Detection of Plasmodium falciparum in Saliva and Stool Samples from Children Living in Franceville, a Highly Endemic Region of Gabon
Abstract
:1. Introduction
2. Materials and Methods
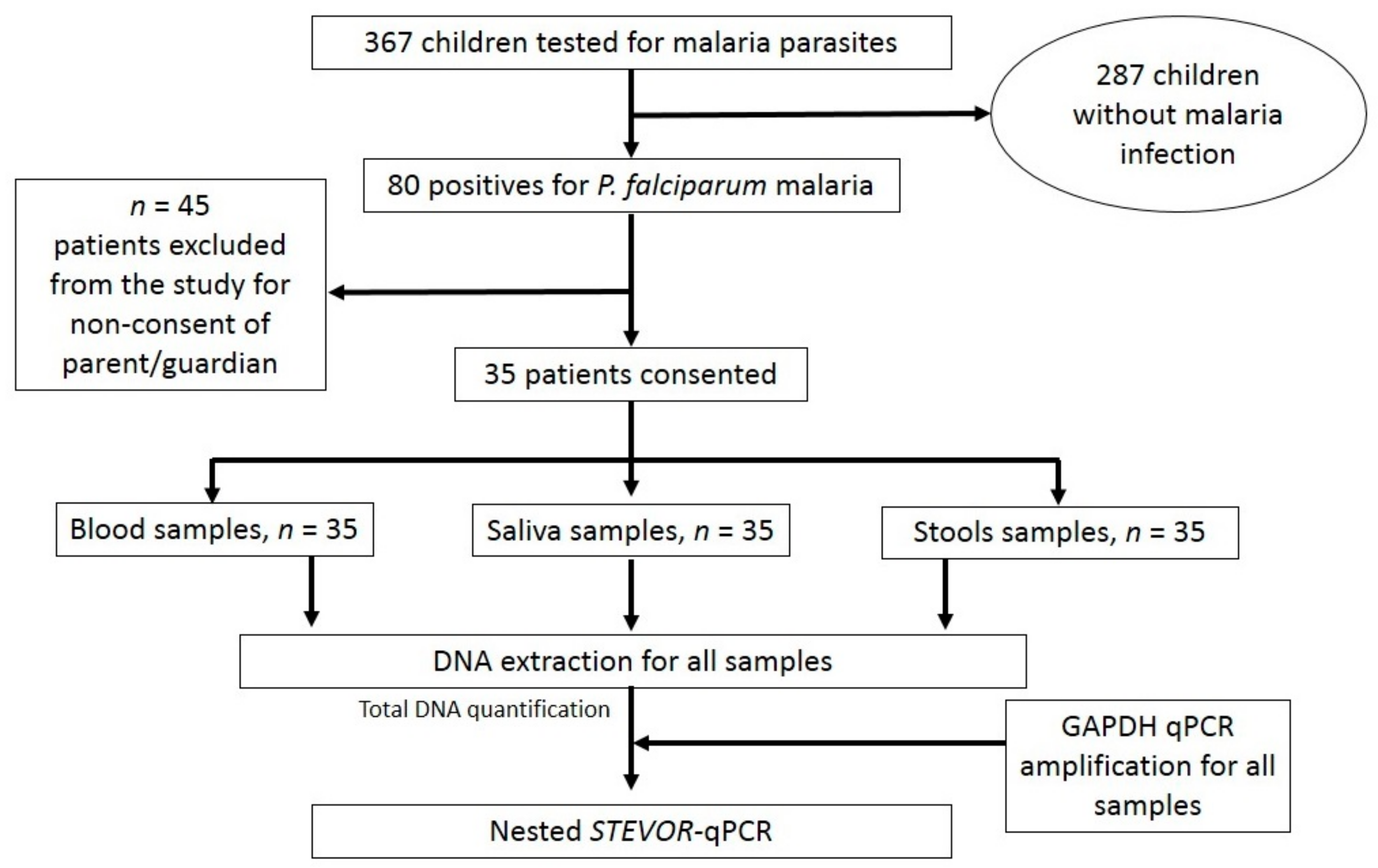
2.1. Study Design and Population
2.2. Malaria Diagnosis
2.3. Saliva and Stool Collection
DNA Extraction
2.4. GAPDH qPCR Amplification
2.5. Nested qPCR Targeting STEVOR Gene
2.6. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Malaria Report 2021; World Health Organization: Geneva, Switzerland, 2021; pp. 5–7. [Google Scholar]
- Thomas, C.J.; Lindsay, S.W. Local-scale variation in malaria infection amongst rural Gambian children estimated by satellite remote sensing. Trans. R. Soc. Trop. Med. Hyg. 2000, 94, 159–163. [Google Scholar] [CrossRef] [PubMed]
- M’Bondoukwe, N.P.; Kendjo, E.; Mawili-Mboumba, D.P.; Koumba Lengongo, J.V.; Offouga Mbouoronde, C.; Nkoghe, D.; Toure, F.; Bouyou-Akotet, M.K. Prevalence of and risk factors for malaria, filariasis, and intestinal parasites as single infections or co-infections in different settlements of Gabon, Central Africa. Infect. Dis. Poverty 2018, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Maghendji-Nzondo, S.; Nzoughe, H.; Lemamy, G.J.; Kouna, L.C.; Pegha-Moukandja, I.; Lekoulou, F.; Mbatchi, B.; Toure-Ndouo, F.; Lekana-Douki, J.B. Prevalence of malaria, prevention measures, and main clinical features in febrile children admitted to the Franceville Regional Hospital, Gabon. Parasite 2016, 23, 32. [Google Scholar] [CrossRef] [PubMed]
- Adegnika, A.A.; Ramharter, M.; Agnandji, S.T.; Ateba Ngoa, U.; Issifou, S.; Yazdanbahksh, M.; Kremsner, P.G. Epidemiology of parasitic co-infections during pregnancy in Lambaréné, Gabon. Trop. Med. Int. Health 2010, 15, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Bouyou-Akotet, M.K.; Mawili-Mboumba, D.P.; Kendjo, E.; Eyang Ekouma, A.; Abdou Raouf, O.; Engohang Allogho, E.; Kombila, M. Complicated malaria and other severe febrile illness in a pediatric ward in Libreville, Gabon. BMC Infect. Dis. 2012, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Imboumy-Limoukou, R.K.; Lendongo-Wombo, J.B.; Nguimbyangue-Apangome, A.F.; Biteghe Bi Essone, J.C.; Mounioko, F.; Oyegue-Libagui, L.S.; Ngoungou, B.E.; Lekana-Douki, J.B. Severe malaria in Gabon: Epidemiological, clinical and laboratory features in Amissa Bongo Hospital of Franceville. Malar. J. 2023, 22, 88. [Google Scholar] [CrossRef] [PubMed]
