Cloud-Based Quad Deep Ensemble Framework for the Detection of COVID-19 Omicron and Delta Variants
Abstract
:1. Introduction
- An ensemble-based deep transfer learning framework is designed for diagnosing Delta and Omicron variants from CT-scan images.
- We used three deep transfer learning models, i.e., VGG-16, Densenet-121, and Inception-v3, and one CapsNet model during the process of stacking and weighed averaging.
- Our proposed approach generated an overwhelming accuracy of 99.93% over the validation and test data.
- The developed framework is deployed in a cloud-based web application.
2. Related Work
2.1. Artificial Intelligence in Healthcare
2.2. Findings from Literature Review
3. Methodology
3.1. Dataset
3.2. The Proposed Architecture
3.3. Model Deployment
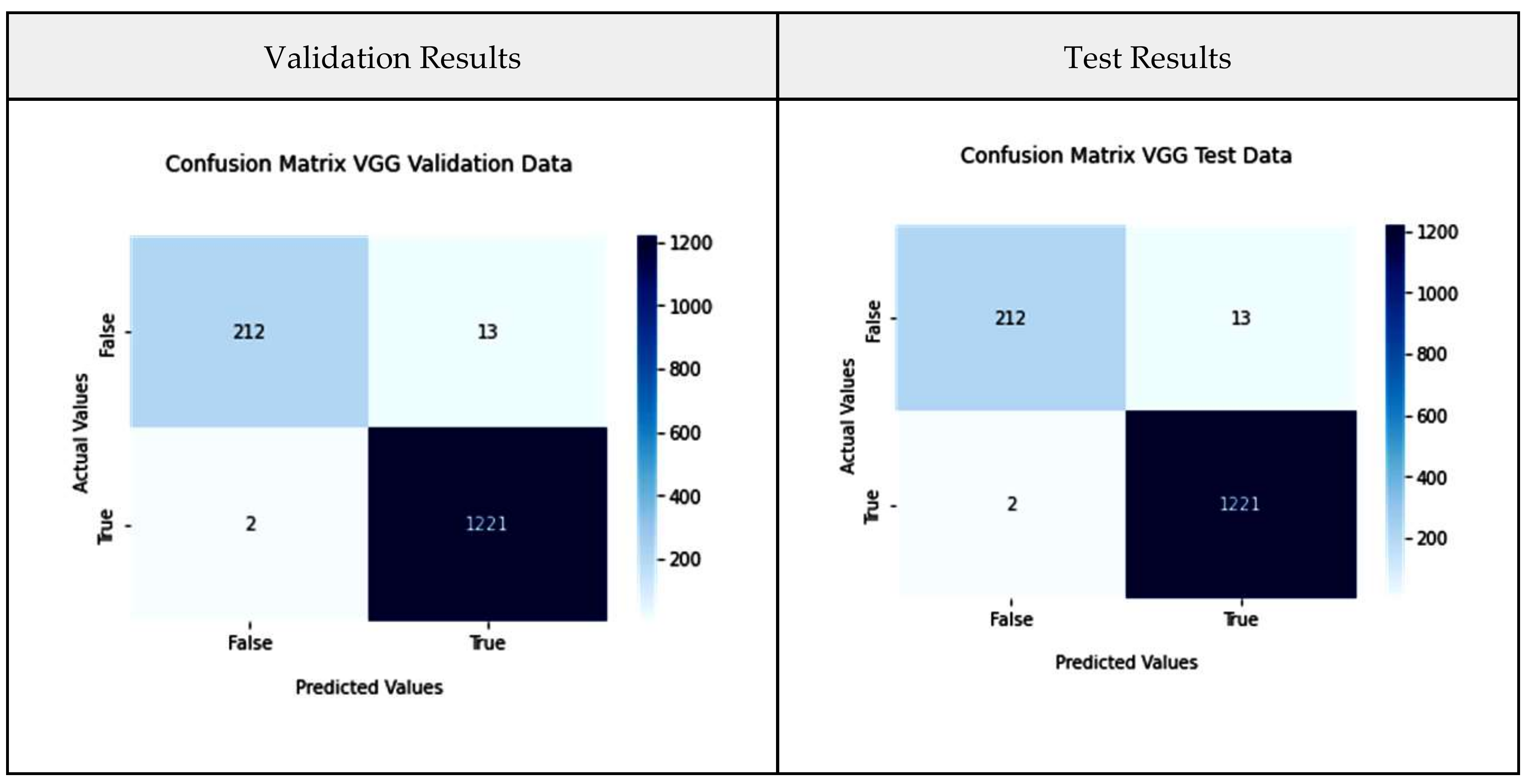
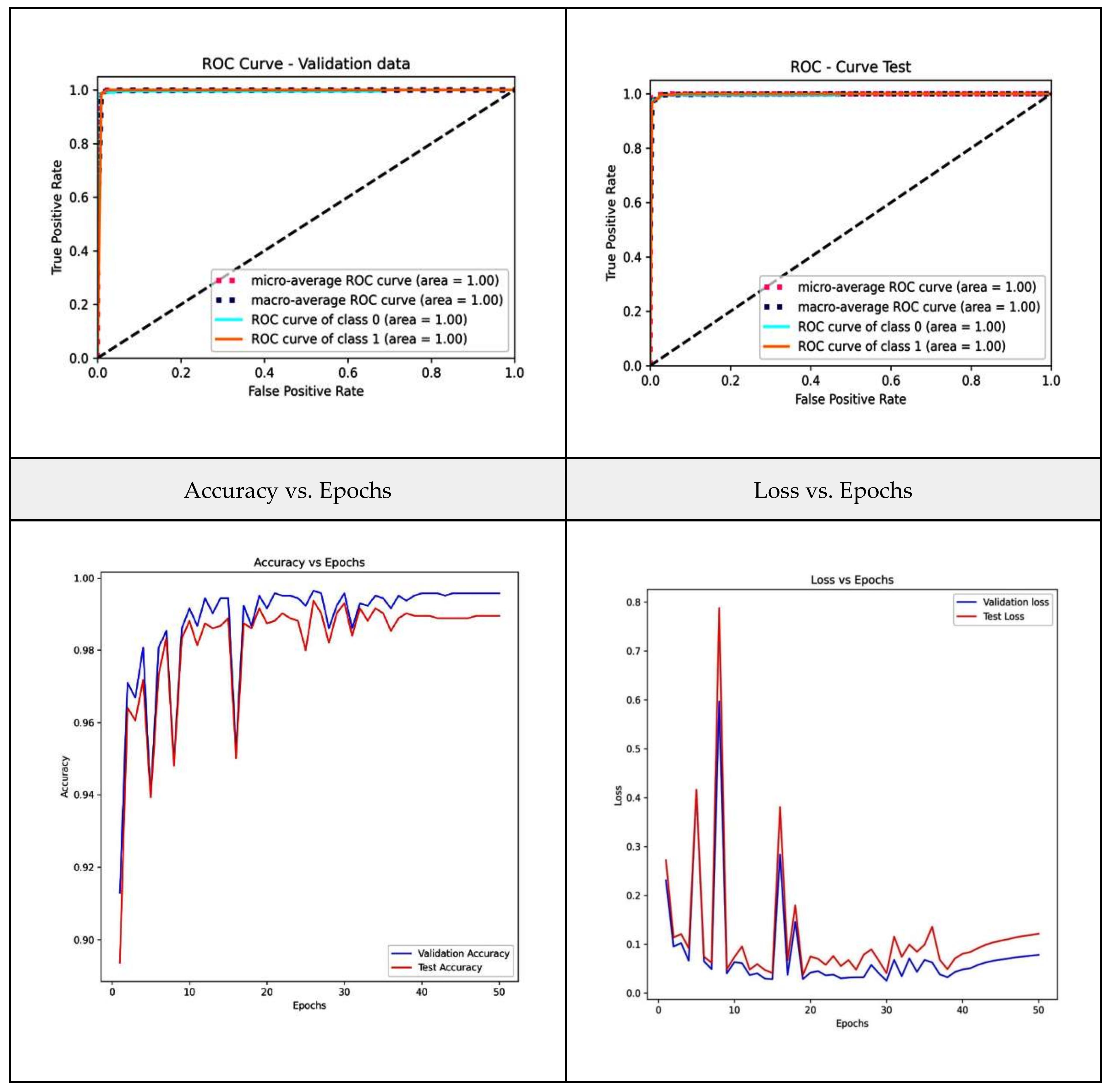
3.4. Validation Metrics
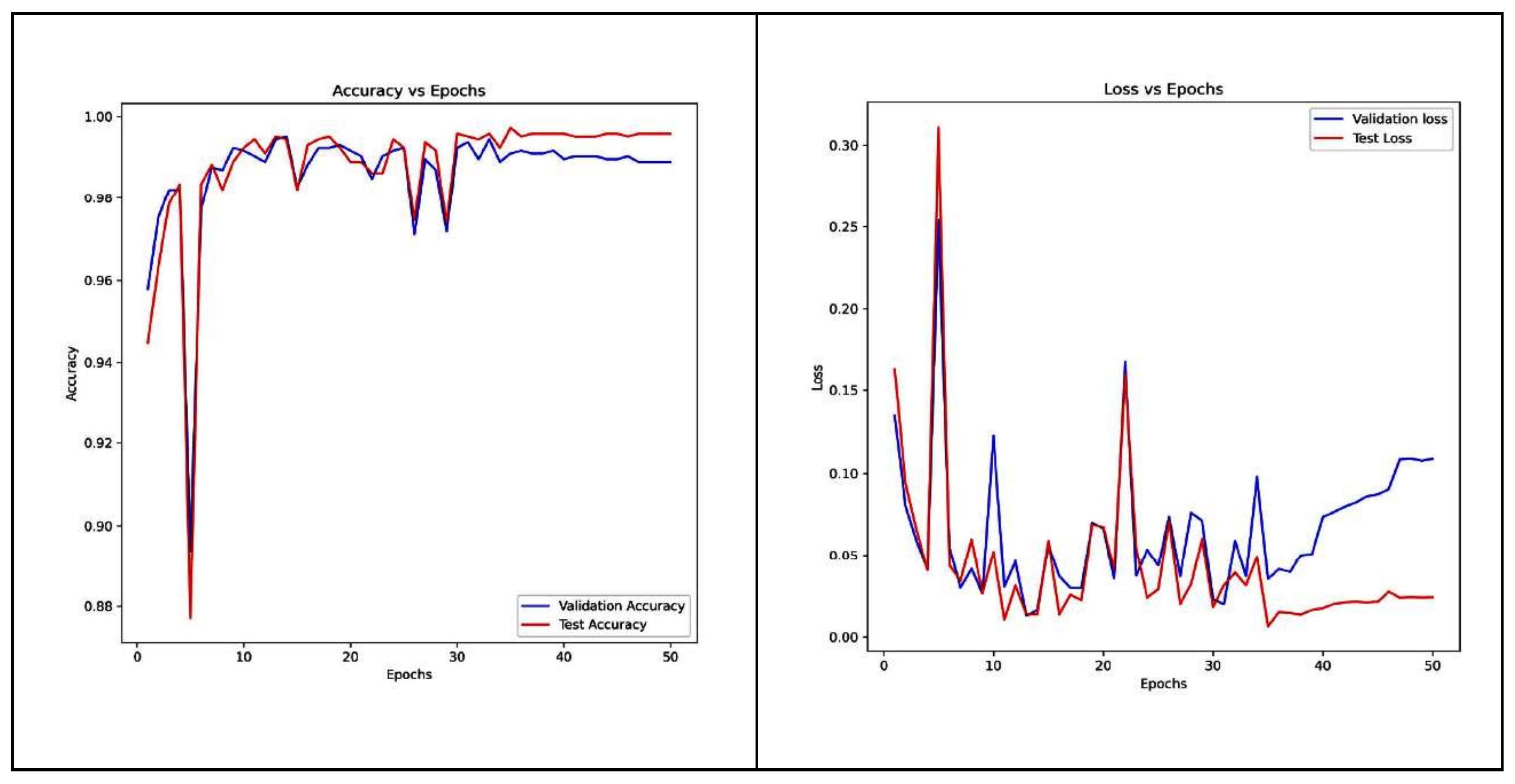
4. Performances of Transfer Learning Models
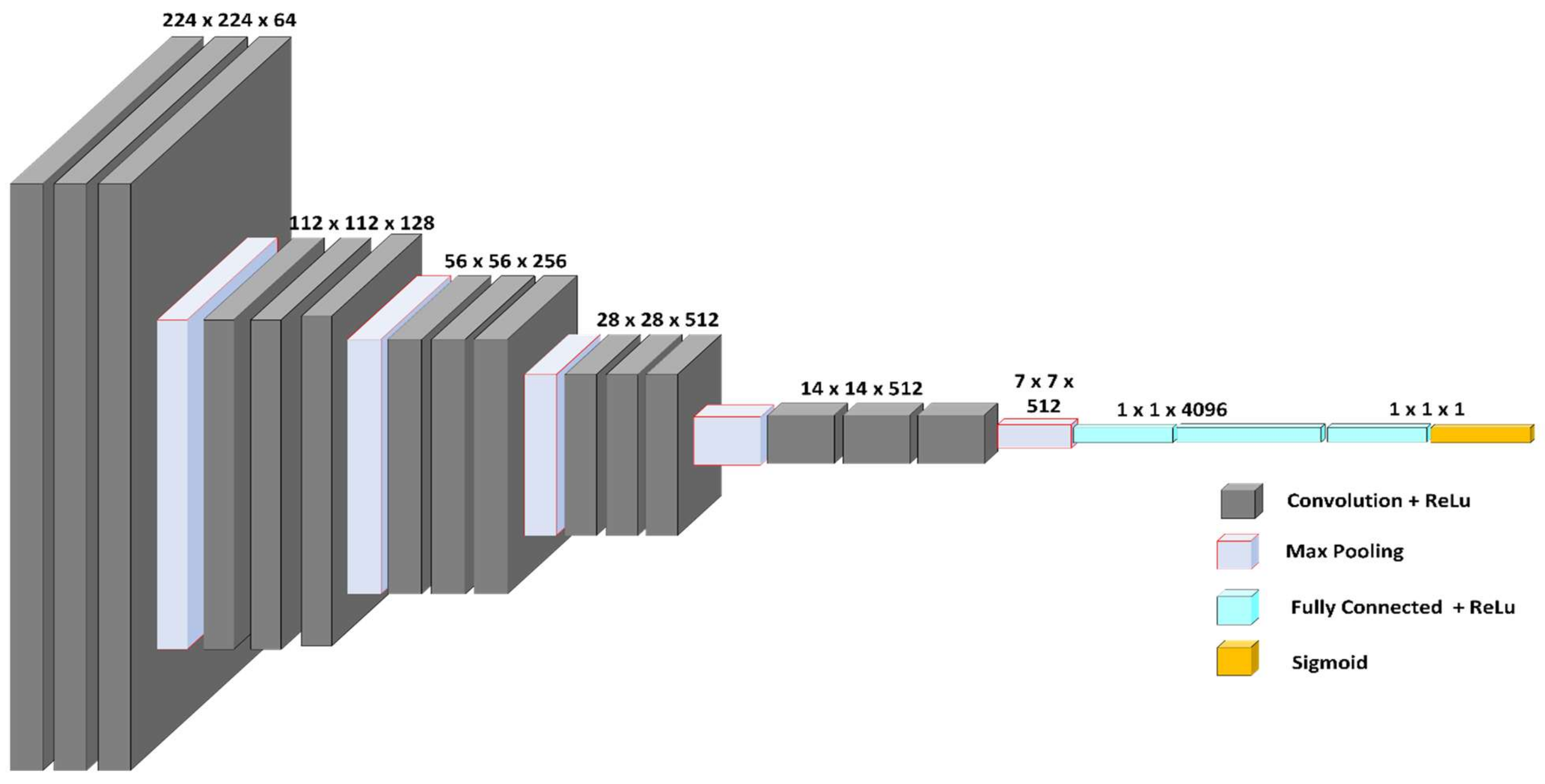
4.1. VGG-16 Model
- Epochs: 8/30.
- Callbacks: Early stopping on val_accuracy with patience 5, restore best weights true.
- Optimizer: Adam with a learning rate of 0.001.
- Loss: Binary Cross Entropy without logits.
- Image_size: (224 × 224 × 3), type: tensor.
- Dense Layer Weight—Initializer: glorot_uniform.
- Activation: Relu.
- Use Bias: True.
- Dense Layer Bias—Initializer: zeros.
- Batch Normalization: a momentum of 0.99 with an epsilon of 0.001 along with a gamma-initializer of ones, a beta-initializer of zeros, and a moving mean initializer of zeros.
