Abstract
Due to its widespread availability, low cost, feasibility at the patient’s bedside and accessibility even in low-resource settings, chest X-ray is one of the most requested examinations in radiology departments. Whilst it provides essential information on thoracic pathology, it can be difficult to interpret and is prone to diagnostic errors, particularly in the emergency setting. The increasing availability of large chest X-ray datasets has allowed the development of reliable Artificial Intelligence (AI) tools to help radiologists in everyday clinical practice. AI integration into the diagnostic workflow would benefit patients, radiologists, and healthcare systems in terms of improved and standardized reporting accuracy, quicker diagnosis, more efficient management, and appropriateness of the therapy. This review article aims to provide an overview of the applications of AI for chest X-rays in the emergency setting, emphasizing the detection and evaluation of pneumothorax, pneumonia, heart failure, and pleural effusion.
1. Introduction
Over recent years, there has been increasing interest in the application of artificial intelligence (AI) techniques to medical imaging examinations. Chest X-ray (CXR) is one of the most frequently performed examinations, particularly in the emergency setting, due to its widespread availability, low costs, and the possibility to be performed at the patient’s bed. It provides significant information on lung parenchyma and the related pathologies, as well as on cardiovascular circulation and pleural disorders.
A correct and rapid CXR report is decisive in choosing the proper treatment and improving the patients’ outcome. Although CXR reading is considered a basic radiological skill, it remains challenging and depends on the radiologist’s experience, workload, and environment. The development of robust and efficient AI algorithms could greatly facilitate CXR readout, particularly in emergency settings, where time spent on CXR interpretation and the accuracy of responses is often of vital importance.
Owing to the high number of CXR examinations performed daily in hospitals around the world, there is a large amount of data available for developing robust AI algorithms. AI-based tools have been shown to facilitate the detection of numerous health-threatening conditions, as well as to prioritize the reporting of patients with critical findings [1,2]. In this narrative review, we provide an overview of the different AI tools used for CXR interpretation and their performance in the emergency setting for the study of: pneumothorax, pneumonia, COVID-19, heart failure and pleural effusion.
1.1. A Quick Introduction to AI
Artificial intelligence (AI) can be defined as technology that mimics human cognitive processes, such as learning, reasoning, and problem-solving. As, in conventional radiology, diagnosis is primarily qualitative, AI-powered assessment could make a significant contribution in this field, reducing the variability in image interpretation and improving diagnostic accuracy [1].
AI applications in radiology are driven by the idea that medical images are a set of data that can be computed by a machine to extract useful information [3]. Therefore, increasing the data storage capacity and computing power is a prerequisite for the development of AI-based tools. However, it is not just a question of making the evaluations routinely expressed by the radiologist faster and more precise, but also of extracting information not visible to the human eye to improve the clinical managment [4]. The systematic application of a quantitative approach (essentially AI-driven) to the problem of interpreting biomedical images is at the base of the so-called radiomics paradigm [4,5].
Machine learning (ML) is a branch of AI that applies concepts and tools from other disciplines, primarily statistics and programming, to build algorithms aimed at the automated detection of meaningful patterns in data, a field closely related to data mining [6]. In radiology, ML can be used to extract information from imaging data [4].
The process of developing ML-based tools embodies several phases, among which the main ones are the training and validation phase [6]. In general, the training phase requires the exposure to a set of data, or cases, which can be variably labeled (supervised learning) or unlabeled (unsupervised learning) [6].
In supervised learning, the simplest form of ML, the system simulates the human cognitive process of “learning by examples”. This type of ML is suitable for very general classification tasks in which new elements need to be labeled according to some predefined categories [7]. Labeled data points might be obtained from human experts that annotate (“label”) data with their corresponding label values (for example, a chest X-ray could be positive for pneumonia “yes = 1” or negative “no = 0”). These methods exploit a training set that consists of tuples (x,y) made of inputs (x), for which we know the corresponding label values, which therefore represent the output (y). The supervised ML algorithm searches for a hypothesis (f) that maps the relationship (x → y) between the data, imitating the human annotator, which allows it to predict the label solely based on the features of a data point.
While radiologists mainly evaluate qualitative features, such as increased or reduced radiopacity, comparing them to a subjective reference standard, ML features are low-level properties, or metrics, of a data point that can be computed or measured easily. There are hand-crafted features (manually defined by data scientists) and automatically extracted features (usually through deep-learning algorithms, see further). The problem of choosing which features to select to build more accurate models is one of the most challenging parts in the radiomic workflow. Even if reproducible features are not necessarily clinically informative, successful AI models must be built upon reproducible and robust features [8].
In unsupervised learning, the system analyzes and extracts significant features from unlabeled data by forming groups or identifying relationships between subgroups [6]. This type of ML is suitable for clustering or associative tasks [7].
At the basis of each ML approach are models that can be trained and tested in data analysis [6]. A model is a theoretical hypothesis that maps a possible relationship between data and it is usually based on statistical assumptions that are computationally feasible, meaning that they can be translated easily into the programming code to perform automated data analysis [6]. Through automation, it is possible to test the feasibility of a model for a certain dataset and compare the performances of different models [6]. Several mapping hypotheses can be used to infer predictions of an amount of interest that satisfies some predefined requirement, as in supervised learning, or to ensure some rules of internal consistency between clusters, as in unsupervised learning. Some models may outperform the others in representing the desired relationship, leading to more accurate predictions. The testing phase, which follows the training phase, is necessary to assess the fitness of the model in mapping the desired relationship (if existing) between the data [4].
Artificial neural networks are a particular learning paradigm inspired by the biological network of the human brain [9]. Their operating principle is not based on statistical hypotheses generated a priori, but on the peculiarities of their structure and on computational properties of the units that compose them. In an artificial neural network, each node represents a cell that operates on the input information, according to certain rules, to obtain an output that it transmitted to the next neuron to be further processed. The computation performed by a single unit is influenced by its interconnections and their weight, which provide a measure of how much each input “counts” in the neuron. In this way, the flow of information is passed through the network while shaping the network itself. The global function (for example image recognition) is obtained through the coordinated activity of smaller units each performing an elementary computational function [9].
Complex artificial neural networks, called Convolutional Neural Networks (CNNs), have been developed and found to be particularly suitable for image analysis and recognition tasks. Deep learning (DL) is a domain of AI that takes advantage of complex artificial neural networks such as CNNs to discover intricate patterns in data. DL networks feature many intermediate layers, where each layer represents increasing levels of abstraction, to the extent that it is unclear exactly how processing the intermediate layers contributes to the overall result [10]. This is also known as the black box phenomenon and contributes to the problem of interpretability of the results of AI tools.
DL models are built to capture the full image context and learn the correlations between the local features, resulting in a superior performance in various radiological tasks, such as interpreting radiographic exams.
1.2. Open Datasets
ML and DL algorithms are trained with datasets and are dependent on the number and quality of the training data. There is a constantly increasing number of publicly available CXR datasets that can be used for image classification and retrieval tasks. Some of the biggest and most commonly used open datasets are listed in Table 1.

Table 1.
The table summarizes the characteristics of the main CXR datasets such as the number of images, number of labels, and labeling technique. The dataset containing more images is the CheXpert with 14 labels. In the last two columns, we have listed the main strengths and weaknesses of the different datasets.
2. Worklist Prioritization
The automatic notification of critical findings is one of the most interesting AI applications in emergency radiology. With the increased demand for imaging studies, delaying the communication of key data to the treating physician can delay critical care and compromise therapeutic efficacy, particularly in urgent scenarios [21].
The priority assigned by the emergency doctor who first examines the patient determines the sequence in which the imaging exams are reported; unfortunately, the precedence is not always consistent with the abnormalities observed. AI-based models may detect and prioritize emergency CXR findings in real time, reduce the report response times for key findings, and optimize therapeutic pathways.
A notification system developed by GE Medical System and Zebra Medical Vision for the evaluation of pneumothorax on CXR demonstrated a significant reduction in the time required for the diagnosis. Three experienced radiologists evaluated 588 CXR with the HealthPNX prioritization software with an average diagnosis time of 8.05 min versus 68.98 min without the software. The time needed to assess the radiograph and send a notification was only 22.1 s [22].
Annarumma et al. created and tested a CNN-based tool to simulate an automatic triage for adult CXRs according to the urgency of the imaging findings. The use of the algorithms resulted in a theoretical reduction in the reporting delay for critical studies, from 11.2 to 2.7 days [23]. Another AI tool developed by Kim et al. allowed the reduction of the time-to-report for CXRs of critical and urgent cases (from 3371.0 to 640.5 s and from 2127.1 to 1840.3 s, respectively) [24].