- Maghendji-Nzondo, S.; Kouna, L.C.; Mourembou, G.; Boundenga, L.; Imboumy-Limoukou, R.K.; Matsiegui, P.B.; Manego-Zoleko, R.; Mbatchi, B.; Raoult, D.; Toure-Ndouo, F.; et al. Malaria in urban, semi-urban and rural areas of southern of Gabon: Comparison of the Pfmdr 1 and Pfcrt genotypes from symptomatic children. Malar. J. 2016, 15, 420. [Google Scholar] [CrossRef]
- Gorret, A.M.; Muhindo, R.; Baguma, E.; Ntaro, M.; Mulogo, E.M.; Deutsch-Feldman, M.; Juliano, J.J.; Nyehangane, D.; Boyce, R.M. Comparison of Capillary Versus Venous Blood for the Diagnosis of Plasmodium falciparum Malaria Using Rapid Diagnostic Tests. J. Infect. Dis. 2021, 224, 109–113. [Google Scholar] [CrossRef]
- Mpina, M.; Stabler, T.C.; Schindler, T.; Raso, J.; Deal, A.; Acuche Pupu, L.; Nyakarungu, E.; Del Carmen Ovono Davis, M.; Urbano, V.; Mtoro, A.; et al. Diagnostic performance and comparison of ultrasensitive and conventional rapid diagnostic test, thick blood smear and quantitative PCR for detection of low-density Plasmodium falciparum infections during a controlled human malaria infection study in Equatorial Guinea. Malar. J. 2022, 21, 99. [Google Scholar]
- Mawili-Mboumba, D.P.; Ndong, R.N.; Rosa, N.B.; Largo, J.L.L.; Lembet-Mikolo, A.; Nzamba, P.; Mbouoronde, C.O.; Kombila, M.; Bouyou Akotet, M.K. Submicroscopic Falciparum Malaria in Febrile Individuals in Urban and Rural Areas of Gabon. Am. J. Trop. Med. Hyg. 2017, 96, 815–818. [Google Scholar] [CrossRef]
- Sutherland, C.J.; Hallett, R. Detecting malaria parasites outside the blood. J. Infect. Dis. 2009, 199, 1561–1563. [Google Scholar] [CrossRef] [PubMed]
- Jirku, M.; Pomajbikova, K.; Petrzelkova, K.J.; Huzova, Z.; Modry, D.; Lukes, J. Detection of Plasmodium spp. in human feces. Emerg. Infect. Dis. 2012, 18, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Mfuh, K.O.; Tassi Yunga, S.; Esemu, L.F.; Bekindaka, O.N.; Yonga, J.; Djontu, J.C.; Mbakop, C.D.; Taylor, D.W.; Nerurkar, V.R.; Leke, R.G.F. Detection of Plasmodium falciparum DNA in saliva samples stored at room temperature: Potential for a non-invasive saliva-based diagnostic test for malaria. Malar. J. 2017, 16, 434. [Google Scholar] [CrossRef] [PubMed]