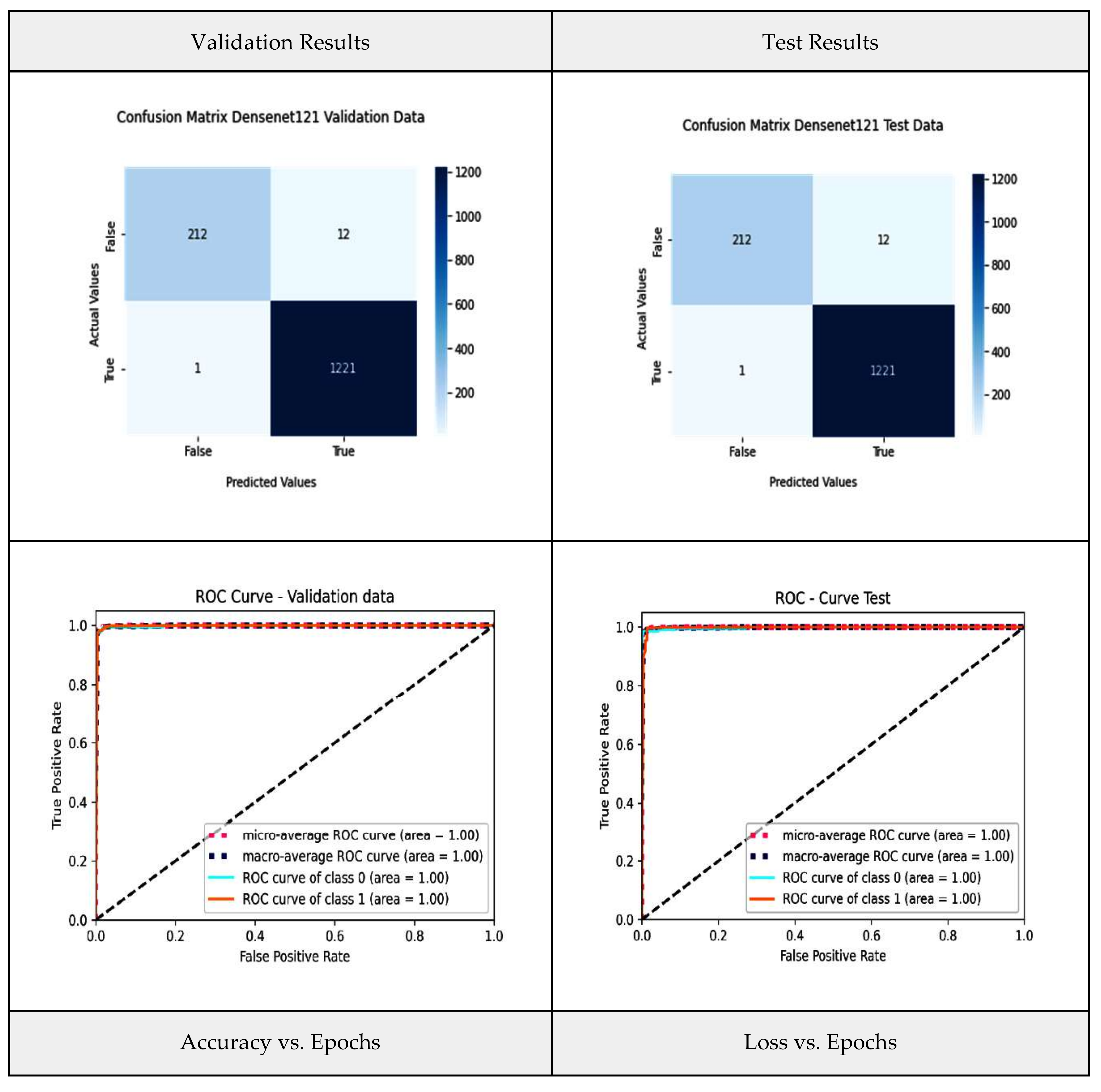
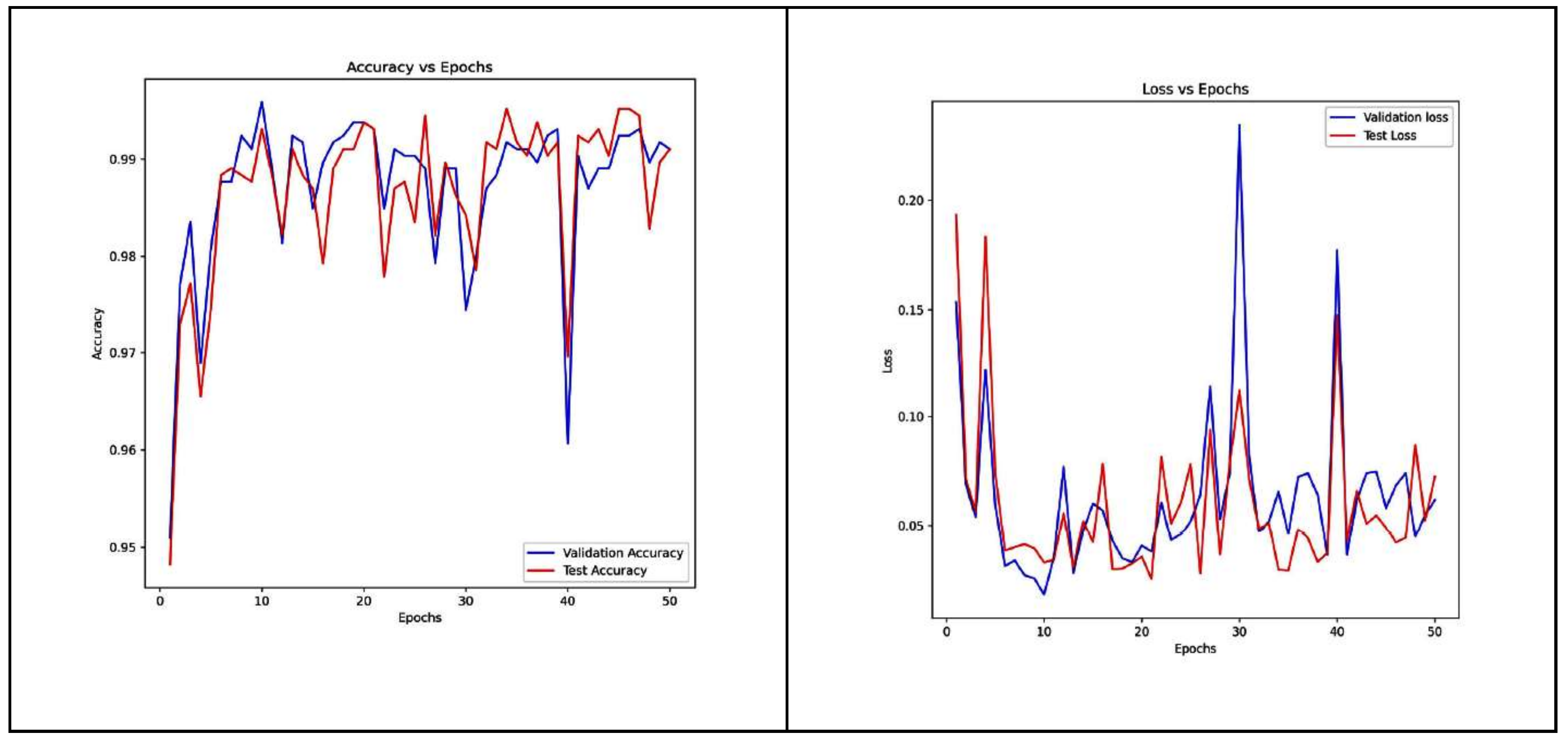
4.2. Densenet-121 Model
- Epochs: 8/30.
- Callbacks: Early stopping on val_accuracy with patience 5, restore best weights true.
- Optimizer: Adam with learning rate of 0.001.
- Loss: Binary Cross Entropy without logits.
- Image_size: (224, 224, 3) type: tensor.
- Dense Layer Weight—Initializer: glorot_uniform.
- Activation: Relu.
- Use Bias: True.
- Dense Layer Bias—Initializer: zeros.
- Batch Normalization: a momentum of 0.99 with an epsilon of 0.001 along with a gamma-initializer of ones, a beta-initializer of zeros and a moving mean initializer of zeros.