In the setting of the COVID-19 pandemic, Tricarico et al. [25] developed an automated tool for the prioritization of patients with the suspicion of the COVID-19 disease based on CXR analysis. This CNN-based system aimed to facilitate the workload in the emergency department by fast-forwarding the testing of suspicious cases. The proposed architecture was reviewed retrospectively on a dataset of cases collected throughout the first months of the pandemic and showed significant improvements for the identification and prioritization of COVID-19 patients. The system’s sensitivity and specificity were 78.23% and 64.2%, respectively. In preliminary real-life testing, the method reached a correlation of 0.873.
3. Pneumothorax
Pneumothorax is a pathological condition in which the pleural cavity fills with air, impairing oxygenation and ventilation. It occurs spontaneously or as a complication of trauma, medical interventions, and infections. Due to the sheer variety of the underlying etiologies and clinical scenarios, pneumothorax represents an important morbidity and mortality factor. Some forms present with a severe progressive hemodynamic compromise, with eventual cardiovascular collapse and respiratory failure if left untreated.
CXR allows for the timely diagnosis and objective quantifying of pneumothorax, which is crucial for the selection of the optimal management strategy. AI could potentially increase the sensitivity for pneumothorax identification and provide quantification through volume segmentation, particularly in low-resource settings where experienced radiologists might be lacking. Automated pneumothorax detection represents a challenging technical task due to the variability of its appearances on CXR. However, a variety of AI solutions have been proposed in recent years, powered by the rapid development of DL and the availability of large CXR datasets.
In 2017, Wang et al. published a large ChestX-ray8 database with image-level labels for eight chest conditions, including pneumothorax. A multilabel deep CNN model was performed on the dataset and demonstrated an accuracy of 0.0816 for pneumothorax detection, with an average false positive rate of only 0.2317 [26]. Smaller datasets have also been used for the training of DL algorithms. For example, Blumenfeld et al. used a dataset of 117 CXRs with pixel classification and reached a diagnostic accuracy of 0.95 for pneumothorax detection [27].
A model involving CNNs on frontal CXR demonstrated a sensitivity of 0.55, a specificity of 0.90, and an area under the curve (AUC) of 0.82 for the assessment of large and moderate pneumothorax on internal testing, although the performance was lower on external testing (AUC = 0.75) [28].
In a pneumothorax segmentation competition organized in 2019 by the Society for Imaging Informatics in Medicine, the winning team achieved a Dice score of 0.8679 by using a deep neural network ensemble and extensive data pre-processing and augmentation [29]. Wang et al. proposed a construct of several modified U-Net convolutional network models, which were validated at the 2019 segmentation competition and reached an area under the curve of 0.9795 and a Dice score of 0.8883 [30]. Another model based on the U-Net CNN architecture showed an accuracy of 97.8% and sensitivity of 69.2%, which was less precise compared to that of an experienced radiologist, but did not differ significantly (p = 0.11). For patients with >21.6% of pneumothorax, the model predicted the need for thoracostomy [31].
In another study, using a fully convolutional network algorithm trained on a large dataset with pixel-level labels, the authors reached 93.45% diagnostic accuracy and high segmentation accuracy with a mean pixel-wise accuracy (MPA) of 0.93 ± 0.13 and dice similarity coefficient of 0.92 ± 0.14 [32].
Several authors have proposed pneumothorax detection algorithms based on ResNet artificial neural networks. Gooßen et al. demonstrated an AUC of 0.96 for a ResNet-50 model [33].
A Deep ResNet-50 model successfully detected pneumothorax with a combined Dice score of 0.82 and allowed for the segmentation of pneumothorax lesions, with a Dice score ranging between 0.72 and 0.79 [34]. In a recent study, two-stage ResNet algorithms trained and validated on a large dataset demonstrated an accuracy of 94.4% and an area under the curve of 97.3% for the detection of pneumothorax [35].
Another CNN-based model correctly recognized pneumothorax and tension pneumothorax cases with an AUC of 0.979 and 0.987, respectively [36].
Yi et al. compared the performance of an algorithm based on ResNet-152 deep CNN with that of first-year radiology residents. Although the model performed faster than the first-year radiology residents, with 1980 and 2 images assessed per minute, respectively, its AUC was significantly lower (0.841 vs. 0.942 and 0.905 (p < 0.01)). The deep CNN identified 9.7% of the pneumothoraxes missed by at least one of the residents [37]. While DL algorithms are not sufficiently robust to independently assess CXR for pneumothorax, recent developments have highlighted their potential role as supportive tools. A multicenter cohort study demonstrated that the AI-aided interpretation of CXR by radiologists showed significant improvement of AUROC for pneumothorax [38].
4. Pneumonia
Pneumonia constitutes a major health hazard despite the advances in its diagnosis and management. A variety of agents can give rise to pneumonia, which translates into the heterogeneity of its epidemiology, signs and symptoms, presentation on diagnostic tests, and clinical course. Pediatric pneumonia is an ongoing global healthcare challenge, which accounts for 14% of all deaths of children under five years old, according to the World Health Organization.
CXR is the first imaging test performed for pneumonia diagnosis. Moreover, the correct interpretation of CXR allows for differentiating between viral and bacterial etiology of pneumonia, with added value for patient management. This is particularly important in developing countries, which account for a large percentage of childhood morbidity and mortality from pneumonia, but have limited access to other diagnostic tests. Not surprisingly, the diagnosis of pneumonia via CXR, particularly in the pediatric setting, has attracted significant interest among AI researchers, with a variety of proposed models.
The multilabel ChestX-ray8 project, which was described before, demonstrated an accuracy of 0.75 for the detection of pneumonia, with an average false positive rate of 0.0691 [26]. A transferable CNN, which demonstrated efficiency in classifying age-related macular degeneration and diabetic macular edema, showed an accuracy of 92.8% and an AUC of 96.8% when applied and trained on the pediatric CXR dataset for pneumonia detection. Moreover, the model was able to reliably differentiate between viral and bacterial pneumonia (accuracy 90.7%, AUC 94.0%) [39].
Similarly, a customized VGG16 model reached 96.2% diagnostic accuracy and 93.6% classification accuracy for distinguishing between bacterial and viral pneumonia, respectively. It also integrated a novel strategy for visualizing the algorithm region of interest on CXR for improved transparency of deep learning inner workings and behavior [40].
Gu et al. developed a full CNN for the segmentation of the lung regions followed by deep CNN for classification, which was evaluated on an internal pediatric CXR dataset with an accuracy of 0.80 and sensitivity of 0.77 [41]. A CNN model by Okeke et al. showed an accuracy of 0.93 for the detection of pneumonia on pediatric CXR [42]. Another CNN reached a similar accuracy, of 96-97%, in the Kaggle pneumonia dataset [43]. A CNN model by Liang et al. showed 96.7% accuracy for the detection of pneumonia in pediatric patients [44].
Rahman et al. assessed four pre-trained CNN models (AlexNet, ResNet18, DenseNet201, and SqueezeNet) on a large CXR dataset and demonstrated the superior performance of DenseNet201, which detected pneumonia with an accuracy of 0.98 and differentiated between viral and bacterial etiologies with an accuracy of 0.95 [45]. In a study by Toğaçar et al., a CNN algorithm with a linear discriminant analysis feature yielded an accuracy of 99.41% for the detection of pneumonia on CXR [46], whereas the novel deep separable residual learning model reached 98.8% accuracy and 0.99 AUC values [47].
A systematic review and meta-analysis by Li et al. demonstrated pooled AUC of 0.99 (95% CI: 0.98–100) across 15 studies assessing the performance of DL algorithms for pneumonia detection on CXR. The pooled sensitivity and specificity were 0.98 and 0.94, respectively. Moreover, it showed a pooled AUC of 0.95 for differentiating between bacterial and viral pneumonia on CXR. However, it was noted that the included studies lacked performance in comparison with healthcare professionals [48].
In a recent study by Kwon et al., an ensemble CNN model demonstrated an AUC of 0.983 for detecting pneumonia on CXR and showed a predictive value in differentiating cases that were improving and those that worsened over seven days of follow-up (p = 0.001), highlighting the potential role of AI in directing management strategies as well as refining diagnosis [49].
5. COVID-19
The outbreak of the COVID-19 pandemic gave rise to a global race toward the development of reliable and accurate diagnostic tools [50]. CXR has been one of the first tools extensively used to screen patients for COVID-19 pneumonia considering its wide availability, considerable prognostic value, and low costs [51,52]. However, interpreting CXR in a COVID-19 setting can be challenging due to its indistinct radiological characteristics, which include consolidation and hazy increased opacities [53]. AI demonstrated the potential to assist radiologists in differentiating COVID-19-positive cases on CXR. The initial lack of wide datasets has been one of the major obstacles to building AI-driven disease detection models. Additionally, CNN training takes a substantial amount of time because of the computational demands and memory constraints. Transfer learning offers an alternative development method that can overcome this issue by using pre-trained models. Utilizing pre-trained networks, such as InceptionV3, VGGNet, InceptionResNetV2, ResNet, etc., the CXR-based identification and detection of COVID-19 has been developed with benchmark accuracies reaching 99% [54].
