DermAI 1.0: A Robust, Generalized, and Novel Attention-Enabled Ensemble-Based Transfer Learning Paradigm for Multiclass Classification of Skin Lesion Images
Abstract
:1. Introduction
2. Literature Review
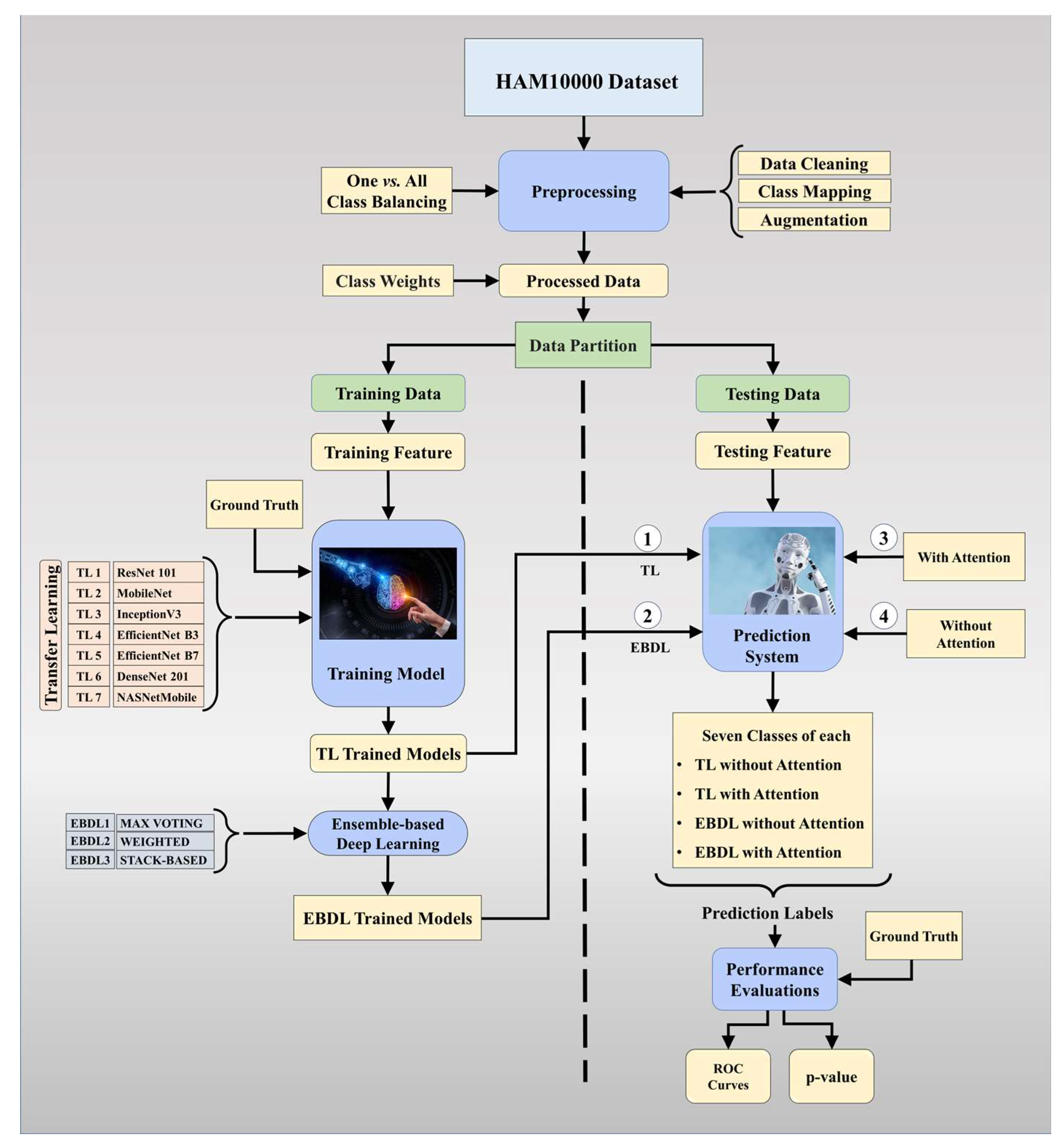
3. Methodology
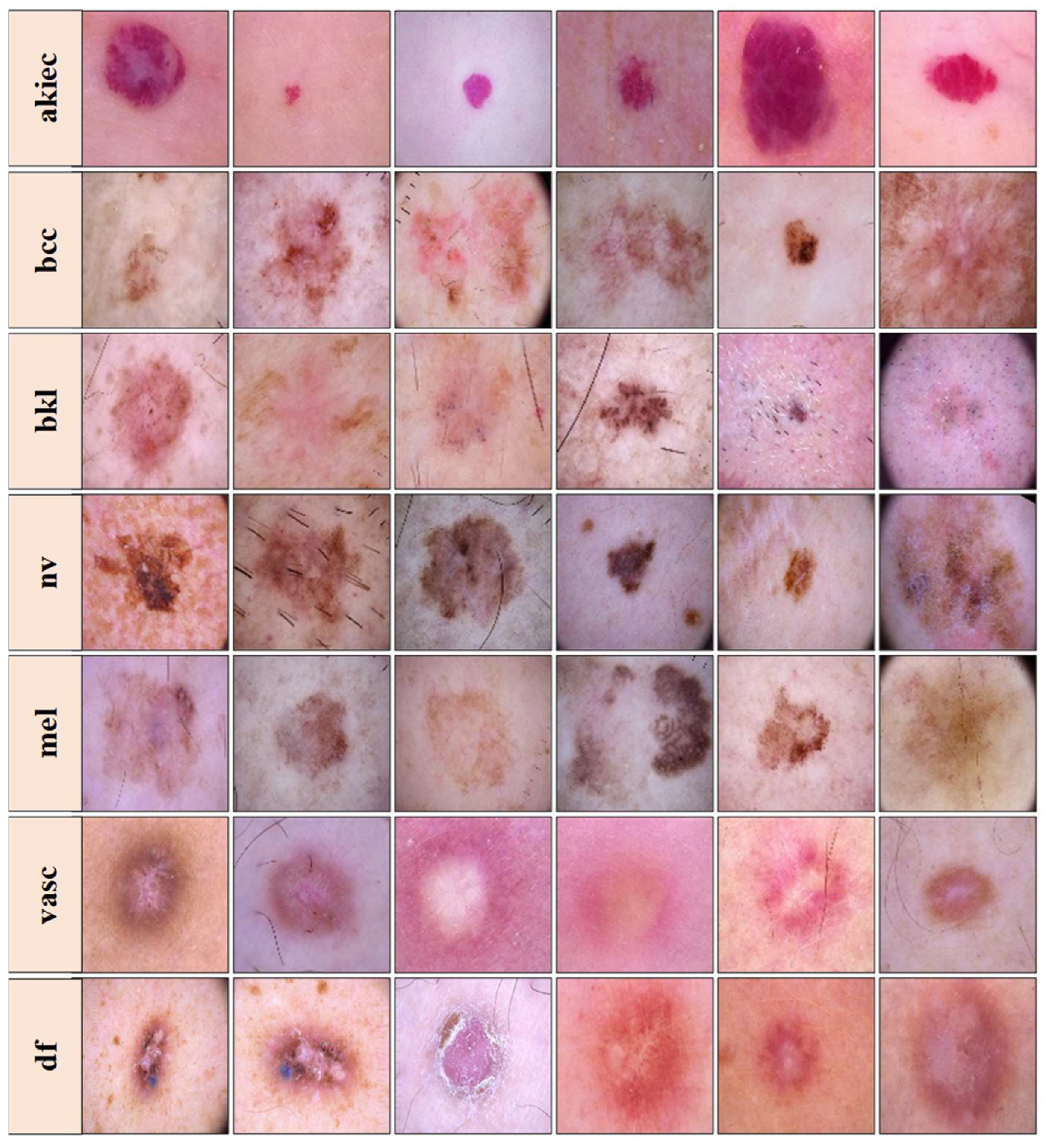
3.1. Dataset and Data Demography
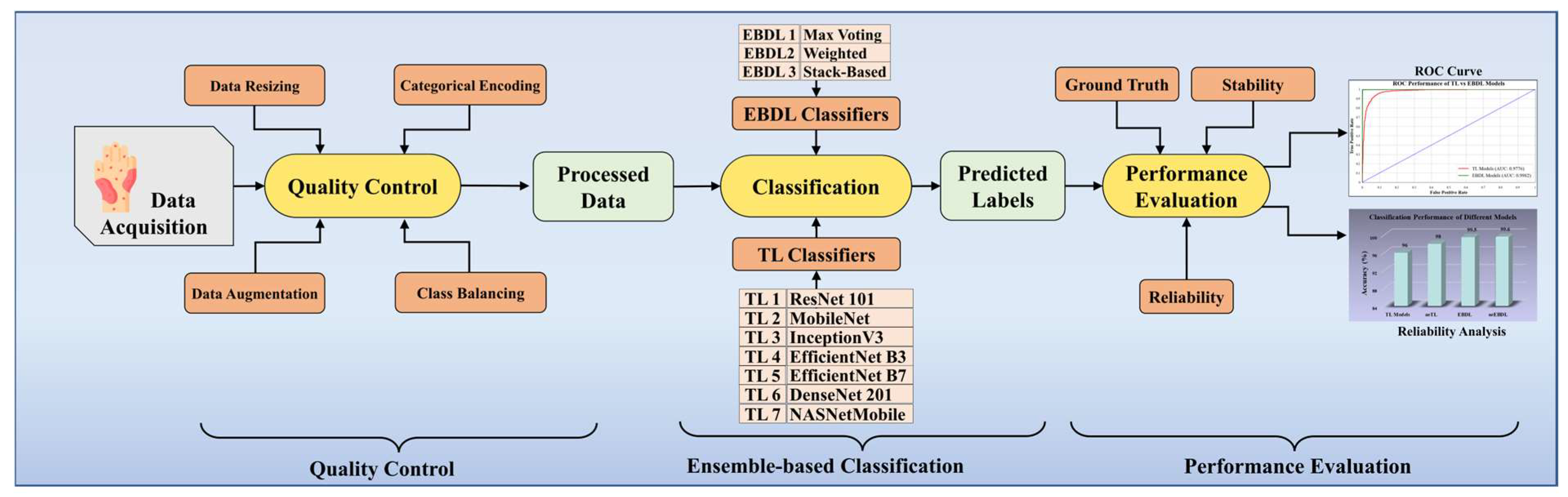
3.2. Quality Control
3.3. Global Architecture of the Proposed System