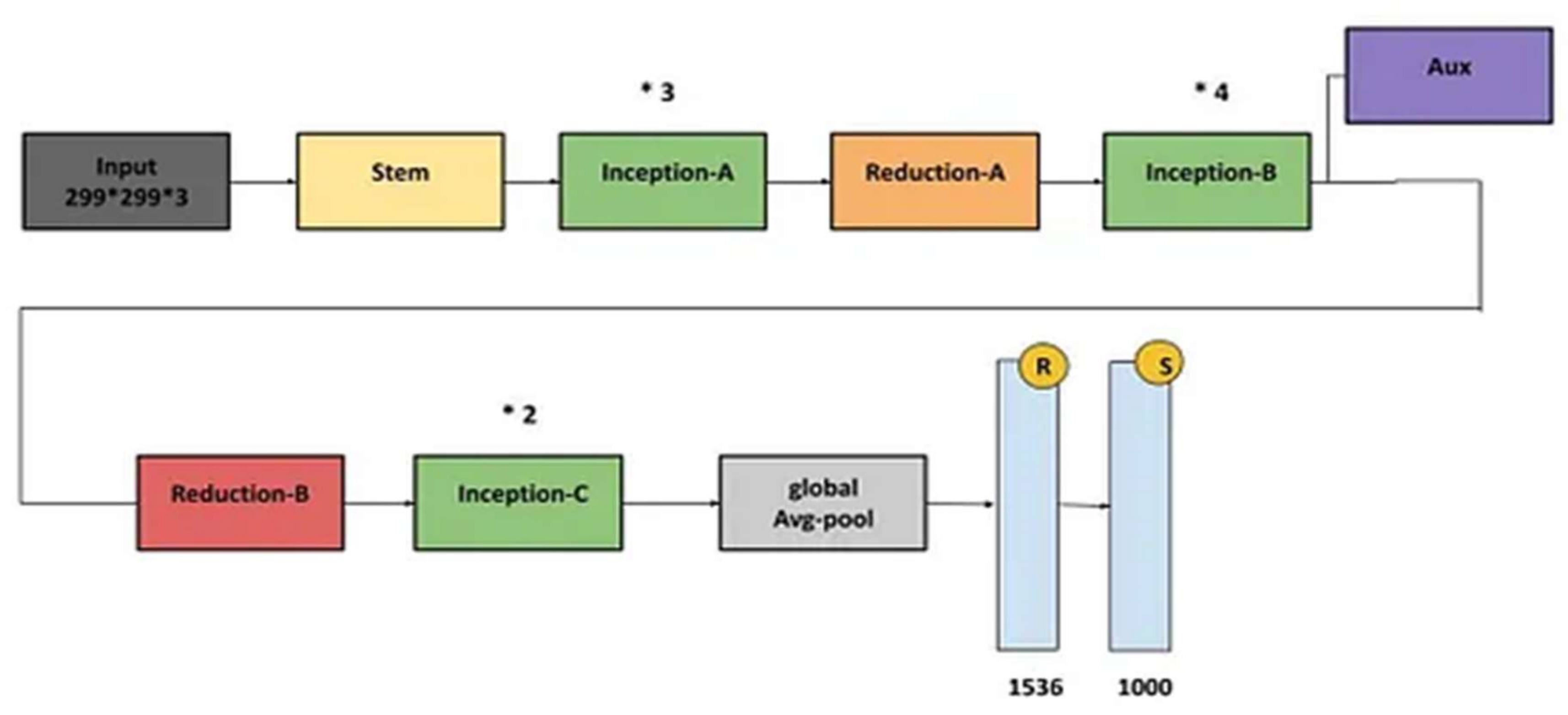
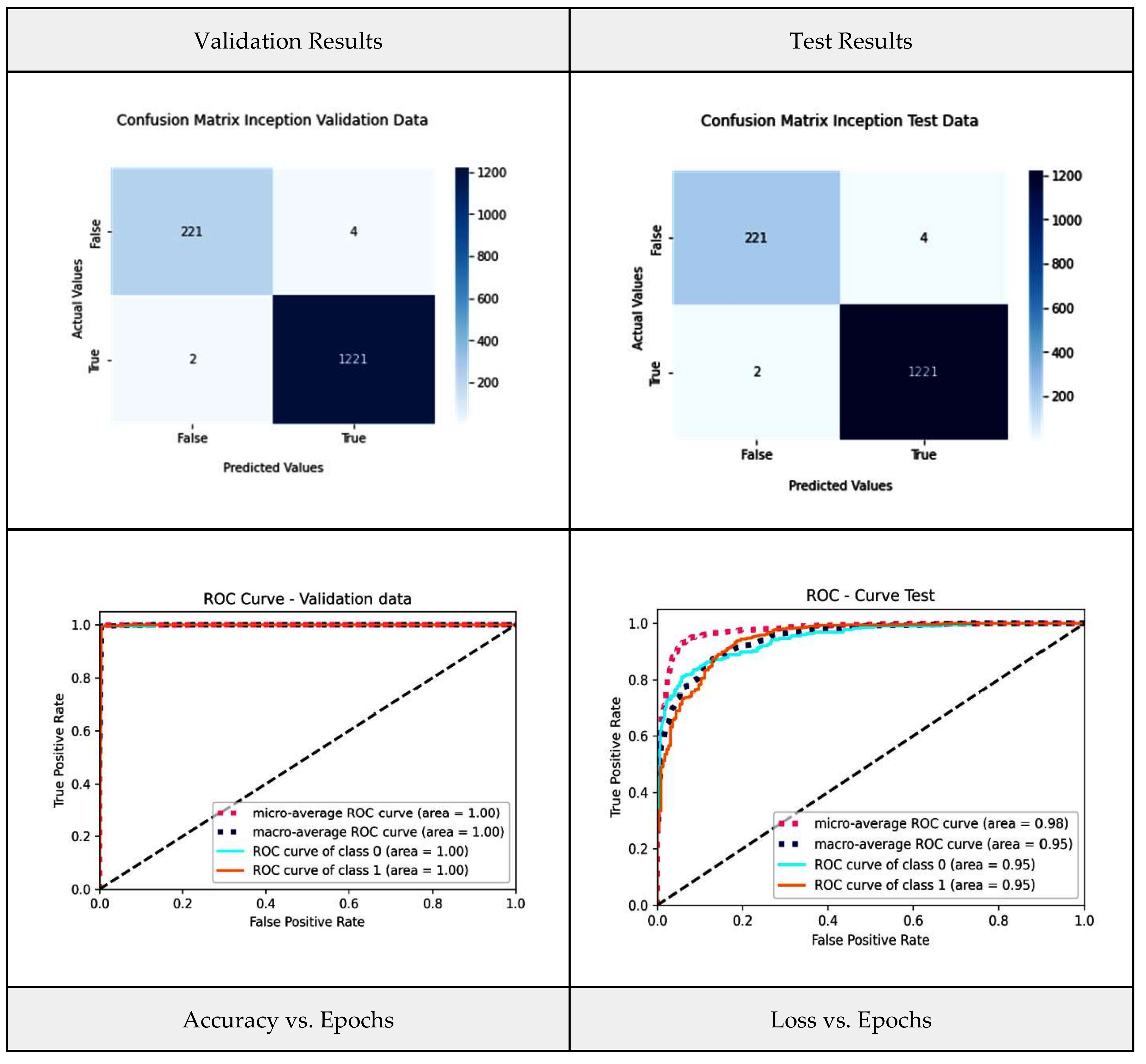
4.3. Inception-V3 Model
- Epochs: 8/30.
- Callbacks: Early stopping on val_accuracy with patience 5, restore best weights true.
- Optimizer: Adam with learning rate of 0.001.
- Loss: BinaryCrossEntropy without logits.
- Image_size: (224 × 224 × 3), type: tensor.
- Dense Layer Weight—Initializer: glorot_uniform.
- Activation: Relu.
- Use Bias: True.
- Dense Layer Bias—Initializer: zeros.
- Batch Normalization: a momentum of 0.99 with an epsilon of 0.001 along with gamma-initializer of ones, a beta-initializer of zeros and a moving mean initializer of zeros.