Baltazar et al. conducted a retrospective clinical analysis on 1171 clinically verified CXR pictures from 821 cohorts that were then made accessible in open-access repositories. Among the five optimized DL architectures, InceptionV3 demonstrated the best performance for COVID-19 pneumonia detection with 86% sensitivity, 99% specificity, 91% positive predictive value, and an AUC of 0.99 in differentiating COVID-19 from negative CXR [55].
Nillmani et al. propose 16 types of segmentation-based classification deep learning-based systems for the automatic detection of COVID-19. The best performing segmentation-based classification model was UNet+Xception, which exhibited the accuracy, precision, recall, F1-score, and AUC of 97.45%, 97.46%, 97.45%, 97.43%, and 0.998 (p-value < 0.0001), respectively [56].
A classifier ensemble strategy using the Choquet fuzzy integral was suggested by Dey et al. It divides CXR scans into three categories: confirmed COVID-19, common pneumonia, and healthy lungs. Using two dense layers and one softmax layer, they extracted characteristics from the CXR pictures and classified them using the pre-trained convolutional neural network models. The accuracy provided by the suggested approach is 99% [57].
The approach presented by Nasiri et al. combines the MobileNet and DenseNet169 Deep Neural Networks to extract the characteristics of patient’s X-ray images. They subsequently used the chosen characteristics as input to the classification algorithm LightGBM (Light Gradient Boosting Machine). The ChestX-ray8 dataset, which comprises 1125 X-ray images, was used to evaluate the performance of the suggested approach. In the two-class (COVID-19, Healthy) and multi-class (COVID-19, Healthy, Pneumonia) classification tasks, they respectively achieved accuracies of 98.54% and 91.11% [58].
Ezzoddin et al. proposed a similar approach using DenseNet169 to extract the features of the patients’ CXR images and LightGBM algorithm in order to classify them. The evaluation of the ChestX-ray8 dataset reached accuracies of 99.20% and 94.22% in the two-class (i.e., COVID-19 and No-findings) and multi-class (i.e., COVID-19, Pneumonia, and No-findings) [59].
In two other studies, Nasiri et al. employed an approach using the DenseNet169 Deep Neural Network (DNN) to extract the features of X-ray images taken from the patients’ chests. In one of the studies, the features were chosen by a feature selection method, i.e., analysis of variance (ANOVA), to reduce the computation and time complexity while overcoming the curse of dimensionality to improve the accuracy. Finally, the extracted features were given as input to the Extreme Gradient Boosting (XGBoost) algorithm in order to perform the classification task. The experiments showed an accuracy of 98.72% and 98.23% for two-class classification (COVID-19, No-findings) and 89.7% to 92% accuracy for multiclass classification (COVID-19, No-findings, and Pneumonia) [60,61].
Other studies have investigated the possibility of determining and classifying images according to the estimated degree of disease severity [62].
Cohen et al. used a DenseNet model to generate a severity score based on CXR imaging. Images from a public COVID-19 database were scored retrospectively in terms of the extent of lung involvement and the degree of opacity. A neural network model that was pre-trained on large (non-COVID-19) CXR datasets was used to construct features for COVID-19 images. As a result, training a regression model on a subset of the outputs from this pre-trained model predicted the geographic extent score with a 1.14 and the lung opacity score with a 0.78 mean absolute error, respectively. The model allowed us to assess the severity of COVID-19 lung infections, which can be used for tracking the effectiveness of the treatment and for escalating or de-escalating care [63]. Jiao et al. evaluated the ability to predict the severity of COVID-19 disease utilizing the CXR as input to an EfficientNet deep neural network together with clinical data. They reported that when CXR was added to clinical data for severity prediction, the AUC increased from 0.82 to 0.84 on internal testing and from 0.73 to 0.79 on external testing. When deep-learning features were added to the clinical data for the progression prediction, the concordance index increased from 0.76 to 0.80 on internal testing and from 0.70 to 0.75 on external testing, concluding that data inferred from CXR through AI applications can augment clinical data in predicting the risk of progression to critical illness in patients with COVID-19 [64].
Khan et al. developed a new CNN architecture, STM-RENet, to interpret radiographic patterns from X-ray images. They proposed a new convolutional block STM that implements the region and edge-based operations both separately and jointly. The learning capacity of STM-RENet was further enhanced by developing a new CB-STM-RENet that exploited channel boosting and learned textural variations to effectively screen the X-ray images. The suggested model performed significantly better than typical CNNs on three datasets, particularly the CoV-NonCoV-15k dataset, with a high detection rate (97%), and accuracy (96.53%) [65].
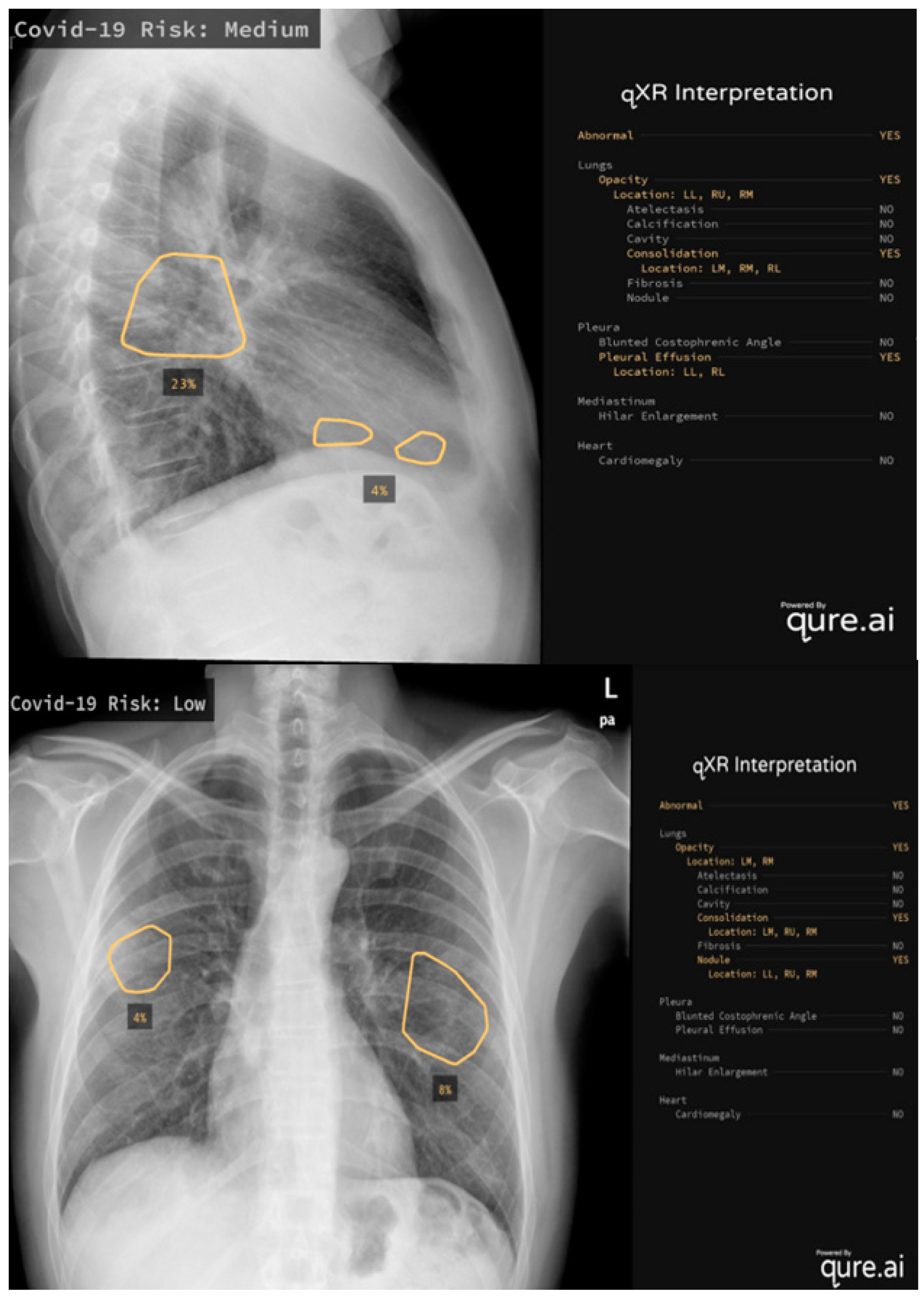
It is also worthwhile to mention some of the real-world applications of AI systems: Mustaq et al. [66] compared the performance of a deep learning AI-based system (qXR v2.1 c2, Qure.ai Technologies) to the RALE score, a radiographic score with strong inter-observer agreement that has been validated to quantify the severity and prediction outcomes in ARDS patients (Figure 1) [67]. This algorithm was initially created for use on TB patients and Mustaq et al. applied it to 694 patients, concluding that a Qure AI score of ≥30 or a RALE score of ≥12 on the CXR at the emergency department presentation was independent of, and equivalent to, the predictors of bad outcomes. Hasani et al. [68] applied an Automated Detection System utilizing X-ray Images (COV-ADSX), which detects COVID-19 using a deep neural network and XGBoost. The accuracy of the model utilized in COV-ADSX based on the ChestX-ray8 dataset was 98.23% in only 10 s, allowing the expert to receive a response rapidly while waiting for the PCR result.