3.4. Architecture of the Classifiers
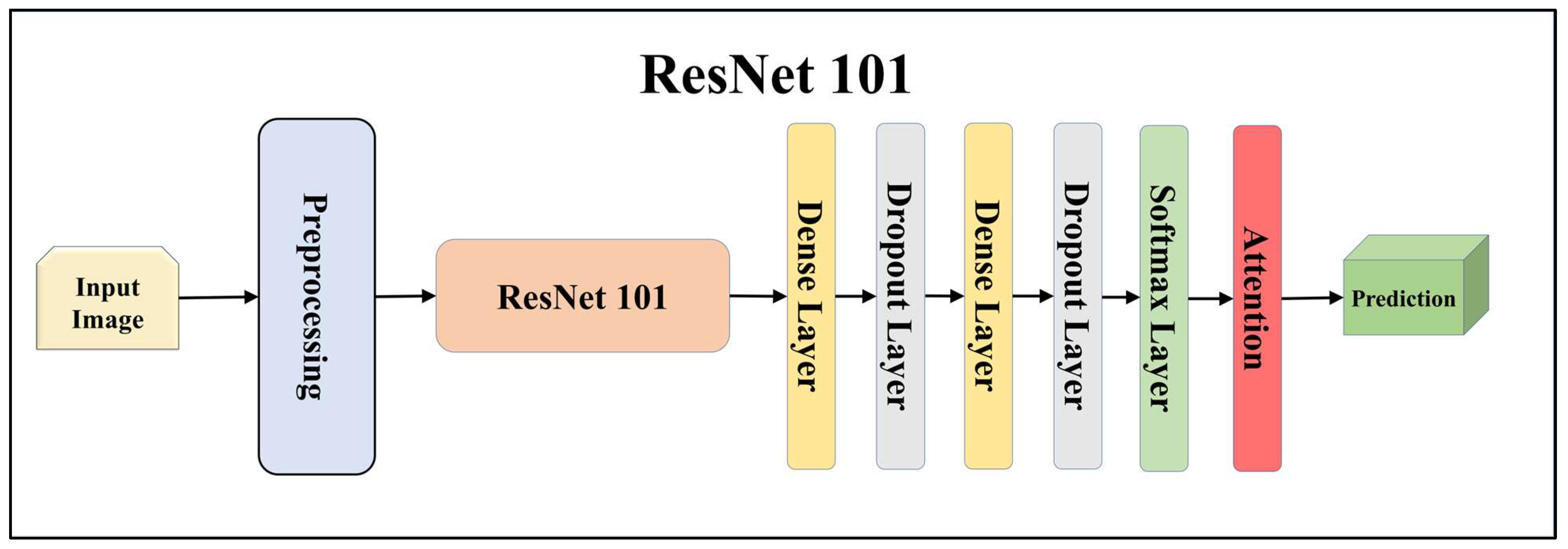
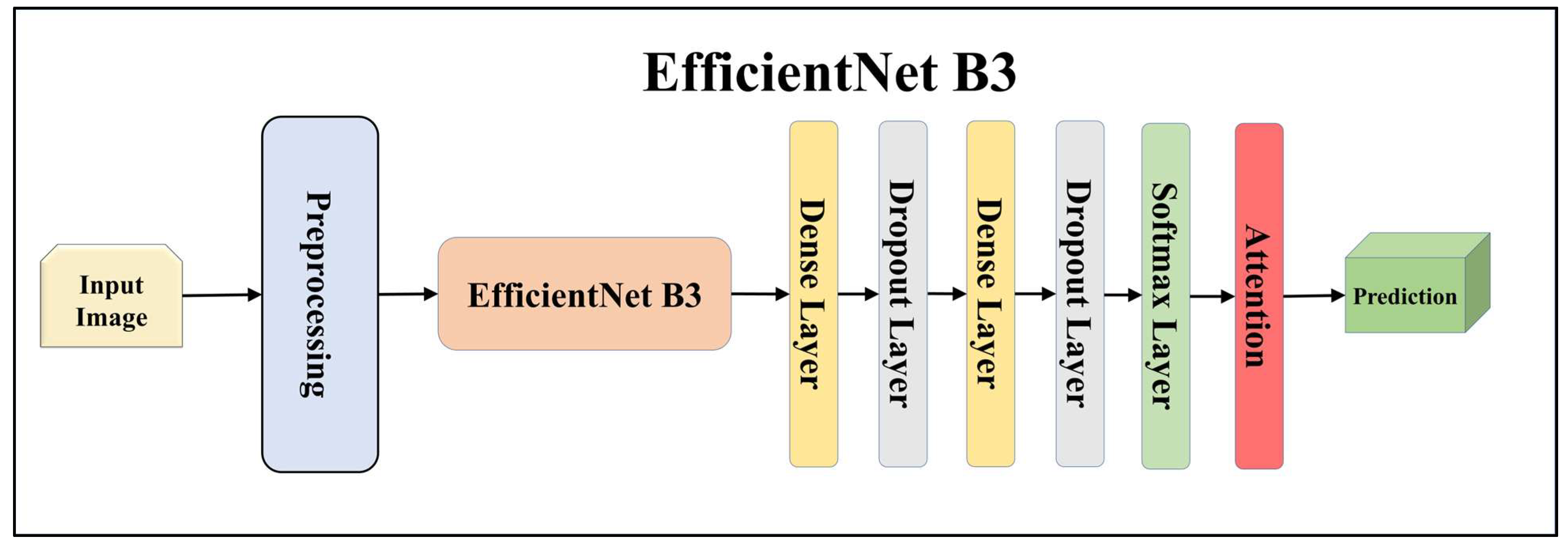
3.4.1. Solo Transfer Learning Models
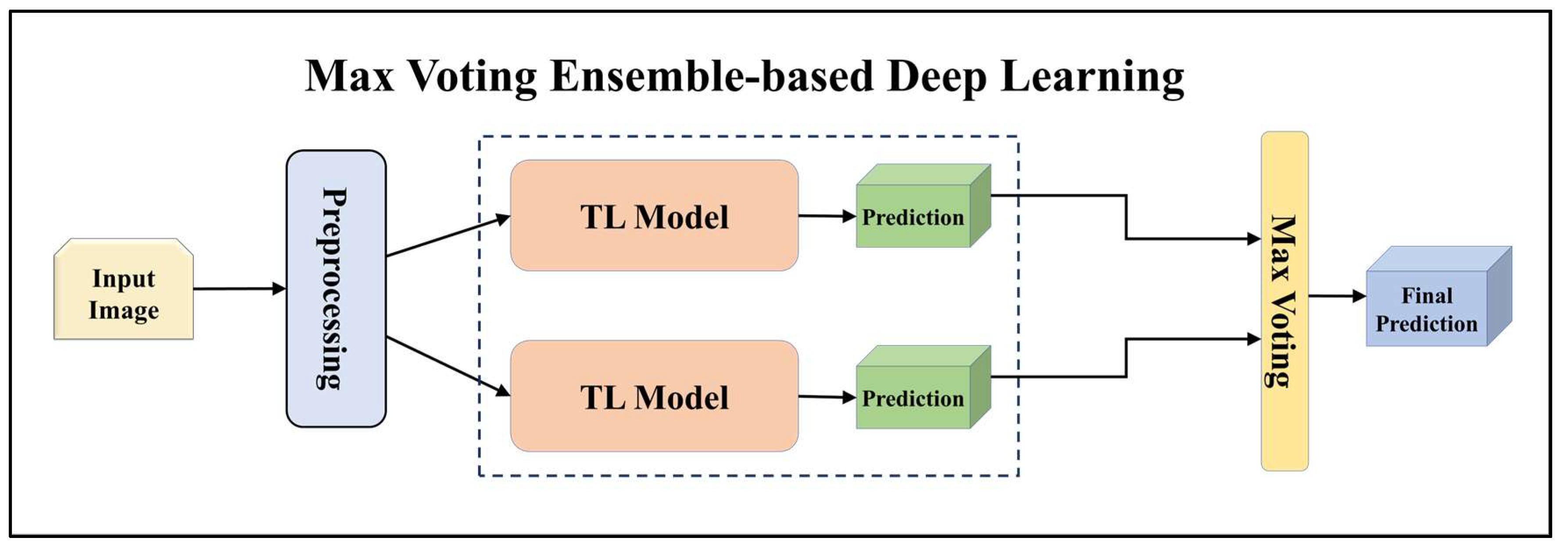
