Predicting Thalassemia Using Feature Selection Techniques: A Comparative Analysis
Abstract
:1. Introduction
- R1
- Which kind of datasets are utilized by ML-based prediction and management techniques for TT?
- R2
- Which ML methods are employed in the TT diagnostic?
- R3
- Which thalassemia variants can be detected using ML-based methods? Or what specific forms of thalassemia are being detected using ML-based methods?
- R4
- What standards are applied to evaluate ML classifiers for illness prediction?
- R5
- Which issues are addressed by ML-based applications in illness management and diagnosis?
- R6
- How effectively ML approaches will work with openly available datasets?
2. Thalassemia
2.1. Alpha (α) Thalassemia
2.1.1. Silent Carrier
2.1.2. Alpha Zero (α0) Thalassemia Carrier
2.1.3. Alpha (α) Intermedia Thalassemia
2.1.4. Hb Constant Spring
2.2. Beta (β) Thalassemia
2.2.1. Beta (β) Thalassemia Minor
2.2.2. Beta (β) Thalassemia Intermedia
2.2.3. Beta (β) Thalassemia Major
2.3. Other Variants of Thalassemia Carrier
3. Systematic Literature Review
3.1. Selection of Articles
3.2. Datasets Review
3.3. Preprocessing Techniques Review
3.4. Classifiers for Detection of Thalassemia
| Ref. | Description | Availability | Features |
|---|---|---|---|
| [7] | Two datasets used are homogenous with 159 records (87 females and 72 males) with ages over 18 years. The other dataset is heterogenous with a record of 1883 as 264 IDA, 27 βT & 1572 are normal. | Private | All CBC parameters, HbA, HbF, MCV, and MCH |
| [44] | Out of 3947 observations, 210 recordings of both classes (βT and non-βT) are taken in equal numbers to lessen bias. The mean age is around 25. | Private | Hb, MCV RBC count, PCV, MCH, RDW-CV, and MCHC |
| [8,38] | A total of 5066 individuals (53% males and 47% females) records are pulled from the PTPP database. Among them, 2015 are β-thalassemia carriers, while 3051 are β thalassemia non-carriers; 54% of the carriers are adults, and 46% are children. | Private | Age, Sex, Hb RBC, MVC, Hct, MCHC, MCH, PLT, RDW, and WBC |
| [45] | The dataset consisted of 594 cases (330 females and 264 males with average age of 29.7 years) with 229 healthy individuals, 160 patients with the α+-trail phenotype, and 205 individuals with a two-allele α-trail metamorphosis. | Private | Age, Gender, RBC count, Hct, Hb, MCH, MCV, RDW, and MCHC |
| [46,47,48,49,50] | The ten-variable dataset 150 has thalassemia, and 68 people are normal. | Private | Basophils, Monocytes, Eosinophils, Segment Neutrophils, Rod Neutrophils, Lymphocytes, PLT, Hb, HCT, and WBC |
| [40] | A total of 379 participants, including 79 positive cases of Thalassemia, made up the Kaggle dataset including a limited amount of data collected by Google form. https://www.kaggle.com/datasets/plenoi/thalassemia (accessed on 5 July 2023) | Public | Age, Gender, RBC, HGB, PCV, RDW, MCHC, PLT, TLC, MCV, and MCH |
| [51] | Out of the 350 patients at the Taipei Veterans General Hospital, 122 (34.8%) had no thalassemia variation, 179 (51.1%) had α-variant, and 49 (14%) had βT. Data are collected from January 2018 to January 2020. | Private | WBC, Hb, RBC, HCT, MCV, MCH, MCHC, RDW, PLT, RDW, RDWI, E&F, S&L, G&K, MDHL, MCHD, HH index, αT, and Βt |
| [52] | The hospital’s Laboratory Information System (LIS) is used to collect data on 1213 Chinese with low HbA2 levels from December 2018 to August 2020. | Private | RDW, Hct, MCV, RBC, Hbf, Hba, Hb, MCH, pregnancy, and age |
| [53,54] | Out of 342, only 152 individuals had thalassemia; data were collected from January 2016 to May 2019, in Elazig Public Health Laboratory. From January to July of 2018, 190 records (2 to 88 years) were declared as anemic. | Private | Hb, HCT, RBC, MCH, MCV, RDW, and MCHC |
| [55] | A total of 907 adults aged 18 and above were included in the study, and 59% had TT, while 41% had IDA. | Private | Hb, MCV, MCH, RDW, MCHC, and RBC |
| [39] | The study analyzed data on 45,498 individuals from 2012 to 2016 by the Palestine Avenir Foundation’s Thalassemia and Hemophilia Middle. 44,360 tests are normal, but 1138 individuals are confirmed to be carriers. | Private | CBC, RBC, Hct, Hb, MCV, MCHC, MCH, plt, RDW, and WBC |
| [56] | As per the opinion of doctors, tests are carried out on four types of data. To increase the precision level, it is vital to test the model with more genuine data. | Private | Hb, MCV, and MCH |
| [57] | CBC testing yielded results from a total of 750 tests conducted on individuals (males = 390 & females = 360) aged between 17 to 32 years old. | Private | RBC, Hb, HCT, and MCV |
| [58] | The dataset comprised a total of 8693 CBC test records along with 2918 genetic test data used for labeling type of thalassemia. | Private | HB, RBC, RDW, MCV, HCT, MCH, PLT, MCHC, WBC, patient’s sex and age |
| [59] | A dataset of 49 patients (31% high, 16% moderate, 10% low risk, and 43% zero signs) is collected from case files in the southwest section hospital of Nigeria. | Private | Age, gender, ethnicity, marital status, family history, social class, diabetes spleen enlargement, parent carriers, and urine color changes |
| [60] | Two datasets are involved as the first dataset had 6058 entities and the second, autonomous dataset had 2637 rows. | Private | Production machine, production date, Hb, Hct, extracted plasma, production site, donor sex, donor age, donor Hb, program and blood bag type |
| [61] | 268 individuals | Private | RBC, HGB, MCV, and MCH |
| [42] | 524 electrophoresis images | Private | Hb variants (HbA, HbA2) |
| [62] | From flow cytometry 302,652 cells and 3289 images | Private | Morphological |
| [63] | 111 TDT patients in Iraq with an age range of 6 to 12 years | Private | Iron grade constraints (transferrin saturation percentage, iron, ferritin) and inflammatory (tumor necrosis factor-α and interleukin-1β) |
