
| Total | 0.09 (0.00–0.37) | 0.07 (0.01–0.23) | 0.20 (−0.00–0.91) | 0.10 (0.00–0.89) | |
| T0 | −0.00 (−0.02–0.01) | 0.00 (−0.02–0.01) | −0.01 (−0.05–0.01) | 0.00 (−0.01–0.01) | 0.020 |
| T1 | 0.14 (0.04–0.42) | 0.11 (0.04–0.30) | 0.18 (0.07–0.68) | 0.22 (0.07–0.72) | <0.001 |
| T2 | 0.21 (0.07–0.65) | 0.14 (0.05–0.30) | 0.67 (0.24–1.90) | 1.14 (0.44–2.88) | <0.001 |
| IGRA, class I/II (IU/mL) | |||||
| Total | 0.16 (0.01–0.67) | 0.13 (0.02–0.41) | 0.30 (0.01–0.58) | 0.20 (0.01–1.48) | |
| T0 | 0.00 (−0.01–0.02) | 0.00 (−0.01–0.02) | 0.00 (−0.03–0.02) | 0.00 (−0.00–0.01) | 0.446 |
| T1 | 0.27 (0.10–0.85) | 0.19 (0.07–0.56) | 0.50 (0.16–1.65) | 0.41 (0.15–1.04) | 0.001 |
| T2 | 0.37 (0.11–1.15) | 0.24 (0.09–0.55) | 1.25 (0.40–3.32) | 2.08 (0.79–4.00) | <0.001 |
| TA Case n/Total n % (95% CI) | PA Case n/Total n % (95% CI) | NA Case n/Total n % (95% CI) | Kappa (95% CI) | |
|---|---|---|---|---|
| RBD Ab_A vs. RBD Ab_B | 87.3 (83.1–91.7) | 81.3 (76.3–86.5) | 99.4 (91.7–100) | 0.74 (0.71–0.77) |
| RBD Ab_A vs. RBD Ab_R | 98.4 (93.9–100) | 99.5 (94.0–100) | 96.1 (88.5–100) | 0.96 (0.95–0.98) |
| RBD Ab_B vs. RBD Ab_R | 86.2 (82.1–90.6) | 99.5 (93.4–100) | 70.4 (64.9–76.3) | 0.72 (0.68–0.75) |
| RBD Ab_A vs. nAb | 89.4 (85.2–93.8) | 99.7 (93.8–100) | 76.1 (70.2–82.4) | 0.78 (0.75–0.81) |
| RBD Ab_B vs. nAb | 91.4 (87.1–95.9) | 90.6 (84.9–96.6) | 92.5 (85.9–99.3) | 0.83 (0.80–0.85) |
| RBD Ab_R vs. nAb | 88.4 (84.2–92.8) | 99.7 (93.8–100) | 73.9 (68.1–80.1) | 0.76 (0.73–0.79) |
| RBD Ab_A vs. IGR | 91.5 (90.0–92.8) | 91.7 (90.2–93.0) | 72.4 (70.1–74.6) | 0.66 (0.62–0.70) |
| RBD Ab_B vs. IGR | 79.4 (77.3–81.4) | 78.5 (76.3–80.5) | 81.6 (79.5–83.5) | 0.55 (0.51–0.60) |
| RBD Ab_R vs. IGR | 86.2 (84.4–87.9) | 92.6 (91.2–93.8) | 71.7 (69.4–73.9) | 0.66 (0.62–0.71) |
| nAb vs. IGR | 87.6 (85.8–89.2) | 90.0 (88.4–91.4) | 66.3 (63.9–68.7) | 0.58 (0.54–0.63) |
| Assay | Vector Vaccine | MM Vaccine | Mrna Vaccine |
|---|---|---|---|
| RBD Ab_A vs. RBD Ab_B | |||
| TA% (95% CI) | 83.9 (78.9–89.1) | 91.1 (81.2–100) | 99.6 (87.6–100) |
| PA% (95% CI) | 75.7 (69.9–81.8) | 88.2 (76.6–100) | 99.4 (84.8–100) |
| NA% (95% CI) | 99.5 (90.4–100) | 98.0 (79.7–100) | 100 (79.7–100) |
| Kappa (95% CI) | 0.68 (0.64–0.72) | 0.80 (0.74–0.87) | 0.99 (0.97–1.00) |
| RBD Ab_A vs. RBD Ab_R | |||
| TA% (95% CI) | 98.3 (93.0–100) | 98.2 (87.9–100) | 99.2 (87.2–100) |
| PA% (95% CI) | 99.8 (93.1–100) | 99.2 (86.9–100) | 98.8 (84.3–100) |
| NA% (95% CI) | 95.4 (86.4–100) | 96.0 (77.9–100) | 100 (79.7–100) |
| Kappa (95% CI) | 0.96 (0.95–0.98) | 0.96 (0.92–0.99) | 0.98 (0.96–1.00) |
| RBD Ab_B vs. RBD Ab_R | |||
| TA% (95% CI) | 82.6 (77.7–87.8) | 90.5 (80.7–100) | 98.8 (86.8–100) |
