A Novel Lightweight CNN Architecture for the Diagnosis of Brain Tumors Using MR Images
Abstract
1. Introduction
- Traditional automatic detection approaches utilized conventional machine learning algorithms, whose performance depends on the choice of appropriate features and learning approaches.
- Some classification methods employed wavelets for image analysis. However, they fail to acquire directional information; the selection of subbands and mother wavelets is also critical.
- Some approaches use handcrafted features but are not robust to noise and exhibit poor discrimination.
- The authors implemented some traditional CNN frameworks such as pre-trained CNN models with transfer learning to classify brain MR images in a few works. However, they demand a large number of parameters and high computational time.
Significant Contributions
- Skull-stripping is performed to enhance the robustness of the segmentation process by eliminating extra-meningeal mater (or dura mater) based on thresholding and morphological operations.
- Image data augmentation is implemented to enhance the sufficiency and diversity of the training database by geometric transformation operators. By this, we significantly reduce the overfitting issues encountered during training progress.
- We proposed a novel lightweight CNN architecture to detect high-level features from brain MR images. We can effectively minimize the parameters, including trainable and non-trainable, compared to the existing CNN models and automatically extract the significant features. Due to this, we limit the influence of human beings in the analysis of brain MR tumor images, which is the considerable benefit of the suggested CNN model.
- Analyze the impact of various optimization algorithms (Stochastic gradient descent with moment (SGDM), Adam, Adagrad, AdaMax, Adadelta, Nadam, and RMSProp) during training of the CNN model with the help of K-fold cross-validation (KFC). It is the fundamental difference between the existing and proposed models.
- The FL-MSCM is employed to separate the foreground (affected regions) and background (non-affected areas) from brain MR images, which can minimize issues of other traditional segmentation algorithms, such as the impact of noise, spurious blobs, and other imaging artifacts, by making each region as uniform as possible. Due to this, we improve the segmentation accuracy, which is a significant advantage of the presented FL-SCM technique.