| [64] | Blood smears from 110 individual cases | Private | Morphological |
| [65] | 1069 MRI images | Private | Morphological |
| [41] | Twenty blood smear images | Private | Morphological |
| [66] | 7108 erythrocytes | Private | Shape, texture feature with gray level co-occurrence matrices (GLCM), color |
| [67] | Data collected from laboratories at Philippine General Hospital compares | Private | Perimeter (P) Area (A) Central Pallor (CP) Diameter (D) Deviation Value (DV) Target Flag (TF) Shape Geometric Factor (SGF) |
| [68] | Standing image datasets from Philippine General Hospital and Mendeley online. | Private | Codocytes and Elliptocytes |
| [43] | Disease-Specific Face (DSF) dataset totals 350 face images https://ieee-dataport.org/documents/disease-specific-faces (accessed on 5 July 2023) | Public | Morphological |
| [69] | 1815 images of affected and normal blood cells https://www.kaggle.com/datasets/kmader/malaria-bounding-boxes (accessed on 5 July 2023) | Public | Morphological |
| Ref. | Field of Use | Preprocessing |
|---|---|---|
| [38] | Classification of βT Carriers | Cleaning (missing value), data transformation (normalization), data reduction (attribute sub-selection) |
| [39] | Identifying βT carriers | Data cleaning (missing values) Normalization, balancing technique: SMOTE |
| [40] | αT Prediction | Data Balance: SMOTE |
| [7] | Differentiation of IDA and βT | Iχ2 feature selection |
| [8] | Prediction of βT | Data balance: SMOTE and ADASYN, Feature Reduction: PCA and SVD |
| [42] | Assessment of Thalassemia | Object detection, filtering and thresholding, erosion and dilation, boundary detection, and lane extraction |
| [41] | Classification of Thalassemia using fusion | PCA to eliminate feature redundancy |
| [43] | Facial diagnosis | DSIFT and DTL2 |
3.4.1. Classifiers for Alpha thalassemia
3.4.2. Classifiers for Beta Thalassemia
3.4.3. Classifiers for Risk Assessment of Thalassemia
| Ref. | Field of Use | Classifier | Compared with | Performance |
|---|---|---|---|---|
| [7] | Differentiation of IDA from βT | MATLAB (R2020a) classification learner toolbox | SVM, ANN, PCA and MLPs, ANFIS, MLPs, Math, Regular over-learning machine | SVM 97.48% accuracy with the 1st dataset & 99.73% accuracy with the 2nd dataset |
| [44] | Recognition of βT among antenatal women. | C4.5 DT, NB and ANN using R studio | Each selected classifier | Accuracy of 85.95% with ANN |
| [8] | Prediction of βT | DT, GBM, ADA, SVC, RF, ETC, LR LSTM, GRU, CNN, and CNN-LSTM. | Each selected classifier with different senarios | SMOTE with PCA and SVD had 96% accuracy |
| [45] | Prediction of the α-T | CNN, SVM, MLP, RF, PLS, LR, ET, LGBM, XGB, DT, and KNNs implemented in Python | Each selected classifier | CNN accuracy of 80.77% |
| [38] | Classification of βT Carriers | SGR-VC ensemble (SVM, GBM, and RF) | Compared to each classifier in the model separately | 93% accuracy |
| [46] | Classification of thalassemia | RF | RF with different percent of training data | 98.99% accuracy |
| [40] | αT Prediction | KNN, NB, RF, LR, SVM, ADA Boosting, Xgboost, DT, MLP, and Gradient Boosting classifier using Google Colab | Each selected classifier | ADA accuracy 100% |
| [47] | Classifier for thalassemia | RBF, Polynomial, and linear kernel functions with SVM | SVM with different kernel and percent of training data | Gaussian RBF kernel with SVM accuracy 99.63% |
| [51] | Discriminating αT and βT | SVM with R. | SVM with 13 indices | NA |
| [52] | αT carrier discrimination | RF with R software version 3.6.2. | 13 built ML models such as DT, KNN, SVM, ADA, LR, NB | Accuracy 91.5% |
| [53] | Discrimination of βT and IDA | LR, KNN, SVM, ELM, and RELM | DT, KNN, NB, DT, MLP, SVM, ANN, PCA, ANFIS, RBF, Math | 96.30% accuracy for females, 94.37% for males, and 95.59% in co-evaluation of males and females with ELM and RELM |
| [48] | Thalassemia Classification | TSVM | TSVM with different kernel and percent of training data | 99.32% accuracy |
| [55] | Finding of βT from IDA | BLTREED and CART | Each selected classifier | BLTREED model 96% accuracy |
| [49] | Thalassemia Classification | Based on density peaks, a hierarchical clustering algorithm with or without a kernel function | With different number of folds | NA |
| [50] | Thalassemia Classification | SVM with hyperparameter optimization using Grid Search | With different values of hyperparameter(C, gemma) | 100% accuracy (90% training) gamma = 0.0000183 and C = 428.13 |
| [39] | Identifying βT carriers | KNN, NB, DT and MLP in Weka (3.8.1) | Each selected classifier | 99.71% Accuraccy |
| [56] | Prediction of thalassemia for children. | Fuzzy-based | NA | NA |
| [57] | Discrimination between IDA and βT | Dynamic Harmony Search (DHS). | ANN, ANFIS, SVM, KNN | Accuracy of approximately 98% |
| [58] | Predicting thalassemia | DNN model with 11 features then removing some of the 11 features | With different Combinations of featutes | 89.7% accuracy |
| [54] | Discrimination of βT and IDA | KNN & SVM | ANNs, specialized ANNs Single Vector Analysis, MLP, LR, ANFIS, DT, NB, Neural Network, PBIS-ANN J48, mathematical method based on SVM and Heuristic algorithm | Accuracy 95.3%, Female, 94.5% Male |
| [59] | Thalassemia risk | NB and MLP in Weka | Each selected classifier | Accuracy 94.12% NB,100% MLP |
| [60] | Iron content and Hemoglobin Estimation | MLR, RF, KNN, DT, SVM, lgbmR, RANSAC and NN using Python | Each selected classifier with different features | NA |
| [61] | Discriminating between IDA and βT | ANN using MATLAB | MLP, SVM, KNN, RBF, PNN, ANFIS (radial basis function) Probabilistic neural network (PNN) adaptive network-based fuzzy inference system | 92.5%, accuracy |