| PA% (95% CI) | 99.8 (92.2–100) | 99.1 (86.1–100) | 98.2 (83.7–100) |
| NA% (95% CI) | 95.6 (86.6–1.05) | 76.4 (61.9–93.2) | 98.8 (78.7–100) |
| Kappa (95% CI) | 0.65 (0.61–0.69) | 0.79 (0.72–0.86) | 0.97 (0.94–1.00) |
| RBD Ab_A vs. nAb | |||
| TA% (95% CI) | 86.9 (84.9–88.7) | 91.4 (87.9–93.9) | 99.2 (97.1–99.8) |
| PA% (95% CI) | 99.7 (99.2–99.9) | 100 (98.9–100) | 98.8 (94.5–99.6) |
| NA% (95% CI) | 72.6 (70.1–75.0) | 77.7 (73.0–81.8) | 99.4 (97.4–99.9) |
| Kappa (95% CI) | 0.73 (0.70–0.77) | 0.81 (0.75–0.88) | 0.98 (0.96–1.00) |
| RBD Ab_B vs. nAb | |||
| TA% (95% CI) | 89.3 (87.5–90.9) | 93.8 (90.7–95.9) | 98.8 (96.5–99.6) |
| PA% (95% CI) | 87.0 (85.0–88.7) | 95.7 (93.0–97.4) | 98.8 (96.5–99.6) |
| NA% (95% CI) | 91.9 (90.3–93.3) | 90.8 (87.2–93.4) | 98.8 (96.5–99.6) |
| Kappa (95% CI) | 0.79 (0.75–0.82) | 0.87 (0.81–0.92) | 0.97 (0.94–1.00) |
| RBD Ab_R vs. nAb | |||
| TA% (95% CI) | 85.8 (83.8–87.6) | 90.2 (86.6–92.9) | 99.2 (97.1–99.8) |
| PA% (95% CI) | 100 (99.2–100) | 99.5 (98.0–99.9) | 98.8 (96.5–99.6) |
| NA% (95% CI) | 69.9 (67.3–72.4) | 75.4 (70.5–79.7) | 100 (98.5–100) |
| Kappa (95% CI) | 0.71 (0.67–0.75) | 0.78 (0.72–0.85) | 0.98 (0.96–1.00) |
| RBD Ab_A vs. IGR | |||
| TA% (95% CI) | 83.9 (81.4–86.2) | 87.7 (83.7–90.8) | 89.8 (85.4–93.0) |
| PA% (95% CI) | 92.1 (90.2–93.7) | 90.4 (868–93.1) | 92.1 (88.1–94.9) |
| NA% (95% CI) | 65.7 (62.5–68.7) | 80.9 (763–84.8) | 85.4 (80.4–89.3) |
| Kappa (95% CI) | 0.61 (0.55–0.66) | 0.70 (0.62–0.79) | 0.77 (0.68–0.86) |
| RBD Ab_B vs. IGR | |||
| TA% (95% CI) | 75.4 (72.5–78.1) | 82.9 (78.5–86.6) | 89.4 (84.9–92.7) |
| PA% (95% CI) | 73.8 (70.8–76.6) | 81.7 (77.2–85.5) | 91.5 (87.4–94.4) |
| NA% (95% CI) | 78.9 (76.1–81.4) | 86.2 (82.1–89.5) | 85.4 (80.4–89.3) |
| Kappa (95% CI) | 0.48 (0.42–0.54) | 0.62 (0.53–0.70) | 0.76 (0.68–0.85) |
| RBD Ab_R vs. IGR | |||
| TA% (95% CI) | 84.2 (81.7–86.4) | 88.3 (84.4–91.3) | 90.7 (86.4–93.7) |
| PA% (95% CI) | 93.3 (94.5–94.8) | 91.3 (87.8–93.9) | 92.1 (88.1–94.9) |
| NA% (95% CI) | 63.9 (60.7–67.0) | 80.9 (76.3–84.8) | 87.8 (83.1–91.3) |
| Kappa (95% CI) | 0.61 (0.55–0.66) | 0.71 (0.63–0.80) | 0.79 (0.71–0.87) |
| nAb vs. IGR | |||
| TA% (95% CI) | 79.1 (76.3–81.6) | 80.8 (76.2–84.7) | 89.8 (85.4–93.0) |
| PA% (95% CI) | 76.1 (73.2–78.8) | 79.6 (75.0–83.6) | 92.1 (88.1–94.9) |
| NA% (95% CI) | 80.4 (77.7–82.9) | 84.0 (79.7–87.5) | 85.4 (80.4–89.3) |
| Kappa (95% CI) | 0.54 (0.48–0.59) | 0.57 (0.48–0.66) | 0.77 (0.69–0.86) |
| Assay | Mean Differences | |||
|---|---|---|---|---|
| Total | Vector Vaccine | MM Vaccine | mRNA Vaccine | |
| RBD Ab_A vs. RBD Ab_B | 99.5 (83.6–115.4) | 34.9 (25.0–44.7) | 253.6 (198.1–309.2) | 218.3 (148.4–288.2) |