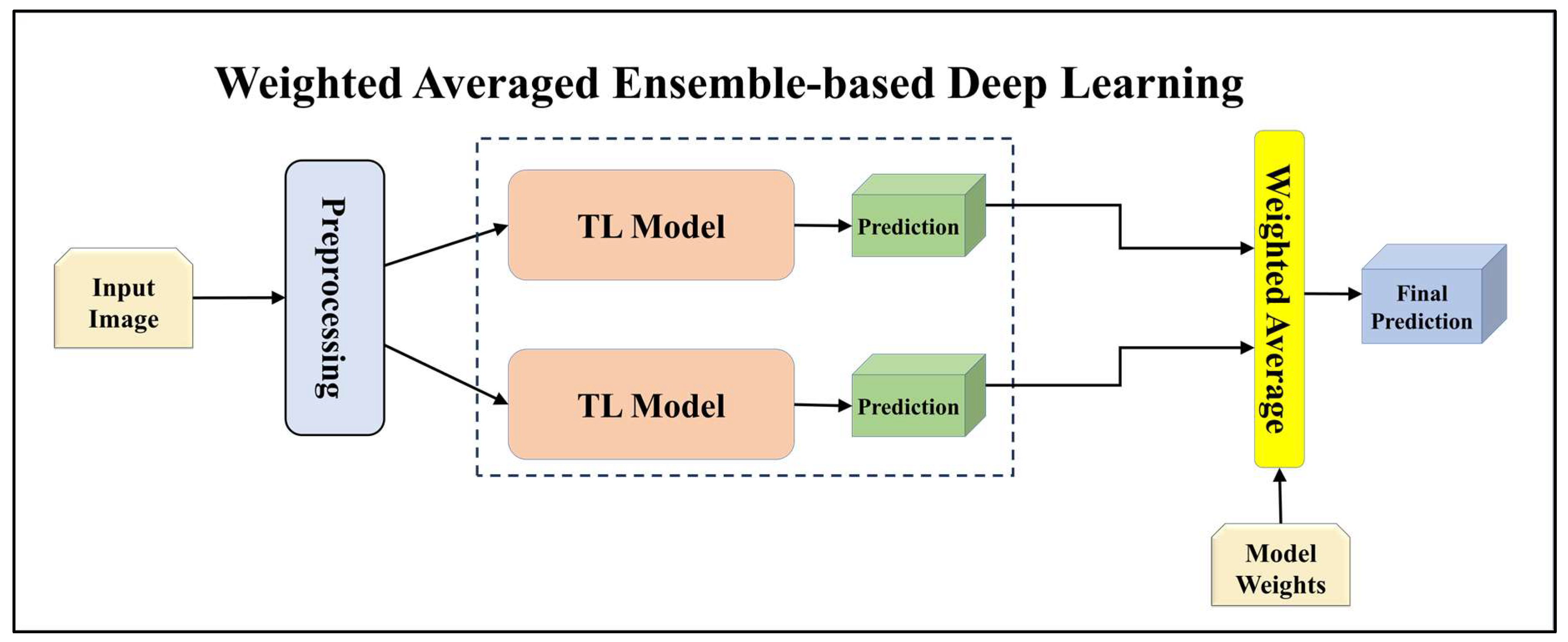
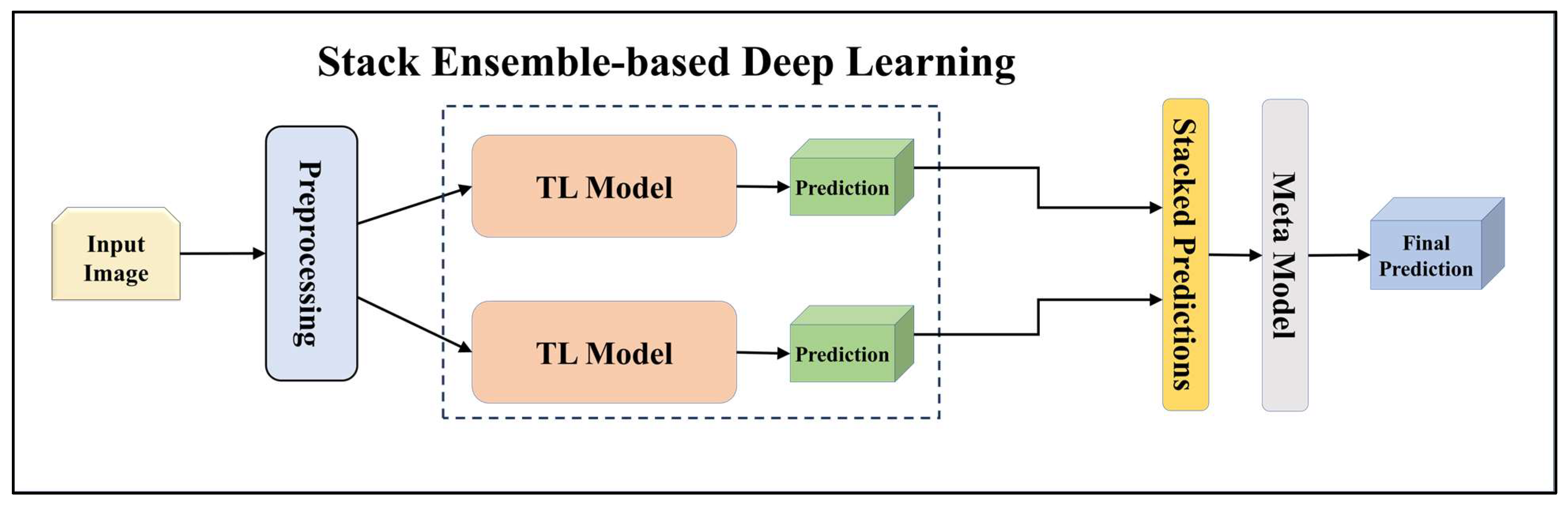
3.4.2. Ensemble-Based Transfer Learning Models
3.4.3. Attention-Enabled Transfer Learning Models