| [42] | Assessment of Thalassemia | MobileNetV2, InceptionV3, Densnet201, ResNet18, ResNet50, ResNet101, SqueezeNet | DT, ANN, KNN, SVM, Naïve Bayes, MLP, genetic programming | Accuracy 95.8% for InceptionV3 |
| [62] | Predicting cell genotypes for βT | MLP, CRISPR genome editing technology. | NA | 82% accuracy |
| [63] | Depression due to transfusion-dependent thalassemia | EFA and Cluster analysis, | NA | NA |
| [64] | Detecting approach for αT | Region-Based Convolutional Network (RCNN) | NA | Accuracy 97.6% |
| [65] | Liver iron content (LIC) evaluation | Deep-learning CNN—HippoNet-2D, 3D, LSTM, and HippoNet-Ensemble | NA | 90% for multiclass accuracy |
| [41] | Classification of Thalassemia using fusion | RF, Naive Bayes, and KNN. | ANN, MLP, CNN, LR, PCA | Accuracy of 99.1% |
| [66] | Thalassemia diagnostic | MLP | With a combination of different features | Accuracy of 98.11% |
| [67] | Identification of Abnormal RBCs | SVM with Raspberry Pi | NA | Accuracy of 93.33% |
| [68] | Early indicators of Thalassemia. | SVM with Raspberry Pi | NA | Accuracy of 94.31 |
| [43] | Facial diagnosis | CNN | CNN and SVM with different variation of features | 90% accuracy |
| [69] | Thalassemia detection | CNN | NA | 93.4% accuracy |
3.5. Thalassemia Applications
| Ref. | Application | Purpose & Link |
|---|---|---|
| [71] | The rule-based chatbot | Thalassemia management support suggests exact scheduling for necessary tests and evaluations https://chats.landbot.io/v3/H-947072-772QZJR6XMJAGCJW/index.html (accessed on 5 July 2023) |
| [72] | Web-based expert system | Management of βT. Real-time patient information, including medical history, medication information, and appointment information. A QR code scanner or smartphone can decode the URL for each patient using a QR code. |
| [73] | Webpage tool of TT@MHA | Prediction based on patients’ provided parameters (RDW-SD, MCHCs, MCV, RBC, Hb, Age group, Sex, Pregnancy) https://dxonline.deepwise.com/prediction/index.html?baseUrl=%2Fapi%2F&id=26408&topicName=undefined&from=share&platformType=wisdom (accessed on 5 July 2023) |
| [74] | Web-based tool ThalPred | Prediction based on patients‘ provided parameters (Hb RBC, MCV Hct, RDW, MCHC, MCH) http://codes.bio/thalpred/ (accessed on 5 July 2023) |
3.6. Performance Measures
| Acc | Recall | Precision/Positive Predictive Values | F1-Score | Sen | Spec | MCC | Negative Predictive Values | FPR | FNR | Youden’s Index | AUC | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ✓ | ✓ | ✓ | ✓ | [7,38] | ||||||||
| ✓ | ✓ | ✓ | ✓ | [39] | ||||||||
| ✓ | ✓ | ✓ | ✓ | [8] | ||||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | [44] | ||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | [45] | |||||||
| ✓ | ✓ | ✓ | [46] | |||||||||
| ✓ | ✓ | ✓ | ✓ | [48] | ||||||||
| ✓ | [49] | |||||||||||
| ✓ | [50] | |||||||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | [51] | |||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | [52] | |||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | [53] | |||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | [54] | ||||
| ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | [55] | |||
| ✓ | ✓ | ✓ | ✓ | ✓ | [57] | |||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | [58] | ||||
| ✓ | ✓ | ✓ | ✓ | [59] | ||||||||
| ✓ | ✓ | ✓ | [60,61,66] | |||||||||
| ✓ | ✓ | ✓ | ✓ | [62] | ||||||||
| ✓ | ✓ | [63] | ||||||||||
| ✓ | ✓ | ✓ | ✓ | ✓ | [64] | |||||||
| ✓ | ✓ | ✓ | [74] | |||||||||
| ✓ | ✓ | ✓ | Our method |
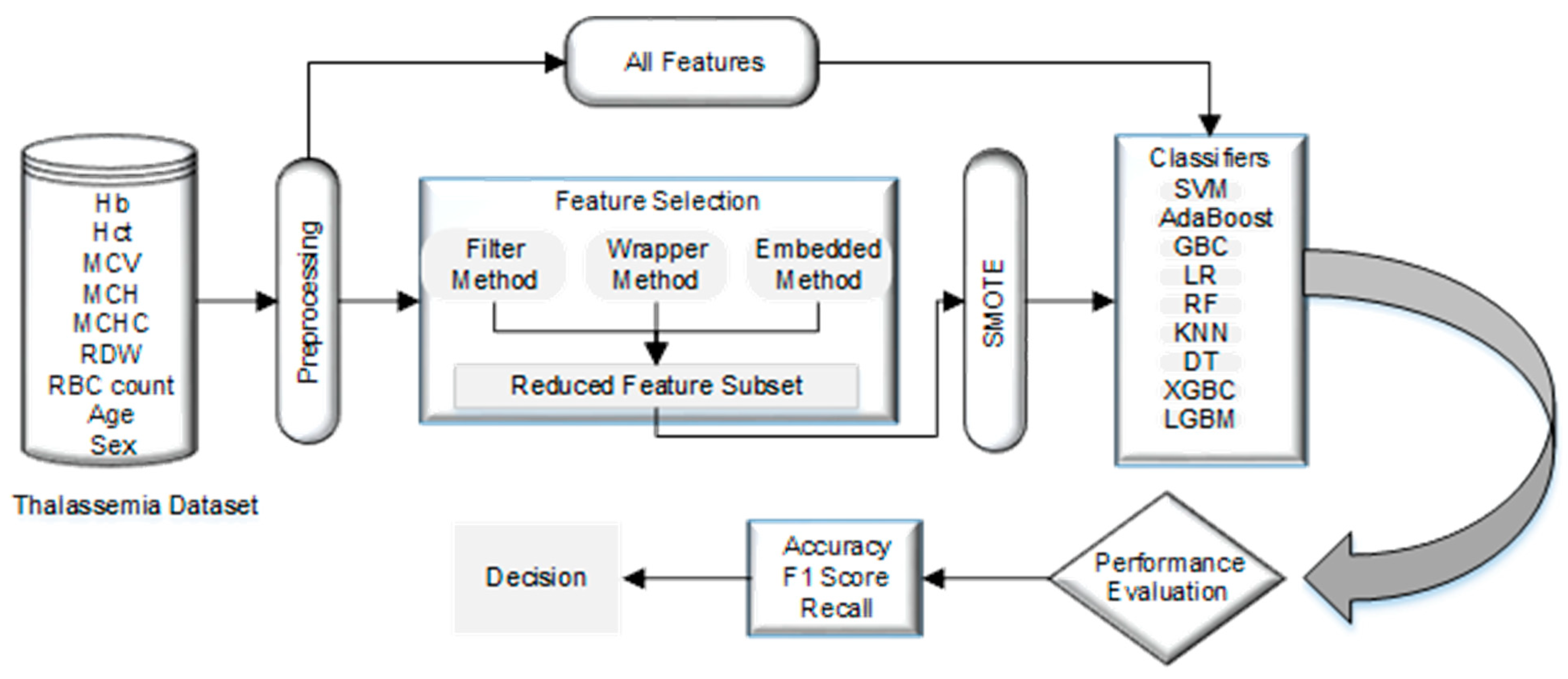
4. Material and Methods
4.1. Dataset
4.2. Data Preparation
Data Cleaning
4.3. Normalization
4.4. Using SMOTE to Address the Unbalanced Data Issue by Data Resampling
4.5. Feature Selection
| Feature Selection | Feature Set (First Dataset) | F# | Feature Set (Second Dataset) | F# |
|---|---|---|---|---|
| Feature importance using RF | MCV, MCH, RDW | F11 | Hb, PCV, RBC, MCV, MCH, WBC, Lymph, HBA2 | F21 |
| Feature importance using GBDT | Age, Hb, MCV, MCH, RDW, RBC count | F12 | Hb, PCV, MCV, MCH, MCHC, RDW, WBC, Neut, Lymph, PLT, HBA2, HBF | F22 |
| Estimation of coefficients using linear regression | Sex, Hb, MCH, RDW | F13 | Hb, RBC, HBA2, HBF, Sex | F23 |
| χ2 | MCV, MCH, RDW, Hb, RBC count, Hct | F14 | PLT, lymph, MCV, MCH, Neut | F24 |
| MCV, MCH, RDW, ‘Hb | F15 | PLT, lymph, MCV, MCH | F25 | |
| MCV, MCH | F16 | PLT, lymph, MCV | F26 | |
| Exhaustive Feature Selection (EFS) | Age, Sex, Hb, MCV | F17 | Hb, PCV, RBC, MCV | F27 |
4.5.1. Filter-Based Feature
4.5.2. Wrapper Methods
4.5.3. Embedded Methods
4.6. Classification Model
4.7. 10-Fold Cross-Validation
5. Result
| Features | Measures (%) | SVM | SVM Hyperparameter (C, gamma, kernel = ‘rbf’) | GBC | ADA | LR | DT | XGBC | RF | LGBM | KNN n = 3 | KNN n = 6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| * C = 100, gamma = 0.0001 | ||||||||||||
| All features (First dataset) | Accuracy | 83.87 | 86.29 | 85.48 | 83.87 | 88.31 | 83.12 | 87.01 | 88.31 | 85.71 | 85.71 | 87.66 |
| F1 Score | 82.55 | 84.607 | 83.6 | 82.19 | 88.02 | 82.88 | 86.74 | 88.02 | 85.3 | 85.42 | 87.33 | |
| Recall | 84.82 | 85.3 | 84.05 | 83.51 | 88.19 | 83.52 | 87.08 | 88.19 | 85.3 | 85.74 | 87.41 | |
| * C = 10, gamma = 0.01 | ||||||||||||
| F11 | Accuracy | 83.87 | 87.1 | 89.52 | 86.29 | 84.42 | 82.47 | 86.36 | 82.47 | 85.71 | 85.06 | 85.71 |
| F1 Score | 82.56 | 85.6 | 88.23 | 84.95 | 83.51 | 80.99 | 85.32 | 81.39 | 84.79 | 84.35 | 84.68 | |
| Recall | 84.82 | 86.55 | 88.99 | 86.61 | 84.65 | 81.11 | 85.75 | 82.32 | 85.66 | 83.84 | 83.72 | |
| * C = 100, gamma = 0.0001 | ||||||||||||
| F12 | Accuracy | 83.06 | 87.1 | 85.48 | 80.64 | 90.91 | 90.9 | 88.96 | 88.31 | 86.36 | 85.71 | 83.76 |
| F1 Score | 81.77 | 85.6 | 83.97 | 78.64 | 89.63 | 89.52 | 86.74 | 86.22 | 83.62 | 82.95 | 80.25 | |