- Al-Shehri, H.; Power, B.J.; Archer, J.; Cousins, A.; Atuhaire, A.; Adriko, M.; Arinaitwe, M.; Alanazi, A.D.; LaCourse, E.J.; Kabatereine, N.B.; et al. Non-invasive surveillance of Plasmodium infection by real-time PCR analysis of ethanol preserved faeces from Ugandan school children with intestinal schistosomiasis. Malar. J. 2019, 18, 109. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, Y.M.; Esemu, L.F.; Antallan, J.; Thomas, B.; Tassi Yunga, S.; Obase, B.; Christine, N.; Leke, R.G.F.; Culleton, R.; Mfuh, K.O.; et al. PCR-based detection of Plasmodium falciparum in saliva using mitochondrial cox3 and varATS primers. Trop. Med. Health 2018, 46, 22. [Google Scholar] [CrossRef] [PubMed]
- Oyedeji, S.I.; Awobode, H.O.; Monday, G.C.; Kendjo, E.; Kremsner, P.G.; Kun, J.F. Comparison of PCR-based detection of Plasmodium falciparum infections based on single and multicopy genes. Malar. J. 2007, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Cloonan, N.; Fischer, K.; Thompson, J.; Waine, G.; Lanzer, M.; Saul, A. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 1998, 97, 161–176. [Google Scholar] [CrossRef]
- Elissa, N.; Karch, S.; Bureau, P.; Ollomo, B.; Lawoko, M.; Yangari, P.; Ebang, B.; Georges, A.J. Malaria transmission in a region of savanna-forest mosaic, Haut-Ogooue, Gabon. J. Am. Mosq. Control Assoc. 1999, 15, 15–23. [Google Scholar]
- Planche, T.; Krishna, S.; Kombila, M.; Engel, K.; Faucher, J.F.; Ngou-Milama, E.; Kremsner, P.G. Comparison of methods for the rapid laboratory assessment of children with malaria. Am. J. Trop. Med. Hyg. 2001, 65, 599–602. [Google Scholar] [CrossRef]
- Reck, M.; Tomasch, J.; Deng, Z.; Jarek, M.; Husemann, P.; Wagner-Dobler, I. Stool metatranscriptomics: A technical guideline for mRNA stabilisation and isolation. BMC Genom. 2015, 16, 494. [Google Scholar] [CrossRef]
- StataCorp. Stata Statistical Software: Release 14; StataCorp LP: College Station, TX, USA, 2015; Available online: https://www.stata.com/ (accessed on 1 March 2023).