2. Preliminaries
2.1. Convolutional Layer
2.2. Batch Normalization Layer
- Improving the training speed of the network.
- Minimizing the internal covariance shift [32].
- Reducing overfitting since it has slight regularization.
| Algorithm 1. Batch normalization |
| Input: Values of over a mini-batch: . Parameters to be learned: . (2) (3) (4) (5) where represents scale; illustrates shift; is the number of feature inputs; and are the mean and variance across the batch, ; is a constant, which is used to enhance the stability when is too small. Output: |
2.3. Activation Functions
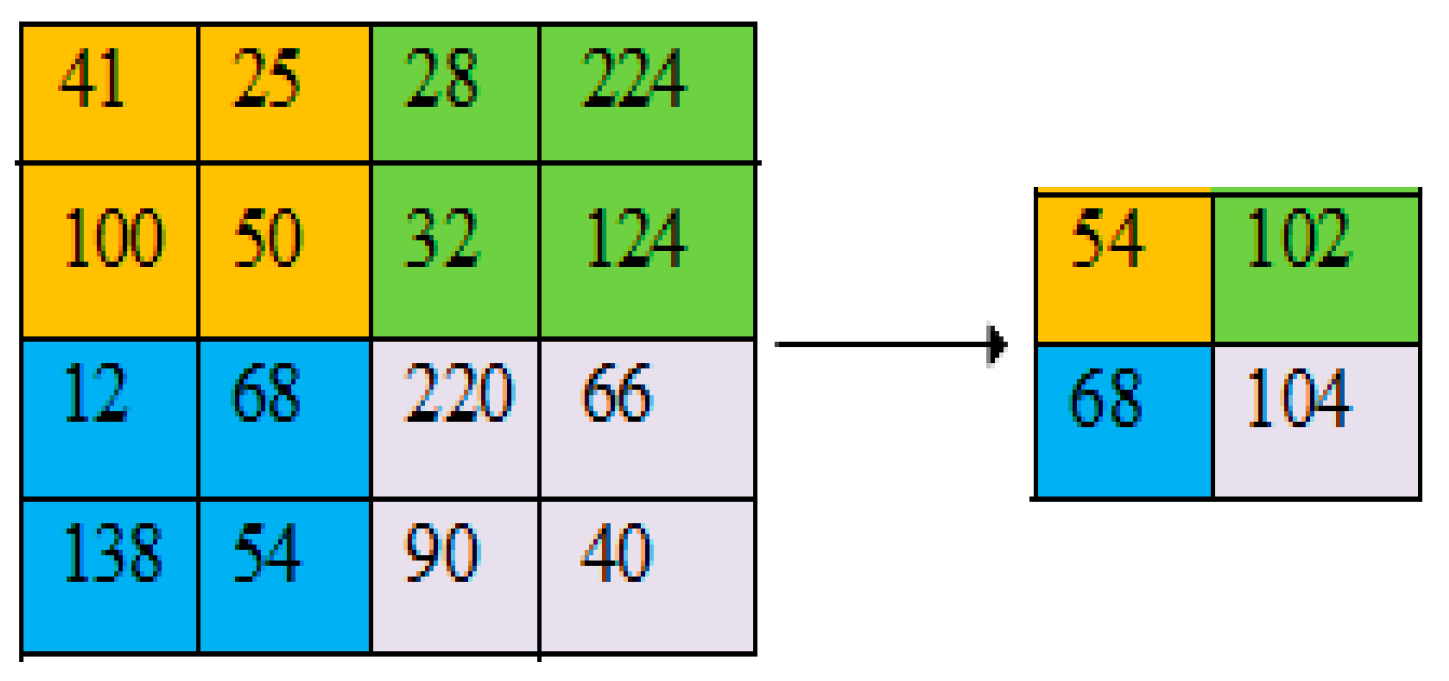
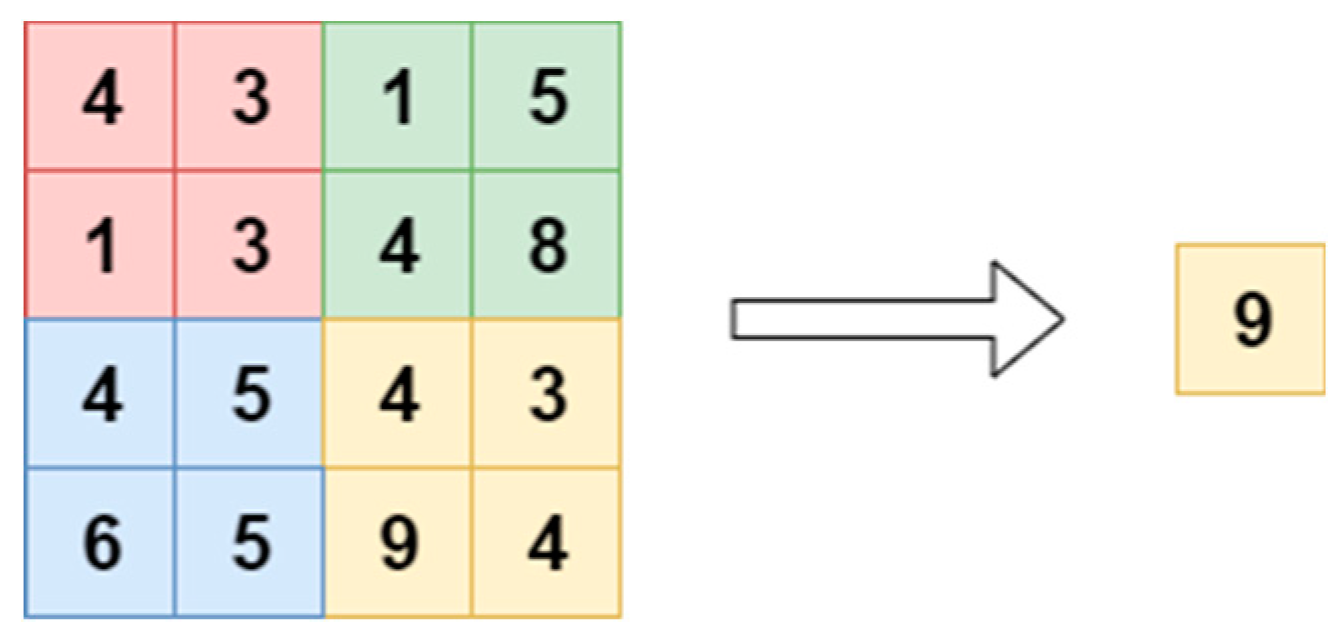
2.4. Pooling Layer
2.5. Softmax
3. Materials and Methods
3.1. Database
3.2. Skull-Stripping
- Initially, we separate the image, into two regions and over an intensity-level of , and . Here, is the number of intensity levels, usually an integer power of 2.
- Obtain the binary image, by setting the optimal thresholding value, which is estimated by the following equationswhere , and , represents the mean and variance of the regions over , and ; define the thresholding.
- Construct a disk-shaped structuring element, with a required radius.
- Eliminate the small peak objects from using a simple area opening operation and then fill the regions with an image filling operation.
- Employ the erosion operation on the outcome of step 3 with the defined . Using this, we can eliminate small objects which appear in the binary image .
- Finally, the binary image obtained in step 5 is superimposed on the original image, and replaces the non-binary region with zeros. With this process, the skull-free brain MR image is obtained, which improves the segmentation accuracy.
3.3. Image Data Augmentation
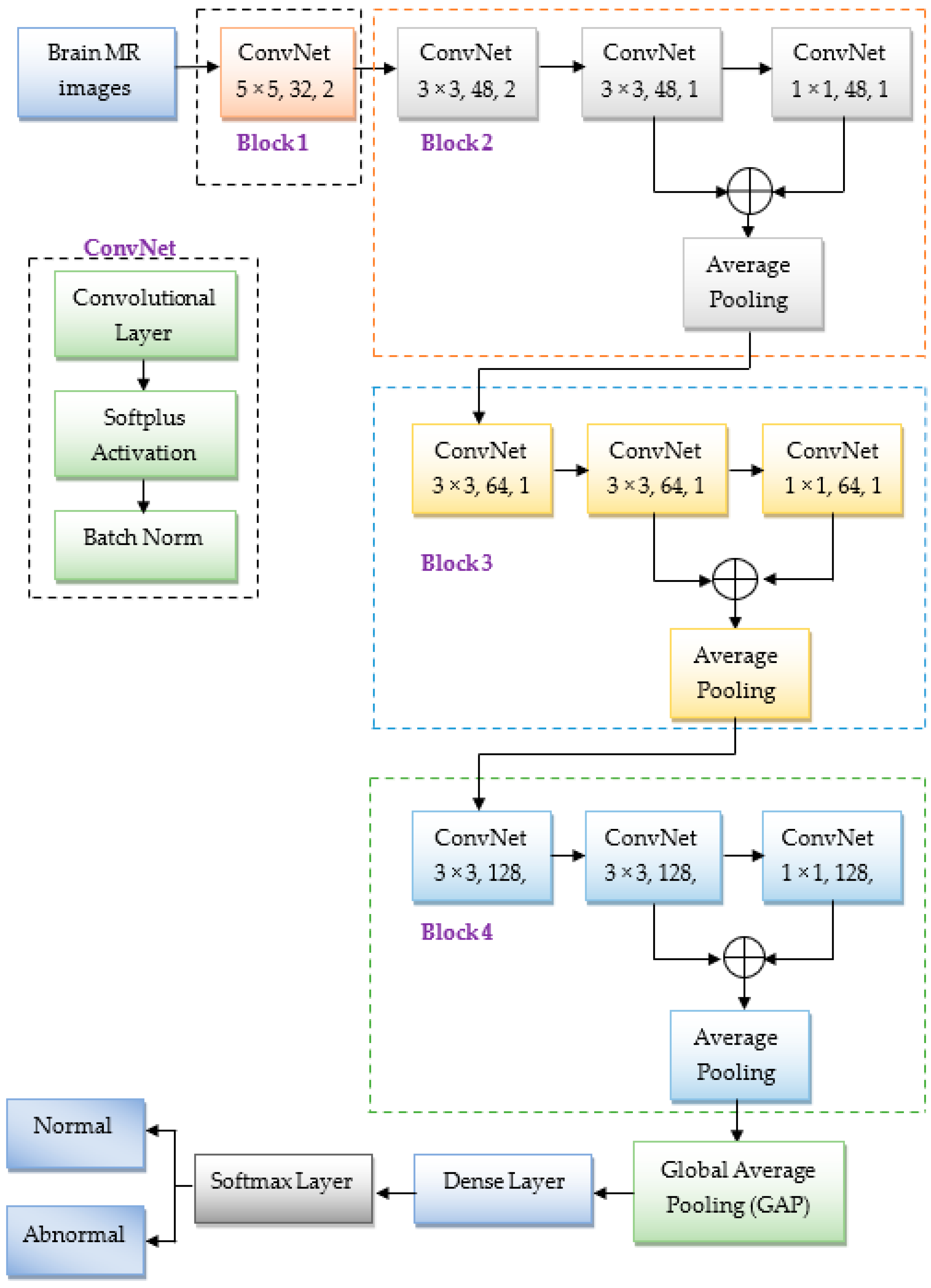
3.4. The Suggested Lightweight CNN Architecture
- In the first block, the ConvNet module has 32 filters with a 5 × 5 kernel size, and the stride is 2. Here, the stride of 2 for the convolutional filter minimizes the input’s size to half, resulting in reduced computational complexity. Usually, the initial convolutional layers extract edge features; therefore, the stride of 2 will not significantly impact the model’s accuracy at initial convolutional layers.