Figure 1.
Two examples of qure.ai application, the algorithm calculates the risk levels, evaluating lungs, pleura, mediastinum and heart on CXR.
By assisting in difficult decision-making, AI has been proven to have the potential to play a pivotal role in the fight against COVID-19. AI algorithms can be trained to automatically detect and classify various features of the disease on CXRs with high accuracy. This could potentially save time and resources in the demanding pandemic setting. There are still some challenges that need to be addressed before AI can be widely used for COVID-19 diagnosis. The principal studies are summarized in Table 2.

Table 2.
Accuracy of most recent studies for COVID-19 diagnosis using artificial intelligence and CXR.
6. Heart Failure
In the context of an increasingly aged patient population, heart failure is one of the major causes of admission to the emergency department. Once again, CXR is one of the first-line tools in the assessment of patients in this setting [69].
Cardiopathic patients may be not compliant and are often bedridden. As a result, CXR is often limited to anterior-posterior projection with the consequent projective enlargement of the heart. In addition, their thorax may present multiple devices, such as electrocardiogram electrodes, pacemakers, or implantable cardioverter-defibrillator, which overlap the lung fields and increase the possibility of erroneous findings [70]. In this context, CXR reporting can become challenging, and a supportive tool could be useful. The Cardio-Thoracic Ratio (CTR) is the most evaluated parameter in these patients, referred to as the ratio between the maximum transverse cardiac diameter and the maximum horizontal thoracic diameter, with cardiomegaly defined as a value higher than 0.50 [71]. An automatic CTR measurement could be a simple and helpful tool to accelerate routine workflow [72]. Precise segmentation is the first step needed for an automatic DL-based calculation of CTR. CardioNet is an automatic segmentation model based on the CNN architecture that can recognize and segment the lungs and heart and generate an intuitive map from CXR, of varying quality, with an accuracy of 98.9%. CardioNet has been trained on 248 images augmented through cropping, horizontal flipping, and translating the initially available images.
A recent clinical evaluation has been performed to compare four DL models (AlbuNet, SegNet, VGG-11, and VGG-16) to find which one would show the highest percentage of automatic results of the cardiac size accepted by users with a measurement variation within ±1.8% of the human-operating range. VGG-16 gave the highest-grade result (68.9%), but the combined AlbuNet + VGG-11 model yielded excellent grades in 77.8% of the images in the evaluation dataset, a coefficient of variation of 1.55%, and reduced the CTR measurement time by almost ten-fold (1.07 ± 2.62 s vs. 10.6 ± 1.5 s) compared with manual operation [73]. Alveolar edema is another sign of heart failure, visible as bilateral perihilar lung shadowing, also known as “bat wing opacities” or “butterfly opacities”. This sign is the result of the hemodynamic pulmonary congestion caused by heart failure that brings high pulmonary capillary pressure and the abnormal transfer of fluid from the vascular to the extravascular compartments of the lungs (interstitium and alveolar spaces) [74]. Blood flow circulation alterations and inflammation result in the abnormal accumulation of fluid, respectively, transudate and exudate, seen as lung opacities on CXR. Despite some radiological similarities, these distinct diseases require different treatments, and early diagnosis of cardiogenic edema is therefore the key to improving patient outcomes.
A reliable AI algorithm should be able to distinguish the most common causes of acute respiratory failure in an emergency, mainly congestive edema and pneumonia [75].
Considering that ML is more efficient in the distinction between localized lesions and lesions with global symmetrical patterns [76], unilateral consolidation pneumonia is easier to identify than diffuse bilateral pulmonary edema. On this basis, Liong-Rung et al. have created an ambitious project using deep CNN models to recognize pneumonia and pulmonary edema in CXR images of older patients. Although the presence of medical devices in the training dataset has brought a decreased predictive performance with an accuracy reduction, from 79.1% to 73.4%, they tried to include those images anyway, attempting to post-process them by cropping, unfortunately, without favorable results [75]. However, they deserve credit for experimenting with DL tools on sub-optimal radiograms in complicated circumstances, those in which radiologists would truly appreciate artificial support. Indeed, the study revealed that using images with explicit signs of edema or pneumonia and without the interference of device overlap for training deep learning models can produce accuracy above 80% in differential diagnosis, while an accuracy of approximately 70% has been achieved in the presence of interference. Lastly, normal CXRs of patients without pneumonia or pulmonary edema had an F1 score over 95% [75].
If CNN models could suit CXR feature recognition, the current scientific literature contains a wide selection of encouraging applications that include more features, including regarding heart failure. For example, Cicero et al. investigated five radiological signs—cardiomegaly, pleural effusion, pulmonary edema, pneumothorax, and consolidation—using the GoogLeNet CNN, trained on a total of 35038 images and tested on a set of 2443 radiographs.
The sensitivity, specificity, and AUC, respectively, were 91%, 91%, and 0.962 for pleural effusion, 82%, 82%, and 0.868 for pulmonary edema, 81%, 80%, and 0.875 for cardiomegaly.
The best results were achieved in classifying a study as normal with an overall sensitivity and specificity of 91% and an AUC of 0.964 [77].
Moreover, another CNN, CheXNeXt, was developed a few years ago to detect the presence of 14 different pathologies in chest radiographs, including cardiomegaly and pleural effusion. In 2018, the model was compared with nine expert radiologists and three senior radiology residents. CheXNeXt did not achieve radiologist-level performance on three pathologies (cardiomegaly, emphysema, hiatal hernia), it performed better than radiologists in detecting atelectasis and there were no statistically significant differences in AUCs for the other ten pathologies, including pleural effusion [76].
7. Pleural Effusion
Pleural effusion is a medical condition characterized by the pathological accumulation of fluid between the two pleural leaflets. Usually, it is used as a generic term to describe any abnormal accumulation of fluid in the pleural cavity, also because most effusions are diagnosed by CXR, which cannot distinguish between different types of fluids.
Recently, and with increasing interest, the world of diagnostic imaging has embraced the use of artificial intelligence, testing its applications in a wide variety of contexts [78].
A few papers have been published to validate AI tools in the diagnosis and/or quantification of pleural effusion via CXR. Zhou et al. [79] developed and validated a DL system for the detection and semi-quantitative analysis of cardiomegaly, pleural effusion, and pneumothorax. They included two datasets: one for detection and one for segmentation. The first dataset (used for detection) consisted of 2838 CXRs from 2638 patients containing findings positive for cardiomegaly, pneumothorax, and pleural effusion; the second dataset (used for segmentation) was from two publicly available datasets, containing 704 CXRs. Based on the accurate detection and segmentation, semiquantitative indexes were calculated. The detection models achieved high accuracy in detecting cardiomegaly, pneumothorax, and pleural effusion. Moreover, the authors believed that semiquantitative analysis could reduce the work of radiologists, improve the objective accuracy of the quantitative measurement, and be a reasonable option to assist clinical diagnosis.
Huang et al. [80] developed a DL system to quantify the severity of pleural effusion in the CXR of patients with chronic obstructive pulmonary disease (COPD). For this purpose, they used the MIMIC-CXR dataset, dividing the patients within it as “with” or “without” COPD. The label of pleural effusion severity was obtained from the extracted COPD radiology reports and classified into four categories: no effusion, small effusion, moderate effusion, and large effusion. The selected reports were re-tagged by a radiologist without knowing their previous tags as a verification cohort; 15,620 CXRs with clearly marked pleural effusion severity were obtained (no effusion, 5685; small effusion, 4877; moderate effusion, 3657; and large effusion, 1401). The highest accuracy rate of the optimized model was 73.07. The micro-average AUCs of the test and validation cohorts were 0.89 and 0.90, respectively, and their macro-average AUCs were 0.86 and 0.89, respectively.