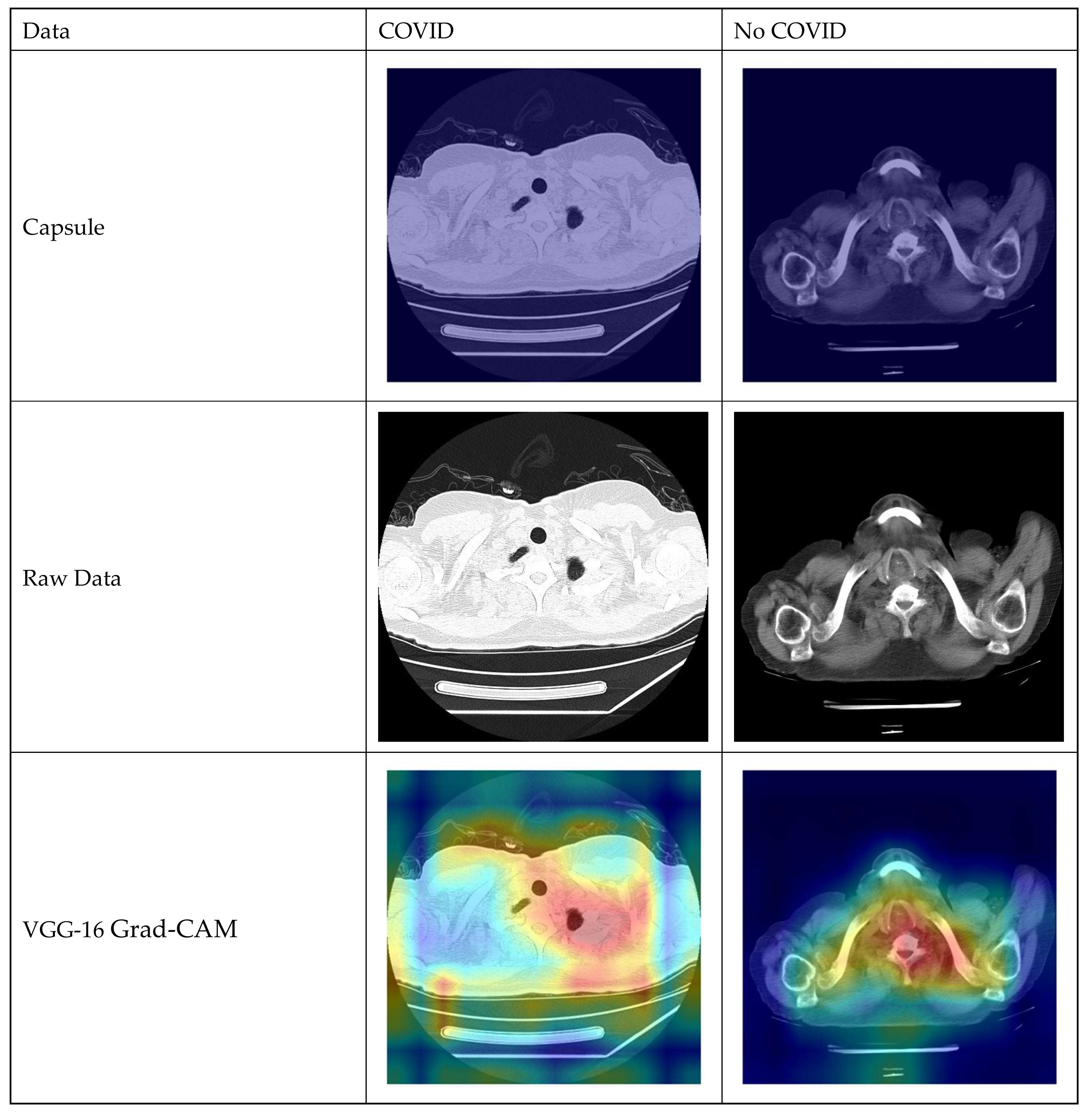
4.4. Capsule Net (CapsNet) Model
- Input: 256 channels (output from the previous convolutional layer).
- Output: 32 capsules.
- Convolutional Operation: 2D convolution with a 9 × 9 kernel and a stride of 2. Each primary capsule performs this operation.
- Digit Capsules: (digit_capsules).
- Decoder
- The decoder component is responsible for reconstructing the input data from the output of the capsule network. It aims to create a reconstruction of the original input.
- Linear Layer 1:
- Input: 160-dimensional capsule output.
- Output: 512 units.
- ReLU Activation: Applies the Rectified Linear Unit activation function.
- Linear Layer 2:
- Input: 512 units.
- Output: 1024 units.
- ReLU Activation: Another ReLU activation.
- Linear Layer 3:
- Input: 1024 units.
- Output: 784 units.
- Sigmoid Activation: Applies the sigmoid activation function to create the final reconstruction.
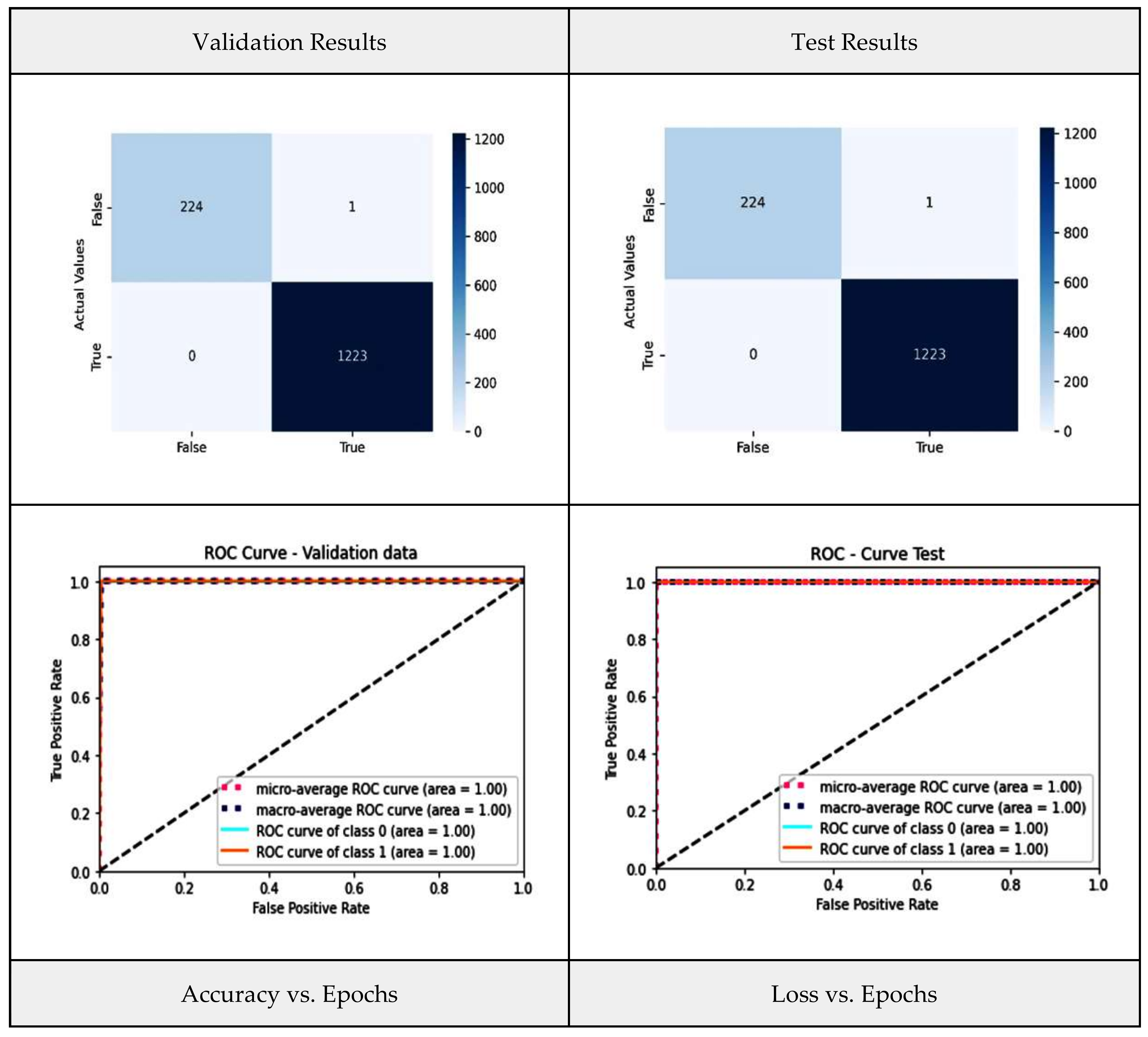
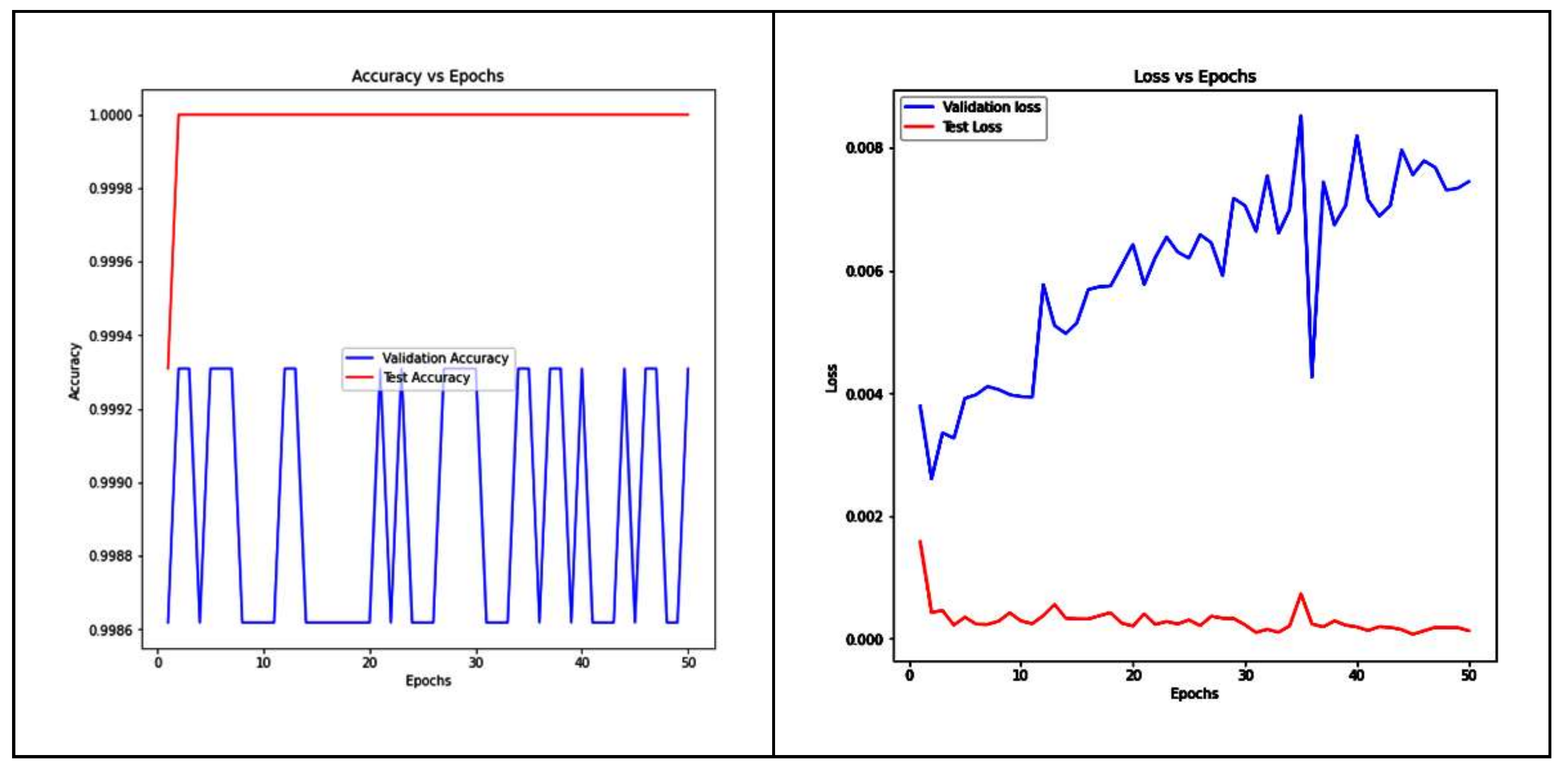
5. Results and Interpretation
5.1. Stacked Generalization (Stacking)
5.2. Weighted-Average Ensemble (WAE)
5.3. Performance Analysis of Proposed Ensemble Models and Base-Learners
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chookajorn, T.; Kochakarn, T.; Wilasang, C.; Kotanan, N.; Modchang, C. Southeast Asia is an emerging hotspot for COVID-19. Nat. Med. 2021, 27, 1495–1496. [Google Scholar] [CrossRef] [PubMed]
- Ghaderzadeh, M.; Eshraghi, M.A.; Asadi, F.; Hosseini, A.; Jafari, R.; Bashash, D.; Abolghasemi, H. Efficient framework for detection of COVID-19 Omicron and delta variants based on two intelligent phases of CNN models. Comput. Math. Methods Med. 2022, 2022, 4838009. [Google Scholar] [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, R.; Wu, Z.; Song, S.; Cheng, L.; Zhu, R. Deep learning-based predictive identification of neural stem cell differentiation. Nat. Commun. 2021, 12, 2614. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Ranaweera, M.; Mahmoud, Q.H. Virtual to Real-World Transfer Learning: A Systematic Review. Electronics 2021, 10, 1491. [Google Scholar] [CrossRef]
- Neena, A.; Geetha, M. Image Classification Using an Ensemble-Based Deep CNN. In Recent Findings in Intelligent Computing Techniques; Springer: Singapore, 2018; Volume 3. [Google Scholar] [CrossRef]
- Korzh, O.; Joaristi, M.; Serra, E. Convolutional neural network ensemble fine-tuning for extended transfer learning. In Proceedings of the International Conference on Big Data, Seattle, WA, USA, 25–30 June 2018. [Google Scholar]
- Hu, H.; Xia, G.S.; Hu, J.; Zhang, L. Transferring deep convolutional neural networks for the scene classification of high-resolution remote sensing imagery. Remote Sens. 2015, 7, 14680. [Google Scholar] [CrossRef]
- Li, Q.; Miao, Y.; Zeng, X.; Tarimo, C.S.; Wu, C.; Wu, J. Prevalence and factors for anxiety during the coronavirus disease 2019 (COVID-19) epidemic among the teachers in China. J. Affect. Disord. 2020, 277, 153–158. [Google Scholar] [CrossRef]
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, D.; Tiwari, P.; Moreira, C.; Damaševičius, R.; de Albuquerque, V.H.C. A Novel Transfer Learning Based Approach for Pneumonia Detection in Chest X-ray Images. Appl. Sci. 2020, 10, 559. [Google Scholar] [CrossRef]
- Jaiswal, A.; Gianchandani, N.; Singh, D.; Kumar, V.; Kaur, M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020, 39, 5682–5689. [Google Scholar] [CrossRef]
- Lu, S.; Yang, J.; Yang, B.; Yin, Z.; Liu, M.; Yin, L.; Zheng, W. Analysis and Design of Surgical Instrument Localization Algorithm. Comput. Model. Eng. Sci. 2023, 137, 669–685. [Google Scholar] [CrossRef]
- Singh, M.; Bansal, S.; Ahuja, S.; Dubey, R.K.; Panigrahi, B.K.; Dey, N. Transfer learning–based ensemble support vector machine model for automated COVID-19 detection using lung computerized tomography scan data. Med. Biol. Eng. Comput. 2021, 59, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Gifani, P.; Shalbaf, A.; Vafaeezadeh, M. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int. J. Comput. Assist. Radiol. Surg. 2020, 16, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Kundu, R.; Singh, P.K.; Ferrara, M.; Ahmadian, A.; Sarkar, R. ET-NET: An ensemble of transfer learning models for prediction of COVID-19 infection through chest CT-scan images. Multimed. Tools Appl. 2021, 81, 31–50. [Google Scholar] [CrossRef] [PubMed]