- Block 2 has three ConvNets, and they have 48 filters with a kernel size of 3 × 3, 3 × 3, and 1 × 1, and the strides of 2, 1, and 1, respectively. Similarly, blocks 3 and 4 contain three ConvNets with 64 and 128 filters. Each filter has a size of 3 × 3, 3 × 3, and 1 × 1, with a stride of 1. Here, the 1 × 1 convolutional filter is mainly used to minimize the computational requirements, i.e., reduce the dimensionality of the feature map. Due to that, the proposed CNN model required significantly fewer learnable parameters to train the model, as illustrated in Table 3. From this table, we observed that the total number of parameters is nearly 0.35 million. This number is much less than the other traditional CNN models discussed in the literature such as AlexNet [18,26], ResNet-50 [29], VGG-19 [30], etc. Hence, we called as a lightweight CNN.
- In each ConvNet, we used a batch norm layer to improve the training speed and minimize overfitting.
3.5. Segmentation
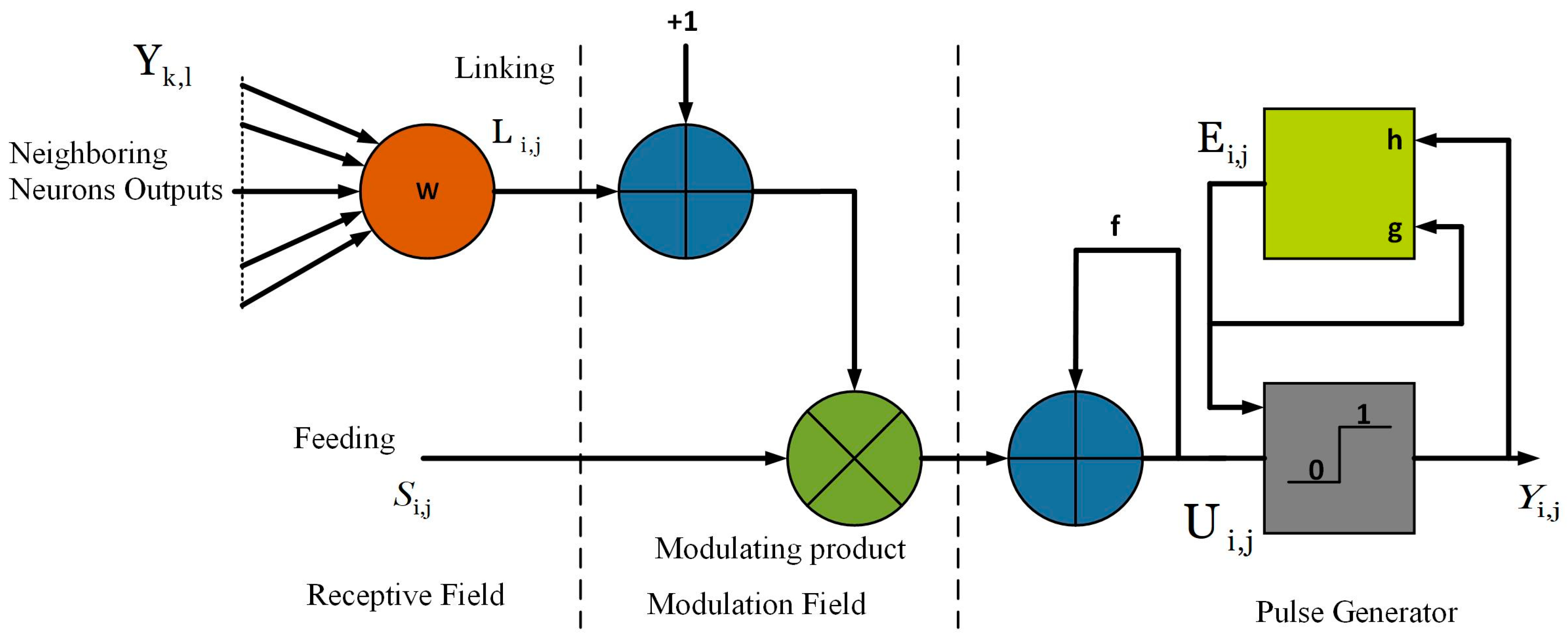
3.5.1. Modified Spiking Cortical Model (MSCM)
3.5.2. Parameter Settings of MSCM
- Firstly, the output, and internal activity, are initialized as ‘zero’.
- Threshold, .
- Decay constant, .
- Magnitude scaling factor, .
- Threshold decay, .
- Due to the position invariant nature can be determined by a 7 × 7 Gaussian filter with standard deviation ‘1′, which is utilized to estimate the precision level of the image pixel.
- The threshold magnitude component, is employed to ensure that each neuron will not fire more than once and is estimated using Equation (16).where the linking strength, is obtained by the following expression:where and , are the central difference gradient of along - direction and - direction.
- The maximum number of iterations can be determined as follows:where is the gray-level thresholding of , estimated from Otsu’s approach [41]. Here, the primary objective of thresholding is to calculate the number of iterations. For better segmentation, we apply the fast-linking algorithm to MSCM.
3.5.3. Fast-Linking
- Internal loop: Here, and are repeated until does not vary.