| Recall | 84.23 | 86.55 | 85.36 | 79.82 | 90.07 | 89.52 | 85.37 | 85.44 | 82.38 | 81.9 | 78.84 | |
| * C = 1000, gamma = 0.0001 | ||||||||||||
| F13 | Accuracy | 84.68 | 86.29 | 86.29 | 84.68 | 86.36 | 79.22 | 79.22 | 84.42 | 78.57 | 81.17 | 84.42 |
| F1 Score | 82.1 | 84.61 | 84.61 | 82.1 | 85.93 | 78.51 | 78.4 | 83.62 | 77.67 | 80.26 | 83.62 | |
| Recall | 84.11 | 85.3 | 85.3 | 84.11 | 87.36 | 79.58 | 79.24 | 84.09 | 78.38 | 80.8 | 84.09 | |
| * C = 10, gamma = 0.001 | ||||||||||||
| F14 | Accuracy | 83.06 | 86.29 | 87.01 | 84.68 | 91.56 | 82.47 | 84.42 | 87.01 | 83.77 | 84.42 | 85.06 |
| F1 Score | 81.77 | 84.78 | 85.6 | 83.18 | 91.04 | 80.31 | 83.03 | 85.86 | 82.01 | 82.6 | 83.38 | |
| Recall | 84.23 | 85.95 | 86.55 | 84.76 | 92.65 | 79.69 | 83.31 | 86.17 | 81.96 | 82.04 | 82.96 | |
| * C = 10, gamma = 0.001 | ||||||||||||
| F15 | Accuracy | 83.87 | 86.29 | 88.71 | 86.29 | 85.71 | 83.12 | 88.31 | 87.01 | 85.06 | 83.12 | 83.76 |
| F1 Score | 82.56 | 84.78 | 87.4 | 84.95 | 84.89 | 82.45 | 87.55 | 86.35 | 84.25 | 82.14 | 82.65 | |
| Recall | 84.82 | 85.95 | 88.39 | 86.61 | 84.51 | 82.64 | 86.93 | 86.14 | 83.97 | 81.79 | 82.05 | |
| * C = 1000, gamma = 0.01 | ||||||||||||
| F16 | Accuracy | 83.87 | 84.68 | 83.87 | 88.71 | 87.66 | 77.92 | 80.52 | 81.89 | 82.47 | 84.42 | 85.06 |
| F1 Score | 82.56 | 82.8 | 81.55 | 87.4 | 86.61 | 76.15 | 78.95 | 80.5 | 81.13 | 82.88 | 83.06 | |
| Recall | 84.82 | 83.45 | 81.55 | 88.39 | 86.76 | 76.36 | 79.19 | 81.01 | 81.51 | 82.63 | 81.92 | |
| * C = 1000, gamma = 0.0001 | ||||||||||||
| F17 | Accuracy | 82.26 | 86.29 | 84.68 | 79.84 | 88.31 | 83.12 | 83.12 | 87.01 | 84.42 | 83.12 | 87.01 |
| F1 Score | 80.99 | 84.95 | 82.99 | 78.84 | 87.71 | 82.02 | 82.35 | 86.5 | 83.8 | 82.25 | 85.85 | |
| Recall | 83.63 | 86.61 | 84.18 | 82.5 | 88.24 | 82.02 | 83.05 | 87.54 | 84.77 | 82.7 | 85.15 | |
| Feature | Measures | SVM | SVM Hyperparameter (C, Gamma, Kernel = ‘rbf’) | GBC | ADA | LR | DT | XGBC | RF | LGBM | KNN n = 3 | KNN n = 6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| * C = 0.1, gamma = 1 | ||||||||||||
| All features (second dataset) | Accuracy | 73.17 | 73.17 | 70.73 | 63.41 | 66.67 | 64.71 | 70.59 | 64.71 | 70.59 | 82.35 | 80.39 |
| F1 Score | 42.25 | 42.25 | 57.29 | 38.81 | 45.02 | 61.36 | 62.22 | 47.84 | 62.22 | 62.32 | 52.78 | |
| Recall | 50.0 | 50.0 | 56.96 | 43.33 | 52.78 | 61.36 | 62.12 | 52.52 | 62.12 | 61.11 | 52.78 | |
| * C = 10, gamma = 0.01 | ||||||||||||
| F21 | Accuracy | 73.17 | 73.17 | 63.41 | 58.54 | 54.90 | 52.94 | 54.90 | 50.47 | 58.82 | 49.02 | 52.94 |
| F1 Score | 42.25 | 65.85 | 44.34 | 45.59 | 35.44 | 49.67 | 46.71 | 44.36 | 51.34 | 36.00 | 34.62 | |
| Recall | 50.0 | 50.17 | 46.21 | 45.76 | 48.28 | 50.39 | 50.47 | 52.82 | 54.46 | 43.65 | 46.55 | |
| * C = 10, gamma = 0.1 | ||||||||||||
| F22 | Accuracy | 73.17 | 73.17 | 75.61 | 70.73 | 66.67 | 58.82 | 64.71 | 72.55 | 66.67 | 68.73 | 70.59 |
| F1 Score | 42.25 | 42.25 | 66.94 | 60.32 | 40.00 | 47.11 | 44.02 | 48.11 | 49.03 | 53.64 | 41.38 | |
| Recall | 50.0 | 50.0 | 66.06 | 59.85 | 45.95 | 47.20 | 46.81 | 52.22 | 50.39 | 53.96 | 48.65 | |
| * C = 0.1, gamma = 1 | ||||||||||||
| F23 | Accuracy | 73.17 | 73.17 | 60.98 | 70.73 | 72.55 | 68.62 | 64.71 | 72.52 | 74.51 | 72.54 | 74.50 |
| F1 Score | 42.25 | 42.25 | 52.87 | 64.66 | 48.11 | 53.62 | 50.96 | 56.46 | 63.51 | 59.43 | 49.19 | |
| Recall | 50.0 | 50.0 | 53.18 | 65.61 | 52.22 | 53.95 | 51.25 | 56.66 | 62.45 | 58.88 | 53.57 | |
| * C = 0.1, gamma = 1 | ||||||||||||
| F24 | Accuracy | 73.17 | 73.17 | 75.61 | 68.29 | 72.55 | 62.75 | 62.75 | 64.71 | 70.59 | 68.63 | 72.55 |
| F1 Score | 42.25 | 42.25 | 61.17 | 51.76 | 42.05 | 43.03 | 38.55 | 39.29 | 47.06 | 46.03 | 42.05 | |
| Recall | 50.0 | 50.0 | 60.30 | 52.42 | 50.0 | 45.46 | 43.24 | 44.59 | 50.87 | 49.52 | 50.0 | |
| * C = 0.1, gamma = 1 | ||||||||||||
| F17 | Accuracy | 73.17 | 73.17 | 70.71 | 65.85 | 88.23 | 64.70 | 70.59 | 76.47 | 74.51 | 82.35 | 80.39 |
| F1 Score | 42.25 | 42.25 | 57.29 | 45.64 | 46.88 | 44.02 | 55.03 | 50.32 | 54.12 | 60.33 | 52.78 | |
| Recall | 50.0 | 50.0 | 56.97 | 47.88 | 50.0 | 43.88 | 61.67 | 50.56 | 56.67 | 61.11 | 52.78 | |
| Feature | Parameter and Value (%) | Classifier | Parameter and Value (%) | Classifier | ||
|---|---|---|---|---|---|---|
| (a) SMOTE and 10-fold | (b) Normalization, SMOTE and 10-fold | |||||
| F11 | Accuracy | 91.03 | ADA | Accuracy | 91.02 | GBC |
| F1 Score | 91 | F1 Score | 91.31 | |||
| Recall | 91.67 | Recall | 93.89 | |||
| F12 | Accuracy | 93.46 | GBC | Accuracy | 91.03 | GBC |
| F1 Score | 93.65 | F1 Score | 91.21 | |||
| Recall | 95.46 | Recall | 94.03 | |||
| F13 | Accuracy | 89.82 | GBC | Accuracy | 91.65 | GBC |
| F1 Score | 89.53 | F1 Score | 91.73 | |||
| Recall | 87.92 | Recall | 94.03 | |||
| F14 | Accuracy | 86.25 | KNN (n = 3) | Accuracy | 89.23 | LGBM |
| F1 Score | 87.63 | F1 Score | 89.47 | |||
| Recall | 95.14 | Recall | 91.53 | |||
| F15 | Accuracy | 89.23 | XGBC | Accuracy | 86.84 | XGBC |
| F1 Score | 89.71 | F1 Score | 86.73 | |||
| Recall | 93.06 | Recall | 86.67 | |||
| F16 | Accuracy | 90.48 | DT | Accuracy | 88.64 | DT |
| F1 Score | 90.73 | F1 Score | 88.6 | |||
| Recall | 91.81 | Recall | 88.19 | |||
| F17 | Accuracy | 90.51 | GBC | Accuracy | 92.79 | RF |
| F1 Score | 90.04 | F1 Score | 92.68 | |||
| Recall | 88.19 | Recall | 93.89 | |||
| Features | Accuracy (%) | Classifier | Feature | Accuracy (%) | Classifier |
|---|---|---|---|---|---|
| First Dataset | Second Dataset | ||||
| All Feature | 88.31 | LR | All Feature | 82.35 | KNN n = 3 |
| F11 | 89.52 | GBC | F21 | 73.17 | SVM |
| F12 | 90.91 | LR | F22 | 75.61 | GBC |
| F13 | 86.36 | LR | F23 | 74.50 | KNN (n = 6) |
| F14 | 91.56 | LR | F24 | 75.61 | GBC |
| F17 | 88.31 | LR | F25 | 88.23 | LR |