- Shaik, N.S.; Cherukuri, T.K. Transfer learning based novel ensemble classifier for COVID-19 detection from chest CT-scans. Comput. Biol. Med. 2021, 141, 105127. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.U.; Khan, S.; Nazir, S. A Novel Deep Learning and Ensemble Learning Mechanism for Delta-Type COVID-19 Detection. Front. Public Health 2022, 10, 875971. [Google Scholar] [CrossRef] [PubMed]
- Vocaturo, E.; Zumpano, E.; Caroprese, L. Convolutional neural network techniques on X-ray images for COVID-19 classification. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 3113–3115. [Google Scholar]
- Gerges, F.; Shih, F.; Azar, D. Automated diagnosis of acne and rosacea using convolution neural networks. In Proceedings of the 2021 4th International Conference on Artificial Intelligence and Pattern Recognition, Xiamen, China, 20–21 September 2023. [Google Scholar] [CrossRef]
- Zumpano, E.; Fuduli, A.; Vocaturo, E.; Avolio, M. Viral pneumonia images classification by Multiple Instance Learning: Preliminary results. In Proceedings of the 25th International Database Engineering & Applications Symposium, Montreal, QC, Canada, 14–16 July 2021; pp. 292–296. [Google Scholar]
- Meskó, B.; Topol, E.J. The imperative for regulatory oversight of large language models (or generative AI) in healthcare. NPJ Digit. Med. 2023, 6, 120. [Google Scholar] [CrossRef] [PubMed]
- Morais, M.; Calisto, F.M.; Santiago, C.; Aleluia, C.; Nascimento, J.C. Classification of Breast Cancer in Mri with Multimodal Fusion. In Proceedings of the 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 18–21 April 2023; pp. 1–4. [Google Scholar]
- Alkan, O.; Celiktutan, O.; Salam, H.; Mahmoud, M.; Buckley, G.; Phelan, N. Interactive Technologies for AI in Healthcare: Diagnosis, Management, and Assistance (ITAH). In Proceedings of the 28th International Conference on Intelligent User Interfaces, Sydney, NSW, Australia, 27–31 March 2023; pp. 193–195. [Google Scholar]
- Calisto, F.M.; Nunes, N.; Nascimento, J.C. BreastScreening: On the use of multi-modality in medical imaging diagnosis. In Proceedings of the International Conference on Advanced Visual Interfaces, Salerno, Italy, 28 September–2 October 2020; pp. 1–5. [Google Scholar]
- Lin, Y.-Y.; Guo, W.-Y.; Lu, C.-F.; Peng, S.-J.; Wu, Y.-T.; Lee, C.-C. Application of artificial intelligence to stereotactic radiosurgery for intracranial lesions: Detection, segmentation, and outcome prediction. J. Neuro-Oncol. 2023, 161, 441–450. [Google Scholar] [CrossRef]
- Diogo, P.; Morais, M.; Calisto, F.M.; Santiago, C.; Aleluia, C.; Nascimento, J.C. Weakly-Supervised Diagnosis and Detection of Breast Cancer Using Deep Multiple Instance Learning. In Proceedings of the 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 18–21 April 2023; pp. 1–4. [Google Scholar]
- Donins, U.; Behmane, D. Challenges and Solutions for Artificial Intelligence Adoption in Healthcare—A Literature Review. In Proceedings of the International KES Conference on Innovation in Medicine and Healthcare, Rome, Italy, 14–16 June 2023; Springer Nature: Singapore, 2023; pp. 53–62. [Google Scholar]
- Calisto, F.M.; Nunes, N.; Nascimento, J.C. Modeling adoption of intelligent agents in medical imaging. Int. J. Hum. Comput. Stud. 2022, 168, 102922. [Google Scholar] [CrossRef]
- Sivaraman, V.; Bukowski, L.A.; Levin, J.; Kahn, J.M.; Perer, A. Ignore, trust, or negotiate: Understanding clinician acceptance of AI-based treatment recommendations in health care. In Proceedings of the 2023 CHI Conference on Human Factors in Computing Systems, Hamburg, Germany, 23–28 April 2023; pp. 1–18. [Google Scholar]
- Calisto, F.M.; Santiago, C.; Nunes, N.; Nascimento, J.C. BreastScreening-AI: Evaluating medical intelligent agents for human-AI interactions. Artif. Intell. Med. 2022, 127, 102285. [Google Scholar] [CrossRef]
- Rani, G.; Misra, A.; Dhaka, V.S.; Zumpano, E.; Vocaturo, E. Spatial feature and resolution maximization GAN for bone suppression in chest radiographs. Comput. Methods Programs Biomed. 2022, 224, 107024. [Google Scholar] [CrossRef] [PubMed]
- Sanghvi, H.A.; Patel, R.H.; Agarwal, A.; Gupta, S.; Sawhney, V.; Pandya, A.S. A deep learning approach for classification of COVID and pneumonia using DenseNet-201. Int. J. Imaging Syst. Technol. 2022, 33, 18–38. [Google Scholar] [CrossRef] [PubMed]