- External loop: Here, the function is iterated.
| Algorithm 2. The fast-linking approach. |
whiledo Update by Equation (14) repeat , Update , and by Equations (11)–(13), until end while |
3.6. Performance Metrics
4. Results and Discussion
4.1. Experimental Outcomes
4.1.1. Classification Analysis
- Fewer parameters to train the model, approximately 0.35 million.
- Minimize the overfitting problems due to the initialization of weights in the layer.
- Significantly achieved high performance due to image data augmentation.
- Low computational time.
- Extraction of complex features without human intervention.
4.1.2. Segmentation Analysis
- Using the proposed skull-stripping process, we significantly isolate the brain tissues from non-brain matters. Due to this, the implemented approach accurately identifies brain-related diseases.
- The proposed FL-MSCM makes each region as homogeneous as possible, with high computational efficiency, simple parameter tuning, low reduction in contrast, and image details. It is a significant advantage of the FL-MSCM.
- The implemented approach access adequately visible edges or boundaries.
5. Conclusions and Future Scope
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Soltaninejad, M.; Yang, G.; Lambrou, T.; Allinson, N.; Jones, T.L.; Barrick, T.R.; A Howe, F.; Ye, X. Supervised learning based multimodal MRI brain tumour segmentation using texture features from supervoxels. Comput. Methods Programs Biomed. 2018, 157, 69–84. [Google Scholar] [CrossRef]
- Hagargi, A.P.; Shubhangi, D.C. Brain tumour detection and ART classification technique in MR brain images using RPCA QT decomposition. Brain 2018, 5, 1717–1725. [Google Scholar]
- Kasban, H. A comparative study of medical imaging techniques. Int. J. Inf. Sci. Intell. Syst. 2015, 4, 37–58. [Google Scholar]
- Amin, J.; Sharif, M.; Yasmin, M.; Fernandes, S.L. A distinctive approach in brain tumor detection and classification using MRI. Pattern Recognit. Lett. 2017, 139, 118–127. [Google Scholar] [CrossRef]
- Gordillo, N.; Montseny, E.; Sobrevilla, P. State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 2013, 31, 1426–1438. [Google Scholar] [CrossRef]
- Arabahmadi, M.; Farahbakhsh, R.; Rezazadeh, J. Deep Learning for Smart Healthcare—A Survey on Brain Tumor Detection from Medical Imaging. Sensors 2022, 22, 1960. [Google Scholar] [CrossRef]
- Xie, Y.; Zaccagna, F.; Rundo, L.; Testa, C.; Agati, R.; Lodi, R.; Manners, D.N.; Tonon, C. Convolutional neural network techniques for brain tumor classification (from 2015 to 2022): Review, chal-lenges, and future perspectives. Diagnostics 2022, 12, 1850. [Google Scholar] [CrossRef]
- Kale, V.V.; Hamde, S.T.; Holambe, R.S. Brain disease diagnosis using local binary pattern and steerable pyramid. Int. J. Multimedia Inf. Retr. 2019, 8, 155–165. [Google Scholar] [CrossRef]
- Singh, R.; Goel, A.; Raghuvanshi, D.K. M.R. brain tumor classification employing ICA and kernel-based support vector machine. Signal Image Video Process. 2020, 15, 501–510. [Google Scholar] [CrossRef]
- Bahadure, N.B.; Ray, A.K.; Thethi, H.P. Comparative Approach of MRI-Based Brain Tumor Segmentation and Classification Using Genetic Algorithm. J. Digit. Imaging 2018, 31, 477–489. [Google Scholar] [CrossRef]
- Gokulalakshmi, A.; Karthik, S.; Karthikeyan, N.; Kavitha, M.S. ICM-BTD: Improved classification model for brain tumor diagnosis using discrete wavelet trans-form-based feature extraction and SVM classifier. Soft Comput. 2020, 24, 18599–18609. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. BrainMRNet: Brain tumor detection using magnetic resonance images with a novel convolutional neural network model. Med. Hypotheses 2019, 134, 109531. [Google Scholar] [CrossRef]
- Neffati, S.; Ben Abdellafou, K.; Taouali, O.; Bouzrara, K. Enhanced SVM–KPCA Method for Brain MR Image Classification. Comput. J. 2019, 63, 383–394. [Google Scholar] [CrossRef]
- Wang, S.; Du, S.; Atangana, A.; Liu, A.; Lu, Z. Application of stationary wavelet entropy in pathological brain detection. Multimedia Tools Appl. 2016, 77, 3701–3714. [Google Scholar] [CrossRef]
- Arunkumar, N.; Mohammed, M.A.; Ghani, M.K.A.; Ibrahim, D.A.; Abdulhay, E.; Ramirez-Gonzalez, G.; de Albuquerque, V.H.C. K-means clustering and neural network for object detecting and identifying abnormality of brain tumor. Soft Comput. 2019, 23, 9083–9096. [Google Scholar] [CrossRef]
- Toğaçar, M.; Cömert, Z.; Ergen, B. Classification of brain MRI using hyper column technique with convolu-tional neural network and feature selection method. Expert Syst. Appl. 2020, 149, 113274. [Google Scholar] [CrossRef]
- Chanu, M.M.; Thongam, K. Computer-aided detection of brain tumor from magnetic resonance images using deep learning network. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 6911–6922. [Google Scholar] [CrossRef]
- Lu, S.; Lu, Z.; Zhang, Y.-D. Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 2018, 30, 41–47. [Google Scholar] [CrossRef]
- Vishnuvarthanan, G.; Rajasekaran, M.P.; Subbaraj, P.; Vishnuvarthanan, A. An unsupervised learning method with a clustering approach for tumor identification and tissue segmentation in magnetic resonance brain images. Appl. Soft Comput. 2016, 38, 190–212. [Google Scholar] [CrossRef]