| Features | Accuracy (%) | Classifier | Features | Accuracy (%) | Classifier |
|---|---|---|---|---|---|
| First Dataset with Normalization, SMOTE and 10-Fold | Second Dataset with Normalization, SMOTE and 10-Fold | ||||
| F11 | 91.02 | GBC | F21 | 85.0 | RF |
| F12 | 91.03 | GBC | F22 | 90.0 | ADA |
| F13 | 91.65 | GBC | F23 | 83.33 | SVM |
| F14 | 89.23 | LGBM | F24 | 86.67 | GBC |
| F17 | 92.79 | RF | F25 | 86.67 | RF |
5.1. Experiment I: Classification without Feature Selection
5.2. Experiment II: Classification with Feature Selection
5.3. Experiment III: Classification with SMOTE, Feature Selection, and 10-Fold Cross-Validation
5.4. Experiment IV: Classification with Normalization, SMOTE, Feature Selection, and 10-Fold Cross-Validation
| Ref. | Method | Features | Accuracy |
|---|---|---|---|
| [45] | CNN, SVM, MLP, RF, PLS, LR, ET, LGBM, XGB, DT, and KNNs implemented in Python | Age, Gender, MCHC, RDW, Hct, MCV, RBC, Hb, MCH | 80.77% with CNN |
| [40] | KNN, SVM, LR, NB, MLP, RF, ADA Boosting, Xgboost, DT, and GBC using Google Colab Data Balance: SMOTE | TLC, MCV, MCH, RDW, MCHC, PCV, RBC count, HGB, PLT, age, gender | 100% with ADA |
| [51] | SVM. | Hb, WBC, HCT, RBC, MCH, MCV, RDW, PLT, MCHC RDW, RDWI, E&F, S&L, G&K, MDHL, MCHD, HH index, αT, βT | NA |
| [52] | RF with R software version 3.6.2. | RDW, Hct, MCV, RBC, Hbf, Hba, Hb, MCH, pregnancy and age, | 91.5% |
| [58] | DNN model with 11 features then removing some of the 11 features | HB, RBC, RDW MCV, HCT, WBC, MCH, PLT, MCHC, patient’s sex and age but RDW, age, sex, WBC, and PLT are more important | 89.7% |
| Our Model (First dataset) | LR without feature selection | Age, Sex, Hct, Hb, MCH, MCV, MCHC, RDW, RBC count | 88.31% |
| χ2 feature section with LR Classifier | MCV, MCH, RDW, Hb, RBC count, and Hct | 91.56%, | |
| SMOTE, Feature importance base selection, and GBC | Age, Hb, MCV, MCH, RDW, RBC count | 93.46% | |
| Normalization, SMOTE, Exhaustive Feature Selection with RF | Age, Sex, Hb, MCV | 92.79% | |
| Our Model (Second dataset) | KNN (n = 3) without feature selection | Hb, PCV, RCB, MCV, MCH, MCHC, RDW, WBC, Neut, LYMPH, PLT, HBA, HBA2, HBF, Sex | 82.35% |
| Exhaustive Feature Selection (EFS) with LR classifier | HB, PCV, RBC, MCV | 88.33% | |
| SMOTE, Feature importance base selection, and XGB | Hb, PCV, RBC, MCV, MCH, WBC, Lymph, HBA2 | 88.33% | |
| Normalization, SMOTE Feature importance using GBDT with ADA | Hb, PCV, MCV, MCH, MCHC, RDW, WBC, Neut, LYMPH, PLT, HBA2, HBF | 90.0% |
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Acronyms | Meaning |
| αT | Alpha-thalassemia |
| ADA | AdaBoost |
| ANFIS | Adaptive neuro-fuzzy inference system |
| ADASYN | Adaptive Synthetic |
| ANN | Artificial neural network |
| AUC | Area Under the ROC Curve |
| βT | Beta-thalassemia |
| BLTREED | Bayesian Logit Treed |
| CBC | Complete Blood Count |
| CNN | Convolutional Neural Network |
| CNN-LSTM | CNN Long Short-Term Memory Network |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DSIFT | Dense Scale Invariant Feature Transform |
| DTL2 | CNN as a feature extractor |
| DT | Decision Tree |
| DHS | Dynamic Harmony Search |
| E&F | England and Fraser |
| ETC | Extra Tree Classifier |
| ET | Extremely Randomized Trees |
| ELM | Extreme Learning Machine |
| FNR | False-negative rate |
| FPR | False-positive rate |
| GBM | Gradient Boosting Machine |
| GBC | Gradient Boosting Classifier |
| GBDT | Gradient Boosting Decision Trees |
| G&K | Green and King |
| GRU | Gated Recurrent Unit |
| Hb/HBA | Hemoglobin |
| HBA2 | Hemoglobin A2 |
| HBF | Fetal hemoglobin |
| HGB | Hemoglobin in grams per deciliter of blood |
| HH index | Huber–Herklotz index |
| IDA | Iron deficiency anemia |
| IoU | Intersection Over Union |
| KNN | K-Nearest Neighbors |
| LR | Logistic Regression |
| LSTM | Long Short-Term Memory |
| LGBM | Light Gradient Boosting Machine |
| L-SVC | Liner SVC |
| lgbmR | Light GBM Regressor |
| Hct | Hematocrit |
| MCV | Mean Cell Volume |
| MCH | Mean Cell Hemoglobin |
| MCHC | Mean Cell Hemoglobin Concentration |
| MCHD | Index, Mean Cell Hemoglobin Density |
| MDHL | Mean Density of Hb/liter of blood |
| MLPs | Multilayer perceptron |
| MLR | Multiple Linear Regression |
| NB | Naive Bayes |
| NCA | Component Analysis Feature Selection |
| NN | Neural Networks |
| NPV | Negative predictive value |
| Neut | Neutrophils |
| PBIS | Pattern-based index selection |
| PCA | Principal component analysis |
| PCV | Polycythemia vera |
| PLT | Platelet |
| PTPP | Punjab Thalassemia Prevention Program |
| PPV | Positive predictive value |
| PLS | partial least squares |
| RANSAC | Random sample consensus |
| RF | Random Forest |
| RBC | Red blood cell |
| RBF | Gaussian radial basis function |
| RDW | Red Cell Distribution Width |
| RDWI | Red Cell Distribution Width Index |
| RELM | Regularized Extreme Learning machine |
| SVD | Singular Value Decomposition |
| SVC | Support Vector Classifier |
| SVM | Support Vector Machine |
| S&L | Shine and Lal |
| SMOTE | Synthetic Minority Oversampling Technique |
| TSVM | Twin Support Vector Machines |
| TT | Thalassemia Trait |
| WBC | White blood cell |
| XGB | Extreme Gradient Boosting |
| Xgboost | Extreme Gradient Boosting |
| χ2 | Chi-square |
| Iχ2 | Iterative Chi-Square |
References
- Baird, D.C.; Batten, S.H.; Sparks, S.K. Alpha- and Beta-thalassemia: Rapid Evidence Review. Am. Fam. Physician 2022, 105, 272–280. Available online: https://pubmed.ncbi.nlm.nih.gov/35289581/ (accessed on 10 May 2023). [PubMed]
- Weatherall, D.J.; Clegg, J.B. Inherited haemoglobin disorders: An increasing global health problem. Bull. World Health Organ. 2001, 79, 704–712. Available online: https://pubmed.ncbi.nlm.nih.gov/11545326/ (accessed on 10 May 2023).