3.4.4. Attention-enabled Ensemble-based Deep Learning Models
3.5. Training Parameters
3.6. Experimental Protocols
3.6.1. Experiment 1: Performance of TL Models Using the HAM10000 Dataset
3.6.2. Experiment 2: Comparison of EBDL Models vs. TL Models
3.6.3. Experiment 3: Comparison of TL Models vs. aeTL Models
3.6.4. Experiment 4: Comparison of aeTL Models vs. aeEBDL Models
3.6.5. Experiment 5: Effect of Training Size on the Performance of TL and aeTL Models
3.7. Performance Metrics
4. Results
4.1. Performance of TL Models Using the HAM10000 Dataset
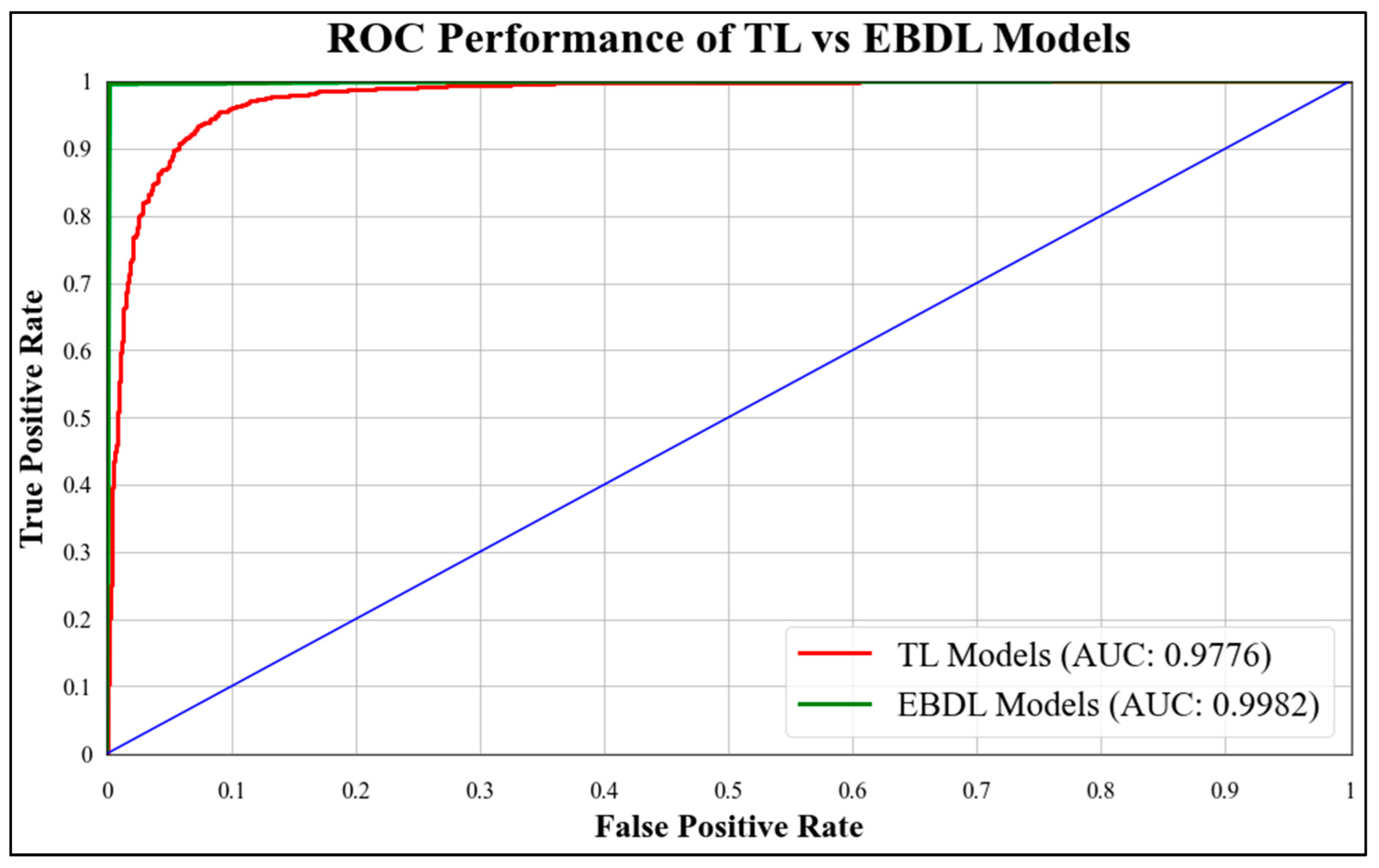
4.2. Comparison of EBDL Models vs. TL Models