Niehues et al. [81] created a DL model used specifically for bedside CXRs to detect clinically relevant findings to help emergency and intensive care physicians to focus on patient care, using the reference standards established by computed tomography (CT) and numerous radiologists. They retrospectively collected 18,361 bedside CXRs of patients treated at a level 1 medical center who had undergone a chest CT within 24 h from the CXR. A DL algorithm was developed to identify eight findings on bedside CXRs (cardiac congestion, pleural effusion, air-space opacification, pneumothorax, central venous catheter, thoracic drain, gastric tube, and tracheal tube/cannula). In case of a disagreement between the CXR and CT, human-in-the-loop annotations were used. The AUC for cardiac congestion, pleural effusion, air-space opacification, pneumothorax, central venous catheter, thoracic drain, gastric tube, and tracheal tube/cannula were 0.90, 0.95, 0.85, 0.92, 0.99, 0.99, 0.98, and 0.99, respectively, showing a similar performance to expert radiologists.
8. Limits and Future Perspective
AI applications for thoracic disorders have shown promising outcomes in terms of enhancing the existing clinical systems and prognostic prediction reasoning. According to a recent study that compared the opinions of thoracic radiologists and computer scientists, 15.6% of computer scientists thought that the radiologist’s position will be obsolete in 10–20 years [82]; despite this, the stakeholders’ opinion is that this scenario will be unlikely, although there is great emphasis put on radiologists’ education and training in the AI [83].
The use of open datasets had a significant impact on the development of AI algorithms, but the absence of standardized and well-defined criteria also led to the creation of flawed datasets. This needs to be addressed to increase the performance of AI models through high-quality training. As a result, it is critical to understand the limitations of the datasets for their effective use; the key limitations are summarized in Table 3.

Table 3.
This Table lists the main limitations related to the datasets used to develop AI algorithms.
Another key aspect would be the integration of AI education into the post-graduation programs for radiology residents. The possibility of increasing performance through personalized learning is the primary motivator for AI-augmented radiological precision education [89]. According to a 2018 poll, 71% of radiologists do not actively use AI. However, 87% of respondents intend to study and 67% are eager to assist in the development and training of such algorithms [90]. Gaube et al. [91] affirm that observers were typically not opposed to preferring AI guidance over human suggestions, showing that when the improvement of radiologists’ performance with AI assistance was lacking, it was likely driven by their confidence in their own opinion rather than their reluctance to trust in the AI-algorithm.
Imaging research is seeing an increase in the number of articles offering novel diagnostic, classification, and prediction tools based on ML that outperform the previous techniques. However, few studies have been conducted to assess the applicability of these models and the tangible advantages they offer to clinical practice in the real-world setting [92]. When such models are evaluated in other patient cohorts or institutions, they demonstrate a lack of reproducibility. Moreover, it is difficult to compare the performance of different ML models. This is due to one of the primary obstacles to effective AI application today: a lack of empirical data confirming the efficacy of AI-based interventions in prospective clinical trials. The existing empirical research is limited and primarily focuses on AI in the general workforce rather than its impact on patient outcomes [93]. Among 516 eligible studies evaluated by Kim et al., only 5% performed external validation and none of these adopted the three design features (diagnostic cohort design, inclusion of multiple institutions, and prospective data collection) [94]. Another critical issue is that most of the existing radiological DL models only incorporate imaging information, without accounting for the background clinical data. We need research on DL models capable of integrating multimodal data, such as clinical and genetic information [95]. The integration of these data has already shown potential for improving prognostic models for other pathologies. For example, Xu et al. used bidirectional deep neural network (BiDNN) DL architecture to combine RNA-Seq and DNA methylation data from a group of gastric cancer patients to identify different prognostic levels [96]. In a more recent study by Cheerla et al., a C-index of 0.78 was found in a pancreatic cancer survival prediction model that used clinical, mRNA, miRNA, and whole-slide imaging data [97]. Emergency departments are a crucial training scenario for determining the benefits and usability of AI-based solutions. In this case, AI assistance might enhance both the performance of a single radiologist and the organization’s management [98].
Last but not the least, the ethical implications of AI integration should be considered before they can see widespread integration into clinical practice. When it comes to AI applications that attempt to enhance patient outcomes, the issue of responsibility becomes considerably more important, particularly when problems and errors occur. At present, it is unclear who should bear responsibility if the system fails. One of the possible solutions includes a joint effort by governments and industries to define the criteria of transparency and accountability, which aspects of the incoming data are influencing the outputs, and utilizing that to deduce what is happening within the “black box” [78]. As described above, an emergency setting is an essential testing ground for the usability and advantages of AI-based solutions. In this scenario, AI systems might assist radiologists with long and highly repetitive activities, minimizing diagnostic mistakes in situations where workload, expectations, and the risk of error are high [98]. Effective AI integration might free up resources that could be committed to other tasks, such as patient communication, that have been overburdened as the workload has increased [99].
9. Conclusions
We have provided an overview of AI applications for CXR in the emergency setting.
We believe that AI will transform the healthcare system, notably, in the imaging sector. Radiologists should know the AI technologies that are currently available and use them when appropriate to improve their diagnostic accuracy and treatment planning in the emergency setting.
Author Contributions
Conceptualization, M.C. (Michaela Cellina); Methodology, M.C. (Maurizio Cè); Literature research, E.C. and G.O.; Data Curation, V.A.; Writing—Original Draft Preparation, M.C. (Michaela Cellina), M.C. (Maurizio Cè), G.I., V.A., E.C., C.M., G.O., N.K. and G.D.P.; Writing—Review & Editing, M.C. (Michaela Cellina), M.C. (Maurizio Cè), N.K. and G.I.; Supervision, S.P. and C.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, D.; Pehrson, L.M.; Lauridsen, C.A.; Tøttrup, L.; Fraccaro, M.; Elliott, D.; Zając, H.D.; Darkner, S.; Carlsen, J.F.; Nielsen, M.B. The Added Effect of Artificial Intelligence on Physicians’ Performance in Detecting Thoracic Pathologies on CT and Chest X-Ray: A Systematic Review. Diagnostics 2021, 11, 2206. [Google Scholar] [CrossRef]