- Taher, A.T.; Weatherall, D.J.; Cappellini, M.D. Thalassaemia. Lancet 2018, 391, 155–167. [Google Scholar] [CrossRef]
- Fibach, E.; Dana, M. Oxidative Stress in β-Thalassemia. Mol. Diagn. Ther. 2019, 23, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Kattamis, A.; Kwiatkowski, J.L.; Aydinok, Y. Thalassaemia. Lancet 2022, 399, 2310–2324. [Google Scholar] [CrossRef] [PubMed]
- Muncie, H.L.; Campbell, J. Alpha and beta thalassemia. Am. Fam. Physician 2009, 80, 339–344. Available online: https://pubmed.ncbi.nlm.nih.gov/19678601/ (accessed on 10 May 2023).
- Erten, M.; Tuncer, T. Automated differential diagnosis method for iron deficiency anemia and beta thalassemia trait based on iterative Chi2 feature selector. Int. J. Lab. Hematol. 2022, 44, 430–436. [Google Scholar] [CrossRef]
- Rustam, F.; Ashraf, I.; Jabbar, S.; Tutusaus, K.; Mazas, C.; Barrera, A.E.P.; de la Torre, D. Prediction of β -Thalassemia carriers using complete blood count features. Sci. Rep. 2022, 12, 19999. [Google Scholar] [CrossRef]
- Musiałek, M.W.; Rybaczek, D. Hydroxyurea—The Good, the Bad and the Ugly. Genes 2021, 12, 1096. [Google Scholar] [CrossRef]
- Bazinet, A.; Popradi, G. A General Practitioner’s Guide to Hematopoietic Stem-cell Transplantation. Curr. Oncol. 2019, 26, 187–191. [Google Scholar] [CrossRef]
- Hatzimichael, E.; Timotheatou, D.; Koumpis, E.; Benetatos, L.; Makis, A. Luspatercept: A New Tool for the Treatment of Anemia Related to β-Thalassemia, Myelodysplastic Syndromes and Primary Myelofibrosis. Diseases 2022, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Jamwal, M.; Sharma, P.; Das, R. Laboratory Approach to Hemolytic Anemia. Indian J. Pediatr. 2020, 87, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, R.A.; Khodeary, A.; Farhan, M.S. Detection of endocrine disorders in young children with multi-transfused thalassemia major. Ital. J. Pediatr. 2021, 47, 165. [Google Scholar] [CrossRef] [PubMed]
- Akiki, N.; Hodroj, M.H.; Bou-Fakhredin, R.; Matli, K.; Taher, A.T. Cardiovascular Complications in β-Thalassemia: Getting to the Heart of It. Thalass. Rep. 2023, 13, 38–50. [Google Scholar] [CrossRef]
- Meloni, A.; Pistoia, L.; Positano, V.; De Luca, A.; Martini, N.; Spasiano, A.; Fotzi, I.; Bitti, P.P.; Visceglie, D.; Alberini, G.; et al. Increased myocardial extracellular volume is associated with myocardial iron overload and heart failure in thalassemia major. Eur. Radiol. 2022, 33, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Dimitroglou, Y.; Anagnostopulous, F.; Aggeli, C.; Delicou, S.; Xydaki, A.; Patsourakos, D.; Tousoulis, D. Severity of heart failure and health-related quality of life in beta-thalassemia patients: A cross-sectional study. Ann. Hematol. 2020, 99, 2037–2046. [Google Scholar] [CrossRef]
- Hoffmann, J.J.M.L.; Urrechaga, E.; Aguirre, U. Discriminant indices for distinguishing thalassemia and iron deficiency in patients with microcytic anemia: A meta-analysis. Clin. Chem. Lab. Med. (CCLM) 2015, 53, 1883–1894. [Google Scholar] [CrossRef]
- Zheng, L.; Huang, H.; Wu, X.; Su, L.; Shen, Q.; Wang, M.; Lin, N.; Xu, L. Screening of Some Indicators for Alpha-Thalassemia in Fujian Province of Southern China. Int. J. Gen. Med. 2021, 14, 7329–7335. [Google Scholar] [CrossRef]
- Husna, N.; Handayani, N.S.N. Molecular and Haematological Characteristics of alpha-Thalassemia Deletions in Yogyakarta Special Region, Indonesia. Rep. Biochem. Mol. Biol. 2021, 10, 346–353. [Google Scholar] [CrossRef]
- Bain, A. Management of Transfusion Dependent Thalassaemia (TDT): A Short Guide; Thalassaemia Internation Federation: Nicosia, Cyprus, 2022. [Google Scholar]
- Gao, J.; Liu, W. Advances in screening of thalassaemia. Clin. Chim. Acta 2022, 534, 176–184. [Google Scholar] [CrossRef]
- Stephens, A. The Diagnosis and Significance of Alpha Thalassaemia. In Practical Management of Haemoglobinopathies; Blackwell Publishing Ltd.: Oxford, UK, 2004; pp. 40–44. [Google Scholar] [CrossRef]
- Galanello, R.; Cao, A. Alpha-thalassemia. Genet. Med. 2011, 13, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Porter, D.; Taher, J. Guidelines for the Management of Transfusion Dependent Thalassaemia (TDT), 4th ed.; Thalassaemia International Federation: Nicosia, Cyprus, 2021. [Google Scholar]
- Cao, A.; Galanello, R. Beta-thalassemia. Genet. Med. 2010, 12, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, V.P. Thalassemia Minor and Major: Current Management. Indian J. Pediatr. 2017, 84, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Musallam, K.M.; Taher, A.T.; Rachmilewitz, E.A. β-thalassemia intermedia: A clinical perspective. Cold Spring Harb. Perspect. Med. 2012, 2, a013482. [Google Scholar] [CrossRef]
- Taher, A.; Isma’eel, H.; Cappellini, M.D. Thalassemia intermedia: Revisited. Blood Cells Mol. Dis. 2006, 37, 12–20. [Google Scholar] [CrossRef]
- Cunningham, M.J. Update on Thalassemia: Clinical Care and Complications. Hematol. Oncol. Clin. N. Am. 2010, 24, 215–227. [Google Scholar] [CrossRef]
- Welcome to the Globin Gene Server. Available online: https://globin.bx.psu.edu/ (accessed on 5 August 2023).