| RBD Ab_A vs. RBD Ab_R | −481.3 (−540.2–−422.4) | −279.0 (−318.3–−239.8) | −1244 (−1479–−1010) | −362.7 (−508.2–−217.2) |
| RBD Ab_B vs. RBD Ab_R | −666.6 (−748.2–−584.9) | −352.2 (−403.9–−300.5) | −1841 (−2180–−1502) | −671.3 (−903.9–438.7) |
| RBD Ab_A vs. nAb | −136.9 (−160.7–−113.2) | −126.7 (−148.4–−105.0) | −369.7 (−447.9–−291.4) | 127.9 (45.5–210.3) |
| RBD Ab_B vs. nAb | −208.9 (−236.1–−181.8) | −156.6 (−180.2–−133.1) | −559.6 (−660.1–−459.0) | −11.1 (−89.0–−66.8) |
| RBD Ab_R vs. nAb | 326.2 (272.5–379.9) | 179.6 (138.3–220.8) | 837.9 (625.2–1051) | 424.0 (240.4–607.6) |
| RBD Ab_A vs. IGR | 302.6 (266.8–338.4) | 104.3 (845.0–123.5) | 400.1 (323.1–477.1) | 898.1 (746.5–1050) |
| RBD Ab_B vs. IGR | 177.7 (156.6–198.8) | 54.4 (48.2–60.6) | 144.0 (121.3–166.7) | 676.3 (576.0–776.5) |
| RBD Ab_R vs. IGR | 1005 (895.4–1115) | 543.8 (469.2–618.3) | 1997 (1638–2357) | 1349 (1048–1651) |
| nAb vs. IGR | 410.1 (371.9–448.4) | 272.6 (238.4–306.7) | 685.3 (570.0–800.6) | 571.9 (462.8–681.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, M.; Cha, J.H.; Kim, S.-W.; Kim, S.B.; Lee, K.-B.; Chung, Y.-S.; Yun, S.G.; Nam, M.-H.; Lee, C.K.; Cho, Y. Performance Evaluation of Three Antibody Binding Assays, a Neutralizing Antibody Assay, and an Interferon-Gamma Release Assay for SARS-CoV-2 According to Vaccine Type in Vaccinated Group. Diagnostics 2023, 13, 3688. https://doi.org/10.3390/diagnostics13243688
Nam M, Cha JH, Kim S-W, Kim SB, Lee K-B, Chung Y-S, Yun SG, Nam M-H, Lee CK, Cho Y. Performance Evaluation of Three Antibody Binding Assays, a Neutralizing Antibody Assay, and an Interferon-Gamma Release Assay for SARS-CoV-2 According to Vaccine Type in Vaccinated Group. Diagnostics. 2023; 13(24):3688. https://doi.org/10.3390/diagnostics13243688
Chicago/Turabian StyleNam, Minjeong, Jae Hyun Cha, Sang-Wook Kim, Sun Bean Kim, Ki-Byung Lee, You-Seung Chung, Seung Gyu Yun, Myung-Hyun Nam, Chang Kyu Lee, and Yunjung Cho. 2023. "Performance Evaluation of Three Antibody Binding Assays, a Neutralizing Antibody Assay, and an Interferon-Gamma Release Assay for SARS-CoV-2 According to Vaccine Type in Vaccinated Group" Diagnostics 13, no. 24: 3688. https://doi.org/10.3390/diagnostics13243688
APA StyleNam, M., Cha, J. H., Kim, S.-W., Kim, S. B., Lee, K.-B., Chung, Y.-S., Yun, S. G., Nam, M.-H., Lee, C. K., & Cho, Y. (2023). Performance Evaluation of Three Antibody Binding Assays, a Neutralizing Antibody Assay, and an Interferon-Gamma Release Assay for SARS-CoV-2 According to Vaccine Type in Vaccinated Group. Diagnostics, 13(24), 3688. https://doi.org/10.3390/diagnostics13243688