- Bizzo, B.C.; Almeida, R.R.; Alkasab, T.K. Artificial Intelligence Enabling Radiology Reporting. Radiol. Clin. N. Am. 2021, 59, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Pirovano, M.; Ciocca, M.; Gibelli, D.; Floridi, C.; Oliva, G. Radiomic Analysis of the Optic Nerve at the First Episode of Acute Optic Neuritis: An Indicator of Optic Nerve Pathology and a Predictor of Visual Recovery? Radiol. Med. 2021, 126, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.M.; Slonimsky, E.; Long, A.D.; Sze, R.W.; Iyer, R.S. Machine Learning Concepts, Concerns and Opportunities for a Pediatric Radiologist. Pediatr. Radiol. 2019, 49, 509–516. [Google Scholar] [CrossRef]
- Jung, A. Machine Learning: The Basics; Springer: Singapore, 2022. [Google Scholar]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial Intelligence in Radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Teng, X.; Zhang, J.; Ma, Z.; Zhang, Y.; Lam, S.; Li, W.; Xiao, H.; Li, T.; Li, B.; Zhou, T.; et al. Improving Radiomic Model Reliability Using Robust Features from Perturbations for Head-and-Neck Carcinoma. Front. Oncol. 2022, 12, 974467. [Google Scholar] [CrossRef]
- Sandino, C.M.; Cole, E.K.; Alkan, C.; Chaudhari, A.S.; Loening, A.M.; Hyun, D.; Dahl, J.; Imran, A.-A.-Z.; Wang, A.S.; Vasanawala, S.S. Upstream Machine Learning in Radiology. Radiol. Clin. N. Am. 2021, 59, 967–985. [Google Scholar] [CrossRef]
- Cellina, M.; Cè, M.; Khenkina, N.; Sinichich, P.; Cervelli, M.; Poggi, V.; Boemi, S.; Ierardi, A.M.; Carrafiello, G. Artificial Intellgence in the Era of Precision Oncological Imaging. Technol. Cancer Res. Treat. 2022, 21, 153303382211417. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.; Peng, Y.; Lu, Z.; Mark, R.G.; Berkowitz, S.J.; Horng, S. MIMIC-CXR-JPG, a Large Publicly Available Database of Labeled Chest Radiographs. arXiv 2019, arXiv:1901.07042. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. Proc. AAAI Conf. Artif. Intell. 2019, 33, 590–597. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Oakden-Rayner, L. Exploring Large-Scale Public Medical Image Datasets. Acad. Radiol. 2020, 27, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.S.; Pinsky, P.F.; Kramer, B.S.; Prorok, P.C.; Purdue, M.P.; Berg, C.D.; Gohagan, J.K. The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial and Its Associated Research Resource. JNCI J. Natl. Cancer Inst. 2013, 105, 1684–1693. [Google Scholar] [CrossRef] [PubMed]
- Bustos, A.; Pertusa, A.; Salinas, J.-M.; de la Iglesia-Vayá, M. PadChest: A Large Chest x-Ray Image Dataset with Multi-Label Annotated Reports. Med. Image Anal. 2020, 66, 101797. [Google Scholar] [CrossRef] [PubMed]
- Reis, E.P.; de Paiva, J.P.Q.; da Silva, M.C.B.; Ribeiro, G.A.S.; Paiva, V.F.; Bulgarelli, L.; Lee, H.M.H.; Santos, P.V.; Brito, V.M.; Amaral, L.T.W.; et al. BRAX, Brazilian Labeled Chest x-Ray Dataset. Sci. Data 2022, 9, 487. [Google Scholar] [CrossRef]
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.; Thoma, G.R.; McDonald, C.J. Preparing a Collection of Radiology Examinations for Distribution and Retrieval. J. Am. Med. Inform. Assoc. 2016, 23, 304–310. [Google Scholar] [CrossRef]
- Kermany, D.; Zhang, K.; Goldbaum, M. Large Dataset of Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images. Physiccs 2018, 172, 1122–1131. [Google Scholar] [CrossRef]
- Shih, G.; Wu, C.C.; Halabi, S.S.; Kohli, M.D.; Prevedello, L.M.; Cook, T.S.; Sharma, A.; Amorosa, J.K.; Arteaga, V.; Galperin-Aizenberg, M.; et al. Augmenting the National Institutes of Health Chest Radiograph Dataset with Expert Annotations of Possible Pneumonia. Radiol. Artif. Intell. 2019, 1, e180041. [Google Scholar] [CrossRef]
- Baltruschat, I.; Steinmeister, L.; Nickisch, H.; Saalbach, A.; Grass, M.; Adam, G.; Knopp, T.; Ittrich, H. Smart Chest X-Ray Worklist Prioritization Using Artificial Intelligence: A Clinical Workflow Simulation. Eur. Radiol. 2021, 31, 3837–3845. [Google Scholar] [CrossRef]
- Adams, S.J.; Henderson, R.D.E.; Yi, X.; Babyn, P. Artificial Intelligence Solutions for Analysis of X-Ray Images. Can. Assoc. Radiol. J. 2021, 72, 60–72. [Google Scholar] [CrossRef]
- Annarumma, M.; Withey, S.J.; Bakewell, R.J.; Pesce, E.; Goh, V.; Montana, G. Automated Triaging of Adult Chest Radiographs with Deep Artificial Neural Networks. Radiology 2019, 291, 196–202. [Google Scholar] [CrossRef]
- Kim, Y.; Park, J.Y.; Hwang, E.J.; Lee, S.M.; Park, C.M. Applications of Artificial Intelligence in the Thorax: A Narrative Review Focusing on Thoracic Radiology. J. Thorac. Dis. 2021, 13, 6943–6962. [Google Scholar] [CrossRef] [PubMed]
- Tricarico, D.; Calandri, M.; Barba, M.; Piatti, C.; Geninatti, C.; Basile, D.; Gatti, M.; Melis, M.; Veltri, A. Convolutional Neural Network-Based Automatic Analysis of Chest Radiographs for the Detection of COVID-19 Pneumonia: A Prioritizing Tool in the Emergency Department, Phase I Study and Preliminary “Real Life” Results. Diagnostics 2022, 12, 570. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 3462–3471. [Google Scholar]
- Blumenfeld, A.; Greenspan, H.; Konen, E. Pneumothorax Detection in Chest Radiographs Using Convolutional Neural Networks. In Proceedings of the Medical Imaging 2018: Computer-Aided Diagnosis, Houston, TX, USA, 10–15 February 2018; Mori, K., Petrick, N., Eds.; SPIE: Bellingham, WA, USA, 2018; p. 3. [Google Scholar]
- Taylor, A.G.; Mielke, C.; Mongan, J. Automated Detection of Moderate and Large Pneumothorax on Frontal Chest X-Rays Using Deep Convolutional Neural Networks: A Retrospective Study. PLoS Med. 2018, 15, e1002697. [Google Scholar] [CrossRef] [PubMed]
- Moses, D.A. Deep Learning Applied to Automatic Disease Detection Using Chest X-rays. J. Med. Imaging Radiat. Oncol. 2021, 65, 498–517. [Google Scholar] [CrossRef]
- Wang, X.; Yang, S.; Lan, J.; Fang, Y.; He, J.; Wang, M.; Zhang, J.; Han, X. Automatic Segmentation of Pneumothorax in Chest Radiographs Based on a Two-Stage Deep Learning Method. IEEE Trans. Cogn. Dev. Syst. 2022, 14, 205–218. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.-H.; Kim, S.-W.; Hong, J.-M.; Kim, S.-J.; Song, M.; Choi, J.-M.; Lee, S.-Y.; Yoon, H.; Yoo, J.-Y. Quantitative Measurement of Pneumothorax Using Artificial Intelligence Management Model and Clinical Application. Diagnostics 2022, 12, 1823. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Q.; Luo, G.; Liu, Z.; Huang, J.; Zhou, Y.; Zhou, Y.; Xu, W.; Cheng, J.-Z. Automated Segmentation and Diagnosis of Pneumothorax on Chest X-Rays with Fully Convolutional Multi-Scale ScSE-DenseNet: A Retrospective Study. BMC Med. Inform. Decis. Mak. 2020, 20, 317. [Google Scholar] [CrossRef]
- Gooßen, A.; Deshpande, H.; Harder, T.; Schwab, E.; Baltruschat, I.; Mabotuwana, T.; Cross, N.; Saalbach, A. Deep Learning for Pneumothorax Detection and Localization in Chest Radiographs. arXiv 2019, arXiv:1907.07324. [Google Scholar]
- Wang, H.; Gu, H.; Qin, P.; Wang, J. CheXLocNet: Automatic Localization of Pneumothorax in Chest Radiographs Using Deep Convolutional Neural Networks. PLoS ONE 2020, 15, e0242013. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wang, J.; Yang, W.; Wang, J.; Qian, D. Deep Multi-instance Transfer Learning for Pneumothorax Classification in Chest X-ray Images. Med. Phys. 2022, 49, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Hillis, J.M.; Bizzo, B.C.; Mercaldo, S.; Chin, J.K.; Newbury-Chaet, I.; Digumarthy, S.R.; Gilman, M.D.; Muse, V.V.; Bottrell, G.; Seah, J.C.Y.; et al. Evaluation of an Artificial Intelligence Model for Detection of Pneumothorax and Tension Pneumothorax in Chest Radiographs. JAMA Netw. Open 2022, 5, e2247172l. [Google Scholar] [CrossRef] [PubMed]
- Yi, P.H.; Kim, T.K.; Yu, A.C.; Bennett, B.; Eng, J.; Lin, C.T. Can AI Outperform a Junior Resident? Comparison of Deep Neural Network to First-Year Radiology Residents for Identification of Pneumothorax. Emerg. Radiol. 2020, 27, 367–375. [Google Scholar] [CrossRef]