- Lekana-Douki, J.B.; Pontarollo, J.; Zatra, R.; Toure-Ndouo, F.S. Malaria in Gabon: Results of a clinical and laboratory study at the Chinese-Gabonese Friendship Hospital of Franceville. Sante 2011, 21, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Ghayour Najafabadi, Z.; Oormazdi, H.; Akhlaghi, L.; Meamar, A.R.; Raeisi, A.; Rampisheh, Z.; Nateghpour, M.; Razmjou, E. Mitochondrial PCR-based malaria detection in saliva and urine of symptomatic patients. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Nwakanma, D.C.; Gomez-Escobar, N.; Walther, M.; Crozier, S.; Dubovsky, F.; Malkin, E.; Locke, E.; Conway, D.J. Quantitative detection of Plasmodium falciparum DNA in saliva, blood, and urine. J. Infect. Dis. 2009, 199, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Buppan, P.; Putaporntip, C.; Pattanawong, U.; Seethamchai, S.; Jongwutiwes, S. Comparative detection of Plasmodium vivax and Plasmodium falciparum DNA in saliva and urine samples from symptomatic malaria patients in a low endemic area. Malar. J. 2010, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Mackeen, M.M.; Cook, M.; Oriero, E.; Locke, E.; Thezenas, M.L.; Kessler, B.M.; Nwakanma, D.; Casals-Pascual, C. Proteomic identification of host and parasite biomarkers in saliva from patients with uncomplicated Plasmodium falciparum malaria. Malar. J. 2012, 11, 178. [Google Scholar] [CrossRef] [PubMed]
- Abkallo, H.M.; Liu, W.; Hokama, S.; Ferreira, P.E.; Nakazawa, S.; Maeno, Y.; Quang, N.T.; Kobayashi, N.; Kaneko, O.; Huffman, M.A.; et al. DNA from pre-erythrocytic stage malaria parasites is detectable by PCR in the faeces and blood of hosts. Int. J. Parasitol. 2014, 44, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Sato, M.; Kato-Hayashi, N.; Kishi, H.; Huffman, M.A.; Maeno, Y.; Culleton, R.; Nakazawa, S. Detection of Plasmodium knowlesi DNA in the urine and faeces of a Japanese macaque (Macaca fuscata) over the course of an experimentally induced infection. Malar. J. 2014, 13, 373. [Google Scholar] [CrossRef] [PubMed]
- Assis, G.M.; Alvarenga, D.A.; Costa, D.C.; Souza, J.C.J.; Hirano, Z.M.; Kano, F.S.; Sousa, T.N.; Brito, C.F. Detection of Plasmodium in faeces of the New World primate Alouatta clamitans. Mem. Inst. Oswaldo Cruz 2016, 111, 570–576. [Google Scholar] [CrossRef]
- Loy, D.E.; Rubel, M.A.; Avitto, A.N.; Liu, W.; Li, Y.; Learn, G.H.; Ranciaro, A.; Mbunwe, E.; Fokunang, C.; Njamnshi, A.K.; et al. Investigating zoonotic infection barriers to ape Plasmodium parasites using faecal DNA analysis. Int. J. Parasitol. 2018, 48, 531–542. [Google Scholar] [CrossRef]
| Primers | |
|---|---|
| P5 | 5′-GGG AAT TCT TTA TTT GAT GAA GAT G-3′ |
| P17 | 5′-ACA TTA TCA TAA TGA (C/T)CC AGA ACT-3′ |
| P18 | 5′-TTT CA(C/T) CAC CAA ACA TTT CTT-3′ |
| P19 | 5′-AAT CCA CAT TAT CAC AAT GA-3′ |
| P20 | 5′-CCG ATT TTA ACA TAA TAT GA-3′ |
| P24 | 5′-GTT TGC AAT AAT TCT TTT TCT AGC-3′ |