- Nigam, N.; Kushwaha, R.; Yadav, G.; Singh, P.K.; Gupta, N.; Singh, B.; Agrawal, M.; Chand, P.; Saxena, S.K.; Bhatt, M.L.B. A demographic prevalence of β Thalassemia carrier and other hemoglobinopathies in adolescent of Tharu population. J. Fam. Med. Prim. Care 2020, 9, 4305–4310. [Google Scholar] [CrossRef]
- Shash, H. Non-Transfusion-Dependent Thalassemia: A Panoramic Review. Medicina 2022, 58, 1496. [Google Scholar] [CrossRef]
- Lama, R.; Yusof, W.; Shrestha, T.R.; Hanafi, S.; Bhattarai, M.; Hassan, R.; Zilfalil, B.A. Prevalence and distribution of major β-thalassemia mutations and HbE/β-thalassemia variant in Nepalese ethnic groups. Hematol. Oncol. Stem Cell. Ther. 2021, 15, 3. [Google Scholar] [CrossRef]
- Porter, J.B.; Garbowski, M. The Pathophysiology of Transfusional Iron Overload. Hematol. Oncol. Clin. N. Am. 2014, 28, 683–701. [Google Scholar] [CrossRef]
- Marsella, M.; Ricchi, P. Thalassemia and hepatocellular carcinoma: Links and risks. J. Blood Med. 2019, 10, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.T.; Yassin, M.A.; De Sanctis, V. Final adult height and endocrine complications in young adults with β-thalassemia major (TM) who received oral iron chelation (OIC) in comparison with those who did not use OIC. Acta Biomed. 2018, 89, 27–32. [Google Scholar] [CrossRef] [PubMed]
- De Sanctis, V.; Soliman, A.T.; Canatan, D.; Tzoulis, P.; Daar, S.; Di Maio, S.; Elsedfy, H.; Yassin, M.A.; Filosa, A.; Soliman, N.; et al. An ICET-A survey on occult and emerging endocrine complications in patients with β-thalassemia major: Conclusions and recommendations. Acta Biomed. 2019, 89, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, S.; Khalid, M.U.; Mui-Zzud-Din; Ullah, S.; Aslam, W.; Mehmood, A.; Choi, G.S.; On, B.-W. Classification of β-Thalassemia Carriers from Red Blood Cell Indices Using Ensemble Classifier. IEEE Access 2021, 9, 45528–45538. [Google Scholar] [CrossRef]
- AlAgha, A.S.; Faris, H.; Hammo, B.H.; Al-Zoubi, A.M. Identifying β-thalassemia carriers using a data mining approach: The case of the Gaza Strip, Palestine. Artif. Intell. Med. 2018, 88, 70–83. [Google Scholar] [CrossRef]
- Devanath, A.; Akter, S.; Karmaker, P.; Sattar, A. Thalassemia Prediction using Machine Learning Approaches. In Proceedings of the 6th International Conference on Computing Methodologies and Communication (ICCMC 2022), Erode, India, 29–31 March 2022; pp. 1166–1174. [Google Scholar] [CrossRef]
- Purwar, S.; Tripathi, R.; Ranjan, R.; Saxena, R. Classification of thalassemia patients using a fusion of deep image and clinical features. In Proceedings of the Confluence 2021: 11th International Conference on Cloud Computing, Data Science and Engineering, Uttar Pradesh, India, 28–29 January 2021; pp. 410–415. [Google Scholar] [CrossRef]
- Khan, M.S.; Ullah, A.; Khan, K.N.; Riaz, H.; Yousafzai, Y.M.; Rahman, T.; Chowdhury, M.E.H.; Abul Kashem, S.B. Deep Learning Assisted Automated Assessment of Thalassaemia from Haemoglobin Electrophoresis Images. Diagnostics 2022, 12, 2405. [Google Scholar] [CrossRef]
- Jin, B.; Cruz, L.; Goncalves, N. Deep Facial Diagnosis: Deep Transfer Learning from Face Recognition to Facial Diagnosis. IEEE Access 2020, 8, 123649–123661. [Google Scholar] [CrossRef]
- Jahan, A.; Singh, G.; Gupta, R.; Sarin, N.; Singh, S. Role of Red Cell Indices in Screening for Beta Thalassemia Trait: An Assessment of the Individual Indices and Application of Machine Learning Algorithm. Indian J. Hematol. Blood Transfus. 2021, 37, 453–457. [Google Scholar] [CrossRef]
- Phirom, K.; Charoenkwan, P.; Shoombuatong, W.; Charoenkwan, P.; Sirichotiyakul, S.; Tongsong, T. DeepThal: A Deep Learning-Based Framework for the Large-Scale Prediction of the α+-Thalassemia Trait Using Red Blood Cell Parameters. J. Clin. Med. 2022, 11, 6305. [Google Scholar] [CrossRef]
- Aszhari, F.R.; Rustam, Z.; Subroto, F.; Semendawai, A.S. Classification of thalassemia data using random forest algorithm. J. Phys. Conf. Ser. 2020, 1490, 012050. [Google Scholar] [CrossRef]
- Wirasati, I.; Rustam, Z.; Aurelia, J.E.; Hartini, S.; Saragih, G.S. Comparison some of kernel functions with support vector machines classifier for thalassemia dataset. IAES Int. J. Artif. Intell. 2021, 10, 430–437. [Google Scholar] [CrossRef]
- Sa’Id, A.A.; Rustam, Z.; Novkaniza, F.; Setiawan, Q.S.; Maulidina, F.; Wibowo, V.V.P. Twin Support Vector Machines for Thalassemia Classification. In Proceedings of the 2021 International Conference on Innovation and Intelligence for Informatics, Computing, and Technologies (3ICT 2021), Zallaq, Bahrain, 29–30 September 2021; pp. 160–164. [Google Scholar] [CrossRef]
- Hartini, S.; Rustam, Z. Hierarchical Clustering Algorithm Based on Density Peaks using Kernel Function for Thalassemia Classification. J. Phys. Conf. Ser. 2019, 1417, 012016. [Google Scholar] [CrossRef]
- Laeli, A.R.; Rustam, Z.; Hartini, S.; Maulidina, F.; Aurelia, J.E. Hyperparameter Optimization on Support Vector Machine using Grid Search for Classifying Thalassemia Data. In Proceedings of the 2020 International Conference on Decision Aid Sciences and Application (DASA 2020), Virtual, 8–9 November 2020; pp. 817–821. [Google Scholar] [CrossRef]
- Fu, Y.K.; Liu, H.-M.; Lee, L.-H.; Chen, Y.-J.; Chien, S.-H.; Lin, J.-S.; Chen, W.-C.; Cheng, M.-H.; Lin, P.-H.; Lai, J.-Y.; et al. The tvgh-nycu thal-classifier: Development of a machine-learning classifier for differentiating thalassemia and non-thalassemia patients. Diagnostics 2021, 11, 1725. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Li, Y.; Liao, Z.; Yao, Z.; Lin, W.; Xie, S.; Hu, B.; Huang, C.; Liu, W.; Xu, H.; et al. An online alpha-thalassemia carrier discrimination model based on random forest and red blood cell parameters for low HbA2 cases. Clin. Chim. Acta 2022, 525, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Çil, B.; Ayyıldız, H.; Tuncer, T. Discrimination of β-thalassemia and iron deficiency anemia through extreme learning machine and regularized extreme learning machine based decision support system. Med. Hypotheses 2020, 138, 109611. [Google Scholar] [CrossRef] [PubMed]
- Ayyıldız, H.; Arslan, S. Tuncer Determination of the effect of red blood cell parameters in the discrimination of iron deficiency anemia and beta thalassemia via Neighborhood Component Analysis Feature Selection-Based machine learning. Chemom. Intell. Lab. Syst. 2020, 196, 103886. [Google Scholar] [CrossRef]
- Jahangiri, M.; Rahim, F.; Saki, N.; Saki, A. Malehi Application of Bayesian Decision Tree in Hematology Research: Differential Diagnosis of β -Thalassemia Trait from Iron Deficiency Anemia. Comput. Math. Methods Med. 2021, 2021, 6401105. [Google Scholar] [CrossRef]
- Susanto, E.R.; Syarif, A.; Muludi, K.; Perdani, R.R.W.; Wantoro, A. Implementation of Fuzzy-based Model for Prediction of Thalassemia Diseases. J. Phys. Conf. Ser. 2021, 1751, 012034. [Google Scholar] [CrossRef]
- Qasem, N.; Mosavi, A. Novel Meta-Heuristic Model for Discrimination between Iron Deficiency Anemia and Β-Thalassemia with CBC Indices Based on Dynamic Harmony Search (DHS). arXiv 2020, arXiv:2004.00480. [Google Scholar]
- Mo, D.; Zheng, Q.; Xiao, B.; Li, L. Predicting thalassemia using deep neural network based on red blood cell indices. Clin. Chim. Acta 2023, 543, 117329. [Google Scholar] [CrossRef]
- Egejuru, N.C.; Olusanya, S.O.; Asinobi, A.O.; Adeyemi, O.J.; Adebayo, V.O.; Idowu, P.A. Using Data Mining Algorithms for Thalassemia Risk Prediction. Int. J. Biomed. Sci. Eng. 2019, 7, 33. [Google Scholar] [CrossRef]