- Ahn, J.S.; Ebrahimian, S.; McDermott, S.; Lee, S.; Naccarato, L.; Di Capua, J.F.; Wu, M.Y.; Zhang, E.W.; Muse, V.; Miller, B.; et al. Association of Artificial Intelligence–Aided Chest Radiograph Interpretation With Reader Performance and Efficiency. JAMA Netw. Open 2022, 5, e2229289. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef]
- Rajaraman, S.; Candemir, S.; Kim, I.; Thoma, G.; Antani, S. Visualization and Interpretation of Convolutional Neural Network Predictions in Detecting Pneumonia in Pediatric Chest Radiographs. Appl. Sci. 2018, 8, 1715. [Google Scholar] [CrossRef]
- Gu, X.; Pan, L.; Liang, H.; Yang, R. Classification of Bacterial and Viral Childhood Pneumonia Using Deep Learning in Chest Radiography. In Proceedings of the 3rd International Conference on Multimedia and Image Processing—ICMIP 2018, Guiyang, China, 16–18 March 2018; ACM Press: New York, NY, USA, 2018; pp. 88–93. [Google Scholar]
- Stephen, O.; Sain, M.; Maduh, U.J.; Jeong, D.-U. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J. Healthc. Eng. 2019, 2019, 1–7. [Google Scholar] [CrossRef]
- Sirazitdinov, I.; Kholiavchenko, M.; Mustafaev, T.; Yixuan, Y.; Kuleev, R.; Ibragimov, B. Deep Neural Network Ensemble for Pneumonia Localization from a Large-Scale Chest x-Ray Database. Comput. Electr. Eng. 2019, 78, 388–399. [Google Scholar] [CrossRef]
- Liang, G.; Zheng, L. A Transfer Learning Method with Deep Residual Network for Pediatric Pneumonia Diagnosis. Comput. Methods Programs Biomed. 2020, 187, 104964. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-Ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z.; Özyurt, F. A Deep Feature Learning Model for Pneumonia Detection Applying a Combination of MRMR Feature Selection and Machine Learning Models. IRBM 2020, 41, 212–222. [Google Scholar] [CrossRef]
- Sarkar, R.; Hazra, A.; Sadhu, K.; Ghosh, P. A Novel Method for Pneumonia Diagnosis from Chest X-Ray Images Using Deep Residual Learning with Separable Convolutional Networks. In Computer Vision and Machine Intelligence in Medical Image Analysis; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–12. [Google Scholar]
- Li, Y.; Zhang, Z.; Dai, C.; Dong, Q.; Badrigilan, S. Accuracy of Deep Learning for Automated Detection of Pneumonia Using Chest X-Ray Images: A Systematic Review and Meta-Analysis. Comput. Biol. Med. 2020, 123, 103898. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Lee, S.P.; Kim, D.; Jang, J.; Lee, M.; Kang, S.U.; Kim, H.; Oh, K.; On, J.; Kim, Y.J.; et al. Diagnostic Performance of Artificial Intelligence Model for Pneumonia from Chest Radiography. PLoS ONE 2021, 16, e0249399. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Orsi, M.; Valenti Pittino, C.; Toluian, T.; Oliva, G. Chest Computed Tomography Findings of COVID-19 Pneumonia: Pictorial Essay with Literature Review. Jpn. J. Radiol. 2020, 38, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Borghesi, A.; Golemi, S.; Scrimieri, A.; Nicosia, C.M.C.; Zigliani, A.; Farina, D.; Maroldi, R. Chest X-Ray versus Chest Computed Tomography for Outcome Prediction in Hospitalized Patients with COVID-19. Radiol. Med. 2022, 127, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Panzeri, M.; Oliva, G. Chest Radiography Features Help to Predict a Favorable Outcome in Patients with Coronavirus Disease 2019. Radiology 2020, 297, E238. [Google Scholar] [CrossRef]
- Cellina, M.; Orsi, M.; Toluian, T.; Valenti Pittino, C.; Oliva, G. False Negative Chest X-Rays in Patients Affected by COVID-19 Pneumonia and Corresponding Chest CT Findings. Radiography 2020, 26, e189–e194. [Google Scholar] [CrossRef]
- Kassania, S.H.; Kassanib, P.H.; Wesolowskic, M.J.; Schneidera, K.A.; Detersa, R. Automatic Detection of Coronavirus Disease (COVID-19) in X-Ray and CT Images: A Machine Learning Based Approach. Biocybern. Biomed. Eng. 2021, 41, 867–879. [Google Scholar] [CrossRef]
- Baltazar, L.R.; Manzanillo, M.G.; Gaudillo, J.; Viray, E.D.; Domingo, M.; Tiangco, B.; Albia, J. Artificial Intelligence on COVID-19 Pneumonia Detection Using Chest Xray Images. PLoS ONE 2021, 16, e0257884. [Google Scholar] [CrossRef]
- Nillmani; Sharma, N.; Saba, L.; Khanna, N.N.; Kalra, M.K.; Fouda, M.M.; Suri, J.S. Segmentation-Based Classification Deep Learning Model Embedded with Explainable AI for COVID-19 Detection in Chest X-Ray Scans. Diagnostics 2022, 12, 2132. [Google Scholar] [CrossRef]
- Dey, S.; Bhattacharya, R.; Malakar, S.; Mirjalili, S.; Sarkar, R. Choquet Fuzzy Integral-Based Classifier Ensemble Technique for COVID-19 Detection. Comput. Biol. Med. 2021, 135, 104585. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, H.; Kheyroddin, G.; Dorrigiv, M.; Esmaeili, M.; Nafchi, A.R.; Ghorbani, M.H.; Zarkesh-Ha, P. Classification of COVID-19 in Chest X-Ray Images Using Fusion of Deep Features and LightGBM. In Proceedings of the 2022 IEEE World AI IoT Congress (AIIoT), Virtual, 6–9 June 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 201–206. [Google Scholar]
- Ezzoddin, M.; Nasiri, H.; Dorrigiv, M. Diagnosis of COVID-19 Cases from Chest X-Ray Images Using Deep Neural Network and LightGBM. arXiv 2022, arXiv:2203.14275. [Google Scholar]
- Nasiri, H.; Alavi, S.A. A Novel Framework Based on Deep Learning and ANOVA Feature Selection Method for Diagnosis of COVID-19 Cases from Chest X-Ray Images. Comput. Intell. Neurosci. 2022, 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, H.; Hasani, S. Automated Detection of COVID-19 Cases from Chest X-Ray Images Using Deep Neural Network and XGBoost. Radiography 2022, 28, 732–738. [Google Scholar] [CrossRef]
- Soda, P.; D’Amico, N.C.; Tessadori, J.; Valbusa, G.; Guarrasi, V.; Bortolotto, C.; Akbar, M.U.; Sicilia, R.; Cordelli, E.; Fazzini, D.; et al. AIforCOVID: Predicting the Clinical Outcomes in Patients with COVID-19 Applying AI to Chest-X-Rays. An Italian Multicentre Study. Med. Image Anal. 2021, 74, 102216. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.P.; Dao, L.; Roth, K.; Morrison, P.; Bengio, Y.; Abbasi, A.F.; Shen, B.; Mahsa, H.K.; Ghassemi, M.; Li, H.; et al. Predicting COVID-19 Pneumonia Severity on Chest X-Ray With Deep Learning. Cureus 2020, 12, e9448. [Google Scholar] [CrossRef]
- Jiao, Z.; Choi, J.W.; Halsey, K.; Tran, T.M.L.; Hsieh, B.; Wang, D.; Eweje, F.; Wang, R.; Chang, K.; Wu, J.; et al. Prognostication of Patients with COVID-19 Using Artificial Intelligence Based on Chest x-Rays and Clinical Data: A Retrospective Study. Lancet Digit. Heal. 2021, 3, e286–e294. [Google Scholar] [CrossRef]
- Khan, S.H.; Sohail, A.; Khan, A.; Lee, Y.-S. COVID-19 Detection in Chest X-Ray Images Using a New Channel Boosted CNN. Diagnostics 2022, 12, 267. [Google Scholar] [CrossRef]
- Mushtaq, J.; Pennella, R.; Lavalle, S.; Colarieti, A.; Steidler, S.; Martinenghi, C.M.A.; Palumbo, D.; Esposito, A.; Rovere-Querini, P.; Tresoldi, M.; et al. Initial Chest Radiographs and Artificial Intelligence (AI) Predict Clinical Outcomes in COVID-19 Patients: Analysis of 697 Italian Patients. Eur. Radiol. 2021, 31, 1770–1779. [Google Scholar] [CrossRef]
- Warren, M.A.; Zhao, Z.; Koyama, T.; Bastarache, J.A.; Shaver, C.M.; Semler, M.W.; Rice, T.W.; Matthay, M.A.; Calfee, C.S.; Ware, L.B. Severity Scoring of Lung Oedema on the Chest Radiograph Is Associated with Clinical Outcomes in ARDS. Thorax 2018, 73, 840–846. [Google Scholar] [CrossRef]
- Hasani, S.; Nasiri, H. COV-ADSX: An Automated Detection System Using X-Ray Images, Deep Learning, and XGBoost for COVID-19. Softw. Impacts 2022, 11, 100210. [Google Scholar] [CrossRef]