- Hasan, A.M.; Meziane, F. Automated screening of MRI brain scanning using grey level statistics. Comput. Electr. Eng. 2016, 53, 276–291. [Google Scholar] [CrossRef]
- Nagarathinam, E.; Ponnuchamy, T. Image registration-based brain tumor detection and segmen-tation using ANFIS classification approach. Int. J. Imaging Syst. Technol. 2019, 29, 510–517. [Google Scholar] [CrossRef]
- Ahmadi, M.; Sharifi, A.; Fard, M.J.; Soleimani, N. Detection of brain lesion location in MRI images using convolutional neural network and robust PCA. Int. J. Neurosci. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Toufiq, D.M.; Sagheer, A.M.; Veisi, H. Brain tumor identification with a hybrid feature extraction method based on discrete wavelet transform and principle component analysis. Bull. Electr. Eng. Inform. 2021, 10, 2588–2597. [Google Scholar] [CrossRef]
- Garg, G.; Garg, R. Brain tumor detection and classification based on hybrid ensemble classifier. arXiv 2021, arXiv:2101.00216. [Google Scholar]
- Pitchai, R.; Supraja, P.; Victoria, A.H.; Madhavi, M. Brain tumor segmentation using deep learning and fuzzy K-Means clustering for magnetic resonance im-ages. Neural Processing Lett. 2021, 53, 2519–2532. [Google Scholar] [CrossRef]
- Lu, S.; Wang, S.-H.; Zhang, Y.-D. Detection of abnormal brain in MRI via improved AlexNet and ELM optimized by chaotic bat algorithm. Neural Comput. Appl. 2021, 33, 10799–10811. [Google Scholar] [CrossRef]
- Murthy, M.Y.B.; Koteswararao, A.; Babu, M.S. Adaptive fuzzy deformable fusion and optimized CNN with ensemble classification for automated brain tumor diagnosis. Biomed. Eng. Lett. 2021, 12, 37–58. [Google Scholar] [CrossRef]
- Dehkordi, A.A.; Hashemi, M.; Neshat, M.; Mirjalili, S.; Sadiq, A.S. Brain Tumor Detection and Classification Using a New Evolutionary Convolutional Neural Network. arXiv 2022, arXiv:2204.12297. [Google Scholar] [CrossRef]
- Sharma, A.K.; Nandal, A.; Dhaka, A.; Koundal, D.; Bogatinoska, D.C.; Alyami, H. Enhanced Watershed Segmentation Algorithm-Based Modified ResNet50 Model for Brain Tumor Detection. BioMed Res. Int. 2022, 2022, 1–14. [Google Scholar] [CrossRef]
- Sharma, S.; Gupta, S.; Gupta, D.; Juneja, A.; Khatter, H.; Malik, S.; Bitsue, Z.K. Deep Learning Model for Automatic Classification and Prediction of Brain Tumor. J. Sensors 2022, 2022, 3065656. [Google Scholar] [CrossRef]
- Haitham, A.; Guesmi, R.; Alshammari, B.M.; Hamrouni, T.; Guesmi, T.; Alzamil, A.; Belguesmi, L. A Novel Data Augmentation-Based Brain Tumor Detection Using Convolutional Neural Network. Appl. Sci. 2022, 12, 3773. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Montavon, G.; Samek, W.; Müller, K.-R. Methods for interpreting and understanding deep neural networks. Digit. Signal Process. 2018, 73, 1–15. [Google Scholar] [CrossRef]
- Harvard Medical School Data. Available online: http://www.med.harvard.edu/AANLIB/ (accessed on 16 March 2021).
- Benson, C.C.; Lajish, V.L. Morphology Based Enhancement and Skull Stripping of MRI Brain Images. In Proceedings of the 2014 International Conference on Intelligent Computing Applications, Coimbatore, India, 6–7 March 2014; pp. 254–257. [Google Scholar] [CrossRef]
- Kalavathi, P.; Prasath, V.B.S. Methods on skull stripping of MRI head scan images—A review. J. Digit. Imag. 2016, 29, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Shi, J.; Li, Q.; Teng, J.; Wang, M. Image segmentation using fast linking SCM. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–17 July 2015. [Google Scholar]
- Zhan, K.; Zhang, H.; Ma, Y. New Spiking Cortical Model for Invariant Texture Retrieval and Image Processing. IEEE Trans. Neural Netw. 2009, 20, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Eckhorn, R.; Frien, A.; Bauer, R.; Woelbern, T.; Kehr, H. High frequency (60–90 Hz) oscillations in primary visual cortex of awake monkey. Neuro Rep. Int. J. Rapid Commun. Res. Neurosci. 1993, 4, 243–246. [Google Scholar] [CrossRef]
- Kuntimad, G.; Ranganath, H. Perfect image segmentation using pulse coupled neural networks. IEEE Trans. Neural Networks 1999, 10, 591–598. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Johnson, J.L.; Padgett, M.L. PCNN models and applications. IEEE Trans. Neural Netw. 1999, 10, 480–498. [Google Scholar] [CrossRef]
- Reddy, K.R.; Dhuli, R. Segmentation and classification of brain tumors from MRI images based on adaptive mechanisms and ELDP feature descriptor. Biomed. Signal Process. Control. 2022, 76, 103704. [Google Scholar] [CrossRef]
- Arlot, S.; Celisse, A. A survey of cross-validation procedures for model selection. Stat. Surv. 2010, 4, 40–79. [Google Scholar] [CrossRef]
- Bottou, L. Stochastic gradient descent tricks. In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2012; pp. 421–436. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Duchi, J.; Hazan, E.; Singer, Y. Adaptive subgradient methods for online learning and stochastic optimization. J. Mach. Learn. Res. 2011, 12, 2121–2159. [Google Scholar]
- Zeiler, M.D. Adadelta: An adaptive learning rate method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Available online: http://www.cs.toronto.edu/~tijmen/csc321/slides/lecture_slides_lec6.pdf (accessed on 12 August 2022).