- Sujatha, K.; Srinivasa Rao, B. Densenet201: A Customized DNN Model for Multi-Class Classification and Detection of Tumors Based on Brain MRI Images. In Proceedings of the 2023 Fifth International Conference on Electrical, Computer and Communication Technologies (ICECCT), Tamil Nadu, India, 22–24 February 2023; pp. 1–7. [Google Scholar]
- Yadlapalli, P.; Kora, P.; Kumari, U.; Gade, V.S.R.; Padma, T. COVID-19 diagnosis using VGG-16 with CT scans. In Proceedings of the 2022 International Conference for Advancement in Technology (ICONAT), Goa, India, 21–22 January 2022; pp. 1–4. [Google Scholar]
- Bohmrah, M.K.; Kaur, H. Classification of Covid-19 patients using efficient fine-tuned deep learning DenseNet model. Glob. Transit. Proc. 2021, 2, 476–483. [Google Scholar] [CrossRef]
- Gülmez, B. A novel deep neural network model based Xception and genetic algorithm for detection of COVID-19 from X-ray images. Ann. Oper. Res. 2022, 328, 617–641. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef]
- COVID-19 Omicron and Delta Variant Lung CT Scans. Available online: https://www.kaggle.com/datasets/mohammadamireshraghi/covid19-omicron-and-delta-variant-ct-scan-dataset (accessed on 30 November 2022).
- Das, T.K.; Chowdhary, C.L.; Gao, X. Chest X-Ray Investigation: A Convolutional Neural Network Approach. J. Biomim. Biomater. Biomed. Eng. 2020, 45, 57–70. [Google Scholar] [CrossRef]
- Das, T.K.; Roy, P.K.; Uddin, M.; Srinivasan, K.; Chang, C.-Y.; Syed-Abdul, S. Early Tumor Diagnosis in Brain MR Images via Deep Convolutional Neural Network Model. Comput. Mater. Contin. 2021, 68, 2413–2429. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Rani, S.; Babbar, H.; Srivastava, G.; Gadekallu, T.R.; Dhiman, G. Security Framework for Internet of Things based Software Defined Networks using Blockchain. IEEE Internet Things J. 2022, 10, 6074–6081. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic routing between capsules. Adv. Neural Inf. Process. Syst. 2017, 30, 1–11. [Google Scholar]
- Hassan, H.F.; Koaik, L.; Khoury, A.E.; Atoui, A.; El Obeid, T.; Karam, L. Dietary exposure and risk assessment of mycotoxins in thyme and thyme-based products marketed in Lebanon. Toxins 2022, 14, 331. [Google Scholar] [CrossRef]
- Lian, Z.; Zeng, Q.; Wang, W.; Gadekallu, T.R.; Su, C. Blockchain-based two-stage federated learning with non-IID data in IoMT system. IEEE Trans. Comput. Soc. Syst. 2022, 10, 1701–1710. [Google Scholar] [CrossRef]
- Caroprese, L.; Vocaturo, E.; Zumpano, E. Argumentation approaches for explanaible AI in medical informatics. Intell. Syst. Appl. 2022, 16, 200109. [Google Scholar] [CrossRef]
- Zumpano, E.; Iaquinta, P.; Caroprese, L.; Dattola, F.; Tradigo, G.; Veltri, P.; Vocaturo, E. Simpatico 3D mobile for diagnostic procedures. In Proceedings of the 21st International Conference on Information Integration and Web-based Applications & Services, Munich, Germany, 2–4 December 2019; pp. 468–472. [Google Scholar]
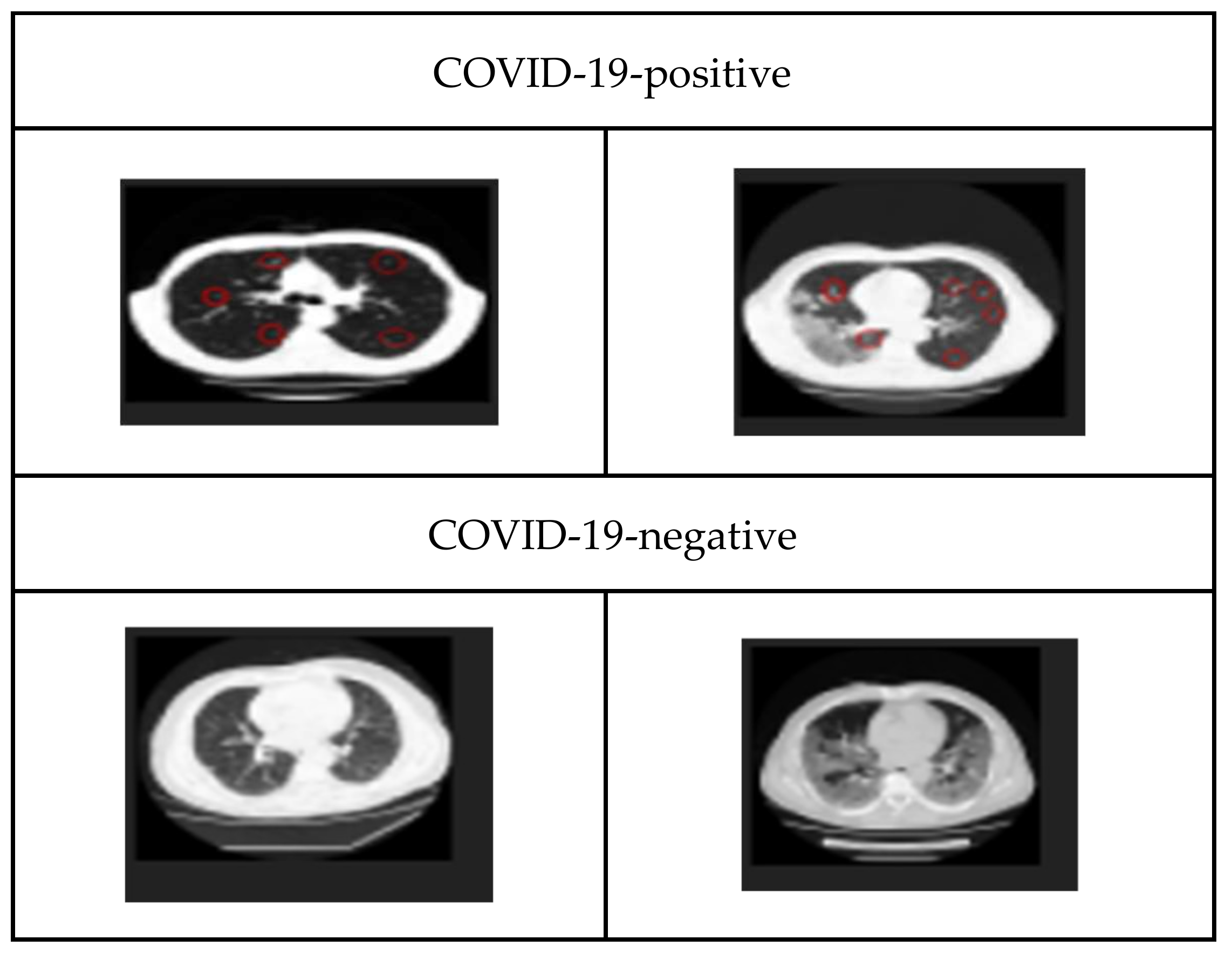










| Studies | Objective | Data Description | Methodology | Key Findings |
|---|---|---|---|---|
| Chouhan et al., 2020 [11] | To increase the accuracy of an ensemble model | Chest X-ray images. | Uses five different transfer learning models and the results are aggregated. | This research has resulted in an accuracy of 96.4% and a recall of 99.62%, respectively. |
| Jaiswal et al., 2021 [12] | To enhance classification accuracy of the model | SARS-CoV-2 CT scan dataset. | Uses a deep transfer learning model DenseNet20. | This research has resulted in an accuracy of 97%. |
| Arora et al., 2021 [13] | The objective of the paper is to increase the accuracy of the classification | SARS-CoV-2 CT scan and COVID-CT scan. | This study fine-tunes a variety of pre-trained models, including VGG (16, and19), MobileNet, and ResNet (50, and 50V2), InceptionResNetV2, Xception, InceptionV3. | Model performance has resulted in a precision of 94.12% and 100%. |
| Singh et al., 2021 [14] | The objective is to predict the early-stage detention of COVID-19 using chest CT. | To maintain the robustness of the model, the dataset is gathered from three independent sources. | DCNN, ELM, online sequential ELM, and bagging ensemble with SVM-these four classifiers are compared for the final classification. | Bagging ensemble with SVM is the leading performer, achieved an F1 score of 0.953, accuracy of 95.7%, precision of 0.958, and AUC of 0.958. |