- Matsumoto, T.; Kodera, S.; Shinohara, H.; Ieki, H.; Yamaguchi, T.; Higashikuni, Y.; Kiyosue, A.; Ito, K.; Ando, J.; Takimoto, E.; et al. Diagnosing Heart Failure from Chest X-Ray Images Using Deep Learning. Int. Heart J. 2020, 61, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.; Curtis, J. Interpreting the Chest Radiograph. BMJ 2012, 344, e988. [Google Scholar] [CrossRef]
- Dimopoulos, K.; Giannakoulas, G.; Bendayan, I.; Liodakis, E.; Petraco, R.; Diller, G.-P.; Piepoli, M.F.; Swan, L.; Mullen, M.; Best, N.; et al. Cardiothoracic Ratio from Postero-Anterior Chest Radiographs: A Simple, Reproducible and Independent Marker of Disease Severity and Outcome in Adults with Congenital Heart Disease. Int. J. Cardiol. 2013, 166, 453–457. [Google Scholar] [CrossRef]
- Jafar, A.; Hameed, M.T.; Akram, N.; Waqas, U.; Kim, H.S.; Naqvi, R.A. CardioNet: Automatic Semantic Segmentation to Calculate the Cardiothoracic Ratio for Cardiomegaly and Other Chest Diseases. J. Pers. Med. 2022, 12, 988. [Google Scholar] [CrossRef]
- Saiviroonporn, P.; Wonglaksanapimon, S.; Chaisangmongkon, W.; Chamveha, I.; Yodprom, P.; Butnian, K.; Siriapisith, T.; Tongdee, T. A Clinical Evaluation Study of Cardiothoracic Ratio Measurement Using Artificial Intelligence. BMC Med. Imaging 2022, 22, 46. [Google Scholar] [CrossRef] [PubMed]
- Dobbe, L.; Rahman, R.; Elmassry, M.; Paz, P.; Nugent, K. Cardiogenic Pulmonary Edema. Am. J. Med. Sci. 2019, 358, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-R.; Chiu, H.-W.; Huang, M.-Y.; Huang, S.-T.; Tsai, M.-F.; Chang, C.-Y.; Chang, K.-S. Using Artificial Intelligence to Establish Chest X-Ray Image Recognition Model to Assist Crucial Diagnosis in Elder Patients With Dyspnea. Front. Med. 2022, 9, 893208. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep Learning for Chest Radiograph Diagnosis: A Retrospective Comparison of the CheXNeXt Algorithm to Practicing Radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef] [PubMed]
- Cicero, M.; Bilbily, A.; Colak, E.; Dowdell, T.; Gray, B.; Perampaladas, K.; Barfett, J. Training and Validating a Deep Convolutional Neural Network for Computer-Aided Detection and Classification of Abnormalities on Frontal Chest Radiographs. Investig. Radiol. 2017, 52, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Khenkina, N.; Toto-Brocchi, M.; Martinenghi, C.; Papa, S.; Carrafiello, G. Artificial Intelligence in Lung Cancer Imaging: Unfolding the Future. Diagnostics 2022, 12, 2644. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yin, X.; Zhang, T.; Feng, Y.; Zhao, Y.; Jin, M.; Peng, M.; Xing, C.; Li, F.; Wang, Z.; et al. Detection and Semiquantitative Analysis of Cardiomegaly, Pneumothorax, and Pleural Effusion on Chest Radiographs. Radiol. Artif. Intell. 2021, 3, e200172. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Yang, R.; Shen, L.; Feng, A.; Li, L.; He, N.; Li, S.; Huang, L.; Lyu, J. Deep Transfer Learning to Quantify Pleural Effusion Severity in Chest X-Rays. BMC Med. Imaging 2022, 22, 100. [Google Scholar] [CrossRef]
- Niehues, S.M.; Adams, L.C.; Gaudin, R.A.; Erxleben, C.; Keller, S.; Makowski, M.R.; Vahldiek, J.L.; Bressem, K.K. Deep-Learning-Based Diagnosis of Bedside Chest X-Ray in Intensive Care and Emergency Medicine. Investig. Radiol. 2021, 56, 525–534. [Google Scholar] [CrossRef]
- Eltorai, A.E.M.; Bratt, A.K.; Guo, H.H. Thoracic Radiologists’ Versus Computer Scientists’ Perspectives on the Future of Artificial Intelligence in Radiology. J. Thorac. Imaging 2020, 35, 255–259. [Google Scholar] [CrossRef]
- Yang, L.; Ene, I.C.; Arabi Belaghi, R.; Koff, D.; Stein, N.; Santaguida, P. Stakeholders’ Perspectives on the Future of Artificial Intelligence in Radiology: A Scoping Review. Eur. Radiol. 2022, 32, 1477–1495. [Google Scholar] [CrossRef]
- Finlayson, S.G.; Subbaswamy, A.; Singh, K.; Bowers, J.; Kupke, A.; Zittrain, J.; Kohane, I.S.; Saria, S. The Clinician and Dataset Shift in Artificial Intelligence. N. Engl. J. Med. 2021, 385, 283–286. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roberts, K.; Lu, L.; Demner-Fushman, D.; Yao, J.; Summers, R.M. Learning to Read Chest X-Rays: Recurrent Neural Cascade Model for Automated Image Annotation. arXiv 2016, arXiv:1603.08486. [Google Scholar]
- Shin, H.-C.; Lu, L.; Summers, R.M. Natural Language Processing for Large-Scale Medical Image Analysis Using Deep Learning. In Deep Learning for Medical Image Analysis; Elsevier: Amsterdam, The Netherlands, 2017; pp. 405–421. [Google Scholar]
- Ganeshan, D.; Duong, P.-A.T.; Probyn, L.; Lenchik, L.; McArthur, T.A.; Retrouvey, M.; Ghobadi, E.H.; Desouches, S.L.; Pastel, D.; Francis, I.R. Structured Reporting in Radiology. Acad. Radiol. 2018, 25, 66–73. [Google Scholar] [CrossRef]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Caloro, E.; Bianchi, L.; Pellegrino, G.; D’Amico, N.; Papa, S.; Carrafiello, G. Artificial Intelligence in Emergency Radiology: Where Are We Going? Diagnostics 2022, 12, 3223. [Google Scholar] [CrossRef] [PubMed]
- Duong, M.T.; Rauschecker, A.M.; Rudie, J.D.; Chen, P.-H.; Cook, T.S.; Bryan, R.N.; Mohan, S. Artificial Intelligence for Precision Education in Radiology. Br. J. Radiol. 2019, 92, 20190389. [Google Scholar] [CrossRef] [PubMed]
- Geis, J.R.; Brady, A.P.; Wu, C.C.; Spencer, J.; Ranschaert, E.; Jaremko, J.L.; Langer, S.G.; Kitts, A.B.; Birch, J.; Shields, W.F.; et al. Ethics of Artificial Intelligence in Radiology: Summary of the Joint European and North American Multisociety Statement. J. Am. Coll. Radiol. 2019, 16, 1516–1521. [Google Scholar] [CrossRef] [PubMed]
- Schutte, K.; Brulport, F.; Harguem-Zayani, S.; Schiratti, J.-B.; Ghermi, R.; Jehanno, P.; Jaeger, A.; Alamri, T.; Naccache, R.; Haddag-Miliani, L.; et al. An Artificial Intelligence Model Predicts the Survival of Solid Tumour Patients from Imaging and Clinical Data. Eur. J. Cancer 2022, 174, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Eche, T.; Schwartz, L.H.; Mokrane, F.-Z.; Dercle, L. Toward Generalizability in the Deployment of Artificial Intelligence in Radiology: Role of Computation Stress Testing to Overcome Underspecification. Radiol. Artif. Intell. 2021, 3, 6. [Google Scholar] [CrossRef]
- Aung, Y.Y.M.; Wong, D.C.S.; Ting, D.S.W. The Promise of Artificial Intelligence: A Review of the Opportunities and Challenges of Artificial Intelligence in Healthcare. Br. Med. Bull. 2021, 139, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Jang, H.Y.; Kim, K.W.; Shin, Y.; Park, S.H. Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers. Korean J. Radiol. 2019, 20, 405. [Google Scholar] [CrossRef]
- Huang, S.-C.; Pareek, A.; Seyyedi, S.; Banerjee, I.; Lungren, M.P. Fusion of Medical Imaging and Electronic Health Records Using Deep Learning: A Systematic Review and Implementation Guidelines. NPJ Digit. Med. 2020, 3, 136. [Google Scholar] [CrossRef]
- Xu, J.; Yao, Y.; Xu, B.; Li, Y.; Su, Z. Unsupervised Learning of Cross-Modal Mappings in Multi-Omics Data for Survival Stratification of Gastric Cancer. Future Oncol. 2022, 18, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Cheerla, A.; Gevaert, O. Deep Learning with Multimodal Representation for Pancancer Prognosis Prediction. Bioinformatics 2019, 35, i446–i454. [Google Scholar] [CrossRef]
- Al-Dasuqi, K.; Johnson, M.H.; Cavallo, J.J. Use of Artificial Intelligence in Emergency Radiology: An Overview of Current Applications, Challenges, and Opportunities. Clin. Imaging 2022, 89, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Jalal, S.; Parker, W.; Ferguson, D.; Nicolaou, S. Exploring the Role of Artificial Intelligence in an Emergency and Trauma Radiology Department. Can. Assoc. Radiol. J. 2021, 72, 167–174. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).