- Timothy, D. Incorporating nesterov momentum into adam. Nat. Hazards 2016, 3, 437–453. [Google Scholar]



| Reference | Methods to Be Used | Accuracy | Pros | Cons |
|---|---|---|---|---|
| Kale et al. [8] | LBP and SP | Accuracy = 96.17% | Significantly extract the directional details of abnormal tissues. | Performance of the model depends upon the selection of orientation bands. |
| Singh et al. [9] | DWT and ICA | Accuracy = 98.87% | Relatively obtain the spatial information that is useful in the classification of brain MR images. | The selection of an appropriate mother wavelet is a major challenge. |
| Bahadure et al. [10] | FCM and BWT | DSC = 82% | In expensive to compute and manipulate. | Relatively required large number of coefficients for approximating the smooth functions. |
| Gokulalakshmi et al. [11] | DWT and GLCM | Accuracy = 92.76% | Low-processing time and easy to implement. | Selection of displacement vector. |
| Toğaçar et al. [12] | BrainMRNet | Accuracy = 96.05% | Substantially abstract the features. | Working on low-resolution images. |
| Neffati et al. [13] | DWT and PCA | Accuracy = 97.02% | Reduce overfitting and improve visualization. | Loose the some significant information. |
| Wang et al. [14] | SWT and Entropy | Accuracy = 96.6% | Significantly highlighting the image edge features. | Irrelevant features might be extracted due to wavelet aliasing. |
| Arunkumar et al. [15] | K-means clustering and ANN | Accuracy = 94.07% | Woks very well on limited data. | Selection of K-value is difficult. |
| Toğaçar et al. [16] | CNN and hyper-column feature selection | Accuracy = 96.77% | Relatively retain the local discriminative features. | High computational time. |
| Chanu et al. [17] | CNN | Accuracy = 97.14 % | Less computational time. | Low performance on normal brain MR images |
| Lu et al. [18] | AlexNet | Accuracy = 95.71 % | Perform well on abnormal brain MR images. | Large number of parameters to be need for training. |
| Vishnuvarthanan et al. [19] | SOM-FKM | DSC = 47% | Faster convergence with significant accuracy. | Requires necessary and sufficient information for developing significant clusters. |
| Hasan et al. [20] | Modified GLCM | Accuracy = 97.8 % | Achieved remarkable accuracy and also independent on atlas registration. | Large memory requirements and computationally expensive. |
| Nagarathinam et al. [21] | GLCM and Morphological operations | DSC = 92% | Does not required any seed points for identification of brain MR tumor Images. | Low classification accuracy on abnormal brain MR images. |
| Ahmadi et al. [22] | CNN | DSC = 91% | Faster segmentation. | Heavily depends upon the network design parameters. |
| Toufiq et al. [23] | DWT, PCA, GLCM | Accuracy = 98 % | Minimizing the overfitting problems. | Loss of essential details due to the feature reduction process. |
| Ginni Garg et al. [24] | SWT, PCA, and Hybrid Ensemble classifier | Accuracy = 97.305% | Relatively improve the robustness of derived texture features. | High time complexity. |
| Pitchai et al. [25] | GLCM, FKM and ANN | Accuracy = 94% | Does not effect by over segmentation. | The accuracy of the ANN classifier entirely depends on the number of hidden neurons (HN). |
| Siyuan et al. [26] | Improved AlexNet, ELM and CBM | Accuracy = 98 % | Minimizing the overfitting problems | High computational complexity. |
| Mantripragada et al. [27] | AFDM and DNN | Accuracy = 96.22% | High convergence rate. | Low training speed and high complexity. |
| Amin et al. [28] | CNN and Nonlinear Lévy Chaotic Moth Flame Optimizer (NLCMFO) | Accuracy = 97.4% | Effective hyper-parameter tuning. | Difficulty in deterinming the ideal control parameters in NLCMFO. |
| Sharma et al. [29] | ResNet 50, EWS | Accuracy = 92 % | Significantly locate the boundary pixels of the tumor. | High computational time and heavily depends on batch normalization layers. |
| Sharma et al. [30] | VGG 19 | Accuracy = 98.04% | Relatively working on a more extensive database. | Only the axial dataset of brain tumors was considered. |
| Haitham et al. [31] | Cascaded CNN | DSC = 85.3% | Relatively achieved good performance in a limited brain MR image database. | Required more time to train the parameters. |
| Augmentation Operator | Value |
|---|---|
| Rotation | Randomly from −30° to 30° |
| Translation | Translate along X (horizontal) and Y (vertical) directions with a range of [−10,10] |
| Reflection | Reflect randomly along X and Y-direction |
| Scale | Uniform scaling with a range of [0.5 to 4] |
| Shear | Shearing along vertical and horizontal with a range of [0° to 30°] |
| Block | Layers and Specifications | Size of the Activations | Parameters |
|---|---|---|---|
| - | Input image | 224 × 224 × 3 | 0 |