| Gifani et al., 2021 [15] | To provide accurate and fast detection of COVID-19 using CT-scan images. | Publicly available datasets. | The overall classification decision is made based on the assembly ensemble of all the models using the majority voting method, using 15 pre-trained CNN architectures. | Accuracy: 0.85, precision: 0.857, recall: 0.852. |
| Kundu et al., 2022 [16] | The objective is to increase the accuracy of an ensemble model consisting of 3 different transfer learning techniques. | Publicly available datasets on GitHub. | Ensembling of three transfer learning models, i.e., DenseNet201, Inception v3, and, ResNet34 has been utilized. | Research impact resulting in a 97.77% accuracy rate. |
| Shaik et al., 2022 [17] | The objective is to improve prediction accuracy and reliability. | SARS-CoV-2 and COVID-CT chest CT-scan image datasets. | The ensemble approach is carried out using 8 different pre-trained models. | The highest accuracy on the SARS-CoV-2 dataset is 98.9% COVID-CT dataset is 93.3%. |
| Khan et al., 2022 [18] | To design a model for predicting any sort of variant and its potential health risks. | Publicly available X-ray images on GitHub. | The VGG16 architecture is used in handling the classification tasks and the SVM is used in handling statistical analysis to determine the severity of the patient’s health condition. | The obtained accuracy is 97.37%. |
| Vocaturo et al., 2021 [19] | COVID-19 classification detection from chest X-rays by CNN | COVID dataset containing 13.800 chest X-ray images. | ResNet50. | Accuracy of 98.66% |
| Rani et al., 2022 [20] | COVID-19 detection using chest X-rays | COVID Pneumonia CXR, which includes bone-suppressed and lung-segmented chest X-rays. | GAN and CNN. | AUC of 96.58% on the validation dataset and 96.48% on the testing dataset |
| Zumpano et al., 2021 [21] | Viral and bacterial pneumonia detection | Chest X-ray dataset from Kaggle. | Multiple instance learning paradigm. | Accuracy about 90% and sensitivity about 94%. |
| Dataset | COVID Images | Non-COVID Images | Total |
|---|---|---|---|
| Kaggle Dataset | 12,231 | 2251 | 14,482 |
| Name of the Model | Number of Layers | Fully CLayer | Classifier Activation | Optimizer |
|---|---|---|---|---|
| Common attributes of all the models: Data Split: Train, Validation, Test Loss Function: Binary Cross Entropy Activation Function: ReLU Epochs: 50 | ||||
| Base Learners | ||||
| VGG-16 | 16 (all trainable) | 256, 64, 32, 1 | Sigmoid | Adam (lr = 0.001) |
| Densenet-121 | 121 (all trainable) | 256, 64, 32, 1 | Sigmoid | Adam (lr = 0.001) |
| Inception-v3 | 22 (all trainable) | 256, 64, 32, 1 | Sigmoid | Adam (lr = 0.001) |
| CapsNet | 14 | 256, 64, 32, 1 | Sigmoid | Rmsprop (lr = 0.001) |
| Ensemble Models | ||||
| Stacking | 7 | 10, 4, 1 | Sigmoid | Adam |
| Weightage Average Aggregator Densenet = 0.8 VGG = 0.6 Inception = 0.4 Capsule = 0.2 | 7 | 10, 4, 1 | Sigmoid | Adam |
| Model Name | Accuracy (%) | Precision (%) | Recall (%) | AUC |
|---|---|---|---|---|
| Base Learners | ||||
| VGG-16 | 98.21 | 98.26 | 98.08 | 0.9924 |
| InceptionV3 | 98.78 | 98.77 | 98.8 | 0.997 |
| DenseNet-121 | 98.66 | 98.67 | 98.64 | 0.996 |
| CapsNet | 92.72 | 92.74 | 92.69 | 0.9747 |
| Ensemble Methods | ||||
| Stacking | 99.93 | 99.93 | 99.93 | 0.9993 |
| WAE | 99.93 | 99.93 | 99.93 | 0.9993 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, R.S.; Dandabani, L.; Das, T.K.; Khan, S.B.; Basheer, S.; Alqahtani, M.S. Cloud-Based Quad Deep Ensemble Framework for the Detection of COVID-19 Omicron and Delta Variants. Diagnostics 2023, 13, 3419. https://doi.org/10.3390/diagnostics13223419
Tiwari RS, Dandabani L, Das TK, Khan SB, Basheer S, Alqahtani MS. Cloud-Based Quad Deep Ensemble Framework for the Detection of COVID-19 Omicron and Delta Variants. Diagnostics. 2023; 13(22):3419. https://doi.org/10.3390/diagnostics13223419
Chicago/Turabian StyleTiwari, Ravi Shekhar, Lakshmi Dandabani, Tapan Kumar Das, Surbhi Bhatia Khan, Shakila Basheer, and Mohammed S. Alqahtani. 2023. "Cloud-Based Quad Deep Ensemble Framework for the Detection of COVID-19 Omicron and Delta Variants" Diagnostics 13, no. 22: 3419. https://doi.org/10.3390/diagnostics13223419
APA StyleTiwari, R. S., Dandabani, L., Das, T. K., Khan, S. B., Basheer, S., & Alqahtani, M. S. (2023). Cloud-Based Quad Deep Ensemble Framework for the Detection of COVID-19 Omicron and Delta Variants. Diagnostics, 13(22), 3419. https://doi.org/10.3390/diagnostics13223419