| Value Parameter | |
|---|---|
| N (number) | 367 |
| Hemoglobin (g/dL) | |
| 8.81 (2.12) |
| 9.03 [8.20–10.3] |
| White blood cells (×103/µL) | |
| 5.80 (3.65) |
| 4.90 [4.05–5.65] |
| Red blood cells (×103/µL) | |
| 3.86 (0.69) |
| 3.78 [3.54–4.44] |
| Platelets (×103/µL) | |
| 185 (107) |
| 162 [100–254] |
| Number | Sensibility [95%CI] | |
|---|---|---|
| qPCR-based assay for blood | ||
| Negative | 0 | 100% |
| Positive | 35 | |
| qPCR-based assay for saliva | ||
| Negative | 27 | 22.86% [12.07–39.02] |
| Positive | 8 | |
| qPCR-based assay for stools | ||
| Negative | 21 | 14.29% [6.26–41.59] |
| Positive | 5 |
| Parasitaemia/µL | n | Nested qPCR Assay Characteristics | |||||
|---|---|---|---|---|---|---|---|
| True Positive | False Negative | Sensitivity (in %) | |||||
| Saliva, n (%) | Stool, n (%) | Saliva, n (%) | Stool, n (%) | Saliva | Stool | ||
| <1000 | 5 | 3 | 0 | 2 | 5 | 60 | 0 |
| 1000–10,000 | 15 | 2 | 3 | 13 | 12 | 13.33 | 20 |
| 10,001–50,000 | 10 | 2 | 1 | 8 | 9 | 20 | 10 |
| 50,001–100,000 | 3 | 1 | 0 | 2 | 3 | 33.33 | 0 |
| >100,000 | 2 | 0 | 1 | 2 | 1 | 0 | 50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imboumy-Limoukou, R.K.; Biteghe-Bi-Essone, J.-C.; Lendongo Wombo, J.B.; Lekana-Douki, S.E.; Rougeron, V.; Ontoua, S.-S.; Oyegue-Liabagui, L.S.; Mbani Mpega Ntigui, C.N.; Kouna, L.C.; Lekana-Douki, J.-B. Detection of Plasmodium falciparum in Saliva and Stool Samples from Children Living in Franceville, a Highly Endemic Region of Gabon. Diagnostics 2023, 13, 3271. https://doi.org/10.3390/diagnostics13203271
Imboumy-Limoukou RK, Biteghe-Bi-Essone J-C, Lendongo Wombo JB, Lekana-Douki SE, Rougeron V, Ontoua S-S, Oyegue-Liabagui LS, Mbani Mpega Ntigui CN, Kouna LC, Lekana-Douki J-B. Detection of Plasmodium falciparum in Saliva and Stool Samples from Children Living in Franceville, a Highly Endemic Region of Gabon. Diagnostics. 2023; 13(20):3271. https://doi.org/10.3390/diagnostics13203271
Chicago/Turabian StyleImboumy-Limoukou, Roméo Karl, Jean-Claude Biteghe-Bi-Essone, Judicael Boris Lendongo Wombo, Sonia Etenna Lekana-Douki, Virginie Rougeron, Steede-Seinnat Ontoua, Lydie Sandrine Oyegue-Liabagui, Cherone Nancy Mbani Mpega Ntigui, Lady Charlène Kouna, and Jean-Bernard Lekana-Douki. 2023. "Detection of Plasmodium falciparum in Saliva and Stool Samples from Children Living in Franceville, a Highly Endemic Region of Gabon" Diagnostics 13, no. 20: 3271. https://doi.org/10.3390/diagnostics13203271
APA StyleImboumy-Limoukou, R. K., Biteghe-Bi-Essone, J.-C., Lendongo Wombo, J. B., Lekana-Douki, S. E., Rougeron, V., Ontoua, S.-S., Oyegue-Liabagui, L. S., Mbani Mpega Ntigui, C. N., Kouna, L. C., & Lekana-Douki, J.-B. (2023). Detection of Plasmodium falciparum in Saliva and Stool Samples from Children Living in Franceville, a Highly Endemic Region of Gabon. Diagnostics, 13(20), 3271. https://doi.org/10.3390/diagnostics13203271