| - | 5 × 5 Convolution with 32 filters | 110 × 110 × 32 | 2432 |
| Batch norm | 110 × 110 × 32 | 128 | |
| 1 | 3 × 3 Convolution with 48 filters | 54 × 54 × 48 | 13,872 |
| Batch norm | 54 × 54 × 48 | 192 | |
| 3 × 3 Convolution with 48 filters | 52 × 52 × 48 | 20,784 | |
| Batch Norm | 52 × 52 × 48 | 192 | |
| 1 × 1 Convolution with 48 filters | 52 × 52 × 48 | 2352 | |
| Batch Norm | 52 × 52 × 48 | 192 | |
| 2 × 2 Average pooling with stride 2 | 26 × 26 × 48 | 0 | |
| 2 | 3 × 3 Convolution with 64 filters | 24 × 24 × 64 | 27,712 |
| Batch Norm | 24 × 24 × 64 | 256 | |
| 3 × 3 Convolution with 64 filters | 22 × 22 × 64 | 36,928 | |
| Batch Norm | 22 × 22 × 64 | 256 | |
| 1 × 1 Convolution with 64 filters | 22 × 22 × 64 | 4160 | |
| Batch Norm | 22 × 22 × 64 | 256 | |
| 2 × 2 Average pooling with stride 2 | 11 × 11 × 64 | 0 | |
| 3 | 3 × 3 Convolution with 128 filters | 9 × 9 × 128 | 73,856 |
| Batch Norm | 9 × 9 × 128 | 512 | |
| 3 × 3 Convolution with 128 filters | 7 × 7 × 128 | 147,584 | |
| Batch Norm | 7 × 7 × 128 | 512 | |
| 1 × 1 Convolution with 128 filters | 7 × 7 × 128 | 16,512 | |
| Batch Norm | 7 × 7 × 128 | 512 | |
| 2 × 2 Average pooling with stride 2 | 3 × 3 × 128 | 0 | |
| Global average pooling | - | 0 | |
| Fully connected layer | - | 258 | |
| Trainable parameters | 347,954 | ||
| Non- trainable parameters | 1504 | ||
| Total parameters | 349,458 |
| Optimizer | Parameters |
|---|---|
| SGDM | = 0.001, momentum = 0.9 |
| Adam | = 0.001, = 0.9, = 0.999, and = 10×10−7 |
| Adamax | = 0.001, = 0.9, =0.999, and = 10×10−7 |
| Adagrad | = 0.001 and = 10×10−7 |
| Adadelta | = 0.001, = 10 × 10−7 and rho = 0.95 |
| RMSprop | = 0.001, = 10×10−7 and rho = 0.9 |
| Nadam | = 0.001, = 0.9, =0.999, and = 10×10−7 |
| 5-FCV | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|
| TPR | TNR | PPV | F-Score | AUC | Accuracy | |
| 1st Fold | 100 | 99.13 | 99.54 | 99.77 | 99.56 | 99.7 |
| 2nd Fold | 99.11 | 98.15 | 99.11 | 99.11 | 98.63 | 98.8 |
| 3rd Fold | 100 | 95.57 | 97.78 | 98.87 | 97.78 | 98.5 |
| 4th Fold | 97.85 | 99 | 99.56 | 98.67 | 98.42 | 98.2 |
| 5th Fold | 99.56 | 96.15 | 98.27 | 98.91 | 97.85 | 98.5 |
| Mean ± SD | 99.3 ± 0.4 | 97.6 ± 0.73 | 98.85 ± 0.35 | 99.06 ± 0.19 | 98.45 ± 0.32 | 98.74 ± 0.52 |
| 5-FCV | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|
| TPR | TNR | PPV | F-Score | AUC | Accuracy | |
| 1st Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| 2nd Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| 3rd Fold | 97.7 | 100 | 100 | 98.84 | 98.9 | 98.5 |
| 4th Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| 5th Fold | 99.57 | 98.98 | 99.57 | 99.57 | 99.4 | 99.4 |
| Mean ± SD | 99.45 ± 0.44 | 99.80 ± 0.2 | 99.91 ± 0.08 | 99.68 ± 0.36 | 99.66 ± 0.22 | 99.58 ± 0.29 |
| 5-FCV | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|
| TPR | TNR | PPV | F-Score | AUC | Accuracy | |
| 1st Fold | 98.68 | 100 | 100 | 99.33 | 99.34 | 99.1 |
| 2nd Fold | 98.58 | 100 | 100 | 99.28 | 99.3 | 99.1 |
| 3rd Fold | 97.4 | 100 | 100 | 98.68 | 98.7 | 98.2 |
| 4th Fold | 99.56 | 99.04 | 99.56 | 99.56 | 99.3 | 99.4 |
| 5th Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| Mean ± SD | 98.84 ± 0.45 | 99.80 ± 0.19 | 99.91 ± 0.08 | 99.37 ± 0.29 | 99.33 ± 0.21 | 99.16 ± 0.29 |
| 5-FCV | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|
| TPR | TNR | PPV | F-Score | AUC | Accuracy | |
| 1st Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| 2nd Fold | 99.56 | 100 | 100 | 99.78 | 99.8 | 99.7 |
| 3rd Fold | 100 | 89.52 | 95.4 | 97.64 | 97.7 | 96.7 |
| 4th Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| 5th Fold | 99.56 | 100 | 100 | 99.78 | 99.8 | 99.7 |
| Mean ± SD | 99.82 ± 0.11 | 97.90 ± 2.1 | 99.08 ± 0.92 | 99.44 ± 0.45 | 99.46 ± 0.44 | 99.22 ± 0.63 |
| 5-FCV | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|
| TPR | TNR | PPV | F-Score | AUC | Accuracy | |
| 1st Fold | 89.57 | 83.60 | 90.43 | 89.99 | 86.6 | 87.38 |
| 2nd Fold | 96.44 | 79.63 | 90.79 | 93.53 | 88.04 | 90.99 |
| 3rd Fold | 99.54 | 80.35 | 90.90 | 95.02 | 89.94 | 93.01 |
| 4th Fold | 96.10 | 81.37 | 92.12 | 94.07 | 88.74 | 91.6 |
| 5th Fold | 99.58 | 58.33 | 85.51 | 92.01 | 78.95 | 87.68 |
| Mean ± SD | 96.25 ± 1.82 | 76.66 ± 4.63 | 89.95 ± 1.14 | 92.92 ± 0.88 | 86.45 ± 1.95 | 90.13 ± 1.1 |
| 5-FCV | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|
| TPR | TNR | PPV | F-Score | AUC | Accuracy | |
| 1st Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| 2nd Fold | 98.68 | 100 | 100 | 99.33 | 99.34 | 99.01 |
| 3rd Fold | 87.66 | 99.06 | 99.5 | 93.20 | 93.36 | 91.3 |
| 4th Fold | 100 | 80.33 | 89.78 | 94.61 | 90.2 | 92.8 |
| 5th Fold | 100 | 100 | 100 | 100 | 100 | 100 |
| Mean ± SD | 97.268 ± 2.41 | 95.88 ± 3.89 | 97.85 ± 2.02 | 97.43 ± 1.46 | 96.58 ± 2.02 | 96.62 ± 1.89 |