- Epah, J.; Gülec, I.; Winter, S.; Dörr, J.; Geisen, C.; Haecker, E.; Link, D.; Schwab, M.; Seifried, E.; Schäfer, R. From Unit to Dose: A Machine Learning Approach for Precise Prediction of Hemoglobin and Iron Content in Individual Packed Red Blood Cell Units. Adv. Sci. 2022, 9, 2204077. [Google Scholar] [CrossRef] [PubMed]
- Kabootarizadeh, L.; Jamshidnezhad, A.; Koohmareh, Z. Differential Diagnosis of Iron-Deficiency Anemia from β-Thalassemia Trait Using an Intelligent Model in Comparison with Discriminant Indexes. Acta Inform. Med. 2019, 27, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zaheri, S.; Nguyen, K.; Liu, L.; Hassanipour, F.; Bleris, L. Machine learning-based approaches for identifying human blood cells harboring CRISPR-mediated fetal chromatin domain ablations. Sci. Rep. 2022, 12, 1481. [Google Scholar] [CrossRef] [PubMed]
- Al-Hakeim, H.K.; Najm, A.H.; Moustafa, S.R.; Maes, M. Construction of an exposure-pathway-phenotype in children with depression due to transfusion-dependent thalassemia: Results of (un)supervised machine learning. J. Affect. Disord. 2021, 282, 644–655. [Google Scholar] [CrossRef]
- Lee, S.Y.; Chen, C.M.; Lim, E.Y.; Shen, L.; Sathe, A.; Singh, A.; Sauer, J.; Taghipour, K.; Yip, C.Y. Image Analysis Using Machine Learning for Automated Detection of Hemoglobin H Inclusions in Blood Smears—A Method for Morphologic Detection of Rare Cells. J. Pathol. Inform. 2021, 12, 18. [Google Scholar] [CrossRef]
- Positano, V.; Meloni, A.; Santarelli, M.F.; Pistoia, L.; Spasiano, A.; Cuccia, L.; Casini, T.; Gamberini, M.R.; Allò, M.; Bitti, P.P.; et al. Deep Learning Staging of Liver Iron Content from Multiecho MR Images. J. Magn. Reason. Imaging 2023, 57, 472–484. [Google Scholar] [CrossRef]
- Tyas, D.A.; Hartati, S.; Harjoko, A.; Ratnaningsih, T. Morphological, Texture, and Color Feature Analysis for Erythrocyte Classification in Thalassemia Cases. IEEE Access 2020, 8, 69849–69860. [Google Scholar] [CrossRef]
- Hortinela, C.C.; Balbin, J.R.; Fausto, J.C.; Divina, P.D.C.; Felices, J.P.T. Identification of Abnormal Red Blood Cells and Diagnosing Specific Types of Anemia Using Image Processing and Support Vector Machine. In Proceedings of the 2019 IEEE 11th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), Laoag, Philippines,, 29 November–1 December 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Pellegrino, R.V.; Tarrobago, A.C.; Zulueta, D.L.B. Development of Anemia Cells Recognition System Using Raspberry Pi. In Proceedings of the 2023 15th International Conference on Computer and Automation Engineering (ICCAE), Sydney, Australia, 3–5 March 2023; pp. 198–203. [Google Scholar] [CrossRef]
- Abdulhay, E.W.; Allow, A.G.; Al-Jalouly, M.E. Detection of Sickle Cell, Megaloblastic Anemia, Thalassemia and Malaria through Convolutional Neural Network. In Proceedings of the 2021 Global Congress on Electrical Engineering (GC-ElecEng), Valencia, Spain, 10–12 December 2021; pp. 21–25. [Google Scholar] [CrossRef]
- Zaylaa, A.J.; Makki, M.; Kassem, R. Thalassemia Diagnosis Through Medical Imaging: A New Artificial Intelligence-Based Framework. In Proceedings of the 2022 International Conference on Smart Systems and Power Management (IC2SPM 2022), Beijing, China, 27–30 October 2022; pp. 41–46. [Google Scholar] [CrossRef]
- Alturaiki, A.M.; Banjar, H.R.; Barefah, A.S.; Alnajjar, S.A.; Hindawi, S. A Smart Chatbot for Interactive Management in Beta Thalassemia Patients. Int. J. Telemed. Appl. 2022, 2022, 9734518. [Google Scholar] [CrossRef]
- Banjar, H.R.; Zaher, G.F.; Almutiry, H.S.; Alshamarni, A.S.A.; Almouhana, G.I.; Alahwal, H.M.; Bahashwan, S.; Barefah, A.S.; Alnajjar, S.A.; Alharbi, H.M. Web-based expert system with quick response code for beta-thalassemia management. Health Inform. J. 2021, 27, 1460458221989397. [Google Scholar] [CrossRef]
- Zhang, F.; Yang, J.; Wang, Y.; Cai, M.; Ouyang, J.; Li, J. TT@MHA: A machine learning-based webpage tool for discriminating thalassemia trait from microcytic hypochromic anemia patients. Clin. Chim. Acta 2023, 545, 117368. [Google Scholar] [CrossRef] [PubMed]
- Laengsri, V.; Shoombuatong, W.; Adirojananon, W.; Nantasenamat, C.; Prachayasittikul, V.; Nuchnoi, P. ThalPred: A web-based prediction tool for discriminating thalassemia trait and iron deficiency anemia. BMC Med. Inform. Decis. Mak. 2019, 19, 212. [Google Scholar] [CrossRef]
- Thalassemia_Deep_07072022. Kaggle. Available online: https://www.kaggle.com/code/plenoi/thalassemia-deep-07072022/input?select=Alpha-2_addition-230622.xlsx (accessed on 5 August 2023).
- Alpha Thalassemia Classifier—1. Kaggle. Available online: https://www.kaggle.com/code/letslive/alpha-thalassemia-classifier-1/input (accessed on 23 October 2023).
- Chawla, N.V.; Japkowicz, N.; Kotcz, A. Editorial: Special issue on learning from imbalanced data sets. ACM SIGKDD Explor. Newsl. 2004, 6, 1–6. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Rahman, M.M.; Davis, D.N. Addressing the Class Imbalance Problem in Medical Datasets. Int. J. Mach. Learn. Comput. 2013, 3, 224–228. [Google Scholar] [CrossRef]
- Visalakshi, S.; Radha, V. A literature review of feature selection techniques and applications: Review of feature selection in data mining. In Proceedings of the 2014 IEEE International Conference on Computational Intelligence and Computing Research, Coimbatore, India, 18–20 December 2014; pp. 1–6. [Google Scholar] [CrossRef]
- Raju, D.N.; Shanmugasundaram, H.; Sasikumar, R. Fuzzy segmentation and black widow–based optimal SVM for skin disease classification. Med. Biol. Eng. Comput. 2021, 59, 2019–2035. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Khaire, U.M.; Dhanalakshmi, R. Stability of feature selection algorithm: A review. J. King Saud Univ.—Comput. Inf. Sci. 2022, 34, 1060–1073. [Google Scholar] [CrossRef]
- Speiser, J.L.; Miller, M.E.; Tooze, J.; Ip, E. A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst. Appl. 2019, 134, 93–101. [Google Scholar] [CrossRef]
- Liu, H.; Setiono, R. Chi2: Feature selection and discretization of numeric attributes. In Proceedings of the 7th IEEE International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995; pp. 388–391. [Google Scholar] [CrossRef]
- Dissanayake, K.; Johar, M.G.M. Comparative Study on Heart Disease Prediction Using Feature Selection Techniques on Classification Algorithms. Appl. Comput. Intell. Soft Comput. 2021, 2021, 5581806. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saleem, M.; Aslam, W.; Lali, M.I.U.; Rauf, H.T.; Nasr, E.A. Predicting Thalassemia Using Feature Selection Techniques: A Comparative Analysis. Diagnostics 2023, 13, 3441. https://doi.org/10.3390/diagnostics13223441
Saleem M, Aslam W, Lali MIU, Rauf HT, Nasr EA. Predicting Thalassemia Using Feature Selection Techniques: A Comparative Analysis. Diagnostics. 2023; 13(22):3441. https://doi.org/10.3390/diagnostics13223441
Chicago/Turabian StyleSaleem, Muniba, Waqar Aslam, Muhammad Ikram Ullah Lali, Hafiz Tayyab Rauf, and Emad Abouel Nasr. 2023. "Predicting Thalassemia Using Feature Selection Techniques: A Comparative Analysis" Diagnostics 13, no. 22: 3441. https://doi.org/10.3390/diagnostics13223441
APA StyleSaleem, M., Aslam, W., Lali, M. I. U., Rauf, H. T., & Nasr, E. A. (2023). Predicting Thalassemia Using Feature Selection Techniques: A Comparative Analysis. Diagnostics, 13(22), 3441. https://doi.org/10.3390/diagnostics13223441