4.3. Comparison of TL Models vs. aeTL Models
4.4. Comparison of aeTL Models vs. aeEBDL Models
4.5. Effect of Training Size on the Performance of TL and aeTL Models
5. Performance Evaluation
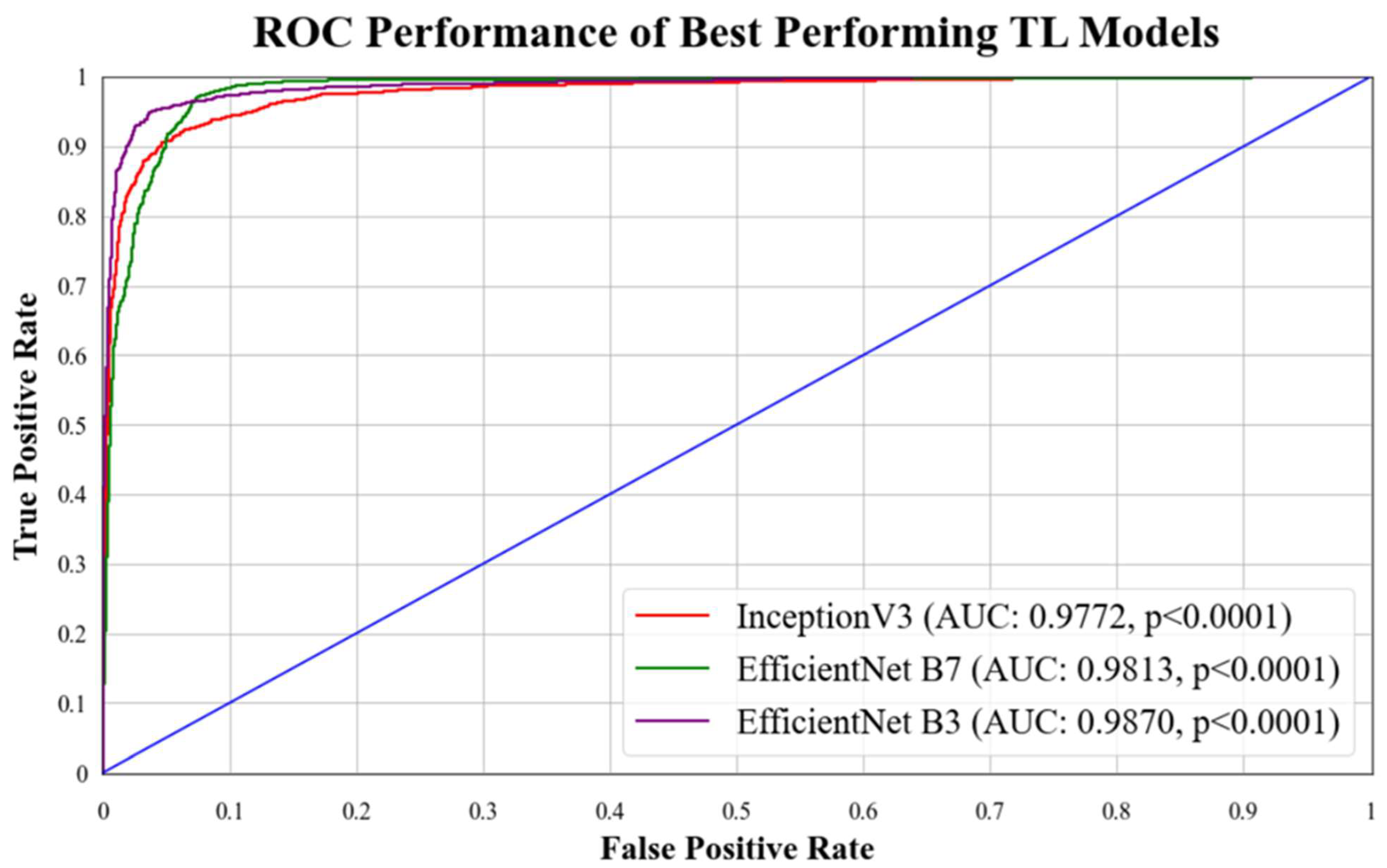