| 5-FCV | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|
| TPR | TNR | PPV | F-Score | AUC | Accuracy | |
| 1st Fold | 98.16 | 100 | 100 | 99.07 | 99.08 | 98.8 |
| 2nd Fold | 97.8 | 95.24 | 97.8 | 97.8 | 96.52 | 96.99 |
| 3rd Fold | 99.54 | 97.34 | 98.65 | 99.09 | 98.44 | 98.8 |
| 4th Fold | 100 | 75.96 | 90.16 | 94.82 | 87.98 | 92.5 |
| 5th Fold | 97.83 | 95.14 | 97.83 | 97.83 | 96.48 | 96.99 |
| Mean ± SD | 98.67 ± 0.46 | 92.74 ± 4.28 | 96.88 ± 1.73 | 96.82 ± 0.77 | 97.72 ± 1.99 | 96.82 ± 1.15 |
| Methodology | Number of Images | Data Augmentation (Yes/No) | Parameters (Millions) | Metrics (%) | ||
|---|---|---|---|---|---|---|
| TPR | TNR | Accuracy | ||||
| MGLCM + MLP [20] | 165 | No | - | 98.1 | 97.6 | 97.8 |
| SW Entropy + RBF-SVM [14] | 255 | No | - | 98.97 | 85 | 96.6 |
| ANN [15] | 230 | No | - | 90.9 | 96.78 | 94.07 |
| DWT + KPCA + SVM [13] | 255 | No | - | 100 | 85 | 97.02 |
| AlexNet [18] | 291 | No | 56.8 | 100 | 75 | 95.71 |
| LBPSPEnerg + BPNN [8] | 612 | No | - | 98.97 | 87.5 | 96.17 |
| DWT + ICA + RBF-SVM [9] | 240 | No | - | 98.97 | 97.68 | 98.87 |
| DWT + GLCM + SVM [11] | 750 | No | - | 99.48 | 60 | 92.76 |
| BrainMRNet [12] | 253 | Yes | 0.605 | 96 | 96.08 | 96.05 |
| AlexNet + VGG-16 + RFE [16] | 310 | Yes | 27.82 | 97.83 | 95.74 | 96.77 |
| 2D CNN [17] | 309 | Yes | - | 100 | 94.11 | 97.14 |
| DWT + PCA + RF [23] | 181 | No | - | 99.2 | 97.8 | 98 |
| SWT-GLCM-Hybrid Classifier [24] | 2556 | No | - | 97.04 | 97.60 | 97.31 |
| FKM-ANN [25] | - | - | - | 98 | 99 | 94 |
| AlexNet-ELM-CBM [26] | 359 | No | 62.3 | 97.14 | 95.71 | 96.43 |
| AFDF-DNN [27] | - | - | - | 98.35 | 50 | 96.44 |
| CNN [28] | 694 | No | - | 96 | 98.6 | 97.4 |
| Modified ResNet50 [29] | 278 | Yes | 23.68 | 83 | 80 | 92 |
| VGG-19 [30] | 257 | No | 143 | 100 | 94.73 | 98.04 |
| The Proposed (lightweight CNN) | 185 | Yes | 0.349 | 99.45 | 99.8 | 99.58 |
| Sample Image | DSC | PPV | TPR | TNR | F-Score | AUC | Accuracy |
|---|---|---|---|---|---|---|---|
| 1 | 96.46 | 99.76 | 99.94 | 94.41 | 99.85 | 97.17 | 99.71 |
| 2 | 94.8 | 99.96 | 99.97 | 94.58 | 99.96 | 97.27 | 99.93 |
| 3 | 96.05 | 99.85 | 99.85 | 96.03 | 99.85 | 97.94 | 99.71 |
| 4 | 99.4 | 99.94 | 99.96 | 99.29 | 99.95 | 99.63 | 99.91 |
| 5 | 89.04 | 99.8 | 99.77 | 89.81 | 99.78 | 94.75 | 99.57 |
| 6 | 88 | 99.86 | 99.73 | 91.53 | 99.79 | 95.63 | 99.59 |
| 7 | 99.53 | 99.99 | 99.95 | 99.8 | 99.97 | 99.8 | 99.94 |
| 8 | 93.16 | 99.37 | 99.53 | 92.23 | 99.45 | 95.88 | 98.98 |
| 9 | 87.98 | 99.4 | 99.86 | 81.91 | 99.62 | 90.88 | 99.28 |
| 10 | 98.58 | 99.82 | 99.98 | 97.51 | 99.89 | 98.74 | 99.81 |
| 11 | 99.94 | 100 | 100 | 99.97 | 99.99 | 99.98 | 99.99 |
| 12 | 98.82 | 99.9 | 99.99 | 97.87 | 99.94 | 98.93 | 99.89 |
| 13 | 97.68 | 99.94 | 99.96 | 97.16 | 99.95 | 98.56 | 99.91 |
| 14 | 89.43 | 99.95 | 99.15 | 98.76 | 99.55 | 98.96 | 99.14 |
| 15 | 95.3 | 99.99 | 99.62 | 99.84 | 99.8 | 99.73 | 99.62 |
| 16 | 95.61 | 99.82 | 99.9 | 94.44 | 99.86 | 97.17 | 99.73 |
| 17 | 91.77 | 100 | 99.74 | 100 | 99.86 | 99.87 | 99.74 |
| 18 | 95.6 | 99.21 | 99.46 | 94.78 | 99.33 | 97.12 | 98.84 |
| 19 | 97.15 | 99.94 | 99.44 | 99.44 | 99.68 | 99.44 | 99.44 |
| 20 | 95.6 | 99.37 | 99.83 | 93.26 | 99.6 | 96.55 | 99.27 |
| 21 | 97.12 | 99.68 | 99.95 | 95.12 | 99.81 | 97.54 | 99.65 |
| 22 | 98.49 | 100 | 99.94 | 100 | 99.97 | 99.97 | 99.94 |
| 23 | 96.29 | 99.99 | 99.82 | 99.57 | 99.9 | 99.69 | 99.81 |
| 24 | 97.22 | 99.98 | 99.64 | 99.75 | 99.81 | 99.7 | 99.65 |
| 25 | 88.74 | 99.51 | 99.46 | 89.3 | 99.78 | 94.75 | 99.01 |
| 26 | 99.56 | 99.98 | 99.98 | 99.56 | 99.98 | 99.77 | 99.96 |
| 27 | 96.36 | 100 | 99.79 | 100 | 99.89 | 99.9 | 99.8 |
| 28 | 98.28 | 99.99 | 99.9 | 99.69 | 99.95 | 99.8 | 99.9 |
| 29 | 99.76 | 100 | 100 | 99.93 | 99.99 | 99.96 | 99.99 |
| 30 | 98.88 | 99.97 | 99.97 | 98.85 | 99.97 | 99.41 | 99.94 |
| Average | 95.7 | 99.83 | 99.8 | 96.5 | 99.82 | 98.15 | 99.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reddy, K.R.; Dhuli, R. A Novel Lightweight CNN Architecture for the Diagnosis of Brain Tumors Using MR Images. Diagnostics 2023, 13, 312. https://doi.org/10.3390/diagnostics13020312
Reddy KR, Dhuli R. A Novel Lightweight CNN Architecture for the Diagnosis of Brain Tumors Using MR Images. Diagnostics. 2023; 13(2):312. https://doi.org/10.3390/diagnostics13020312
Chicago/Turabian StyleReddy, Kamireddy Rasool, and Ravindra Dhuli. 2023. "A Novel Lightweight CNN Architecture for the Diagnosis of Brain Tumors Using MR Images" Diagnostics 13, no. 2: 312. https://doi.org/10.3390/diagnostics13020312
APA StyleReddy, K. R., & Dhuli, R. (2023). A Novel Lightweight CNN Architecture for the Diagnosis of Brain Tumors Using MR Images. Diagnostics, 13(2), 312. https://doi.org/10.3390/diagnostics13020312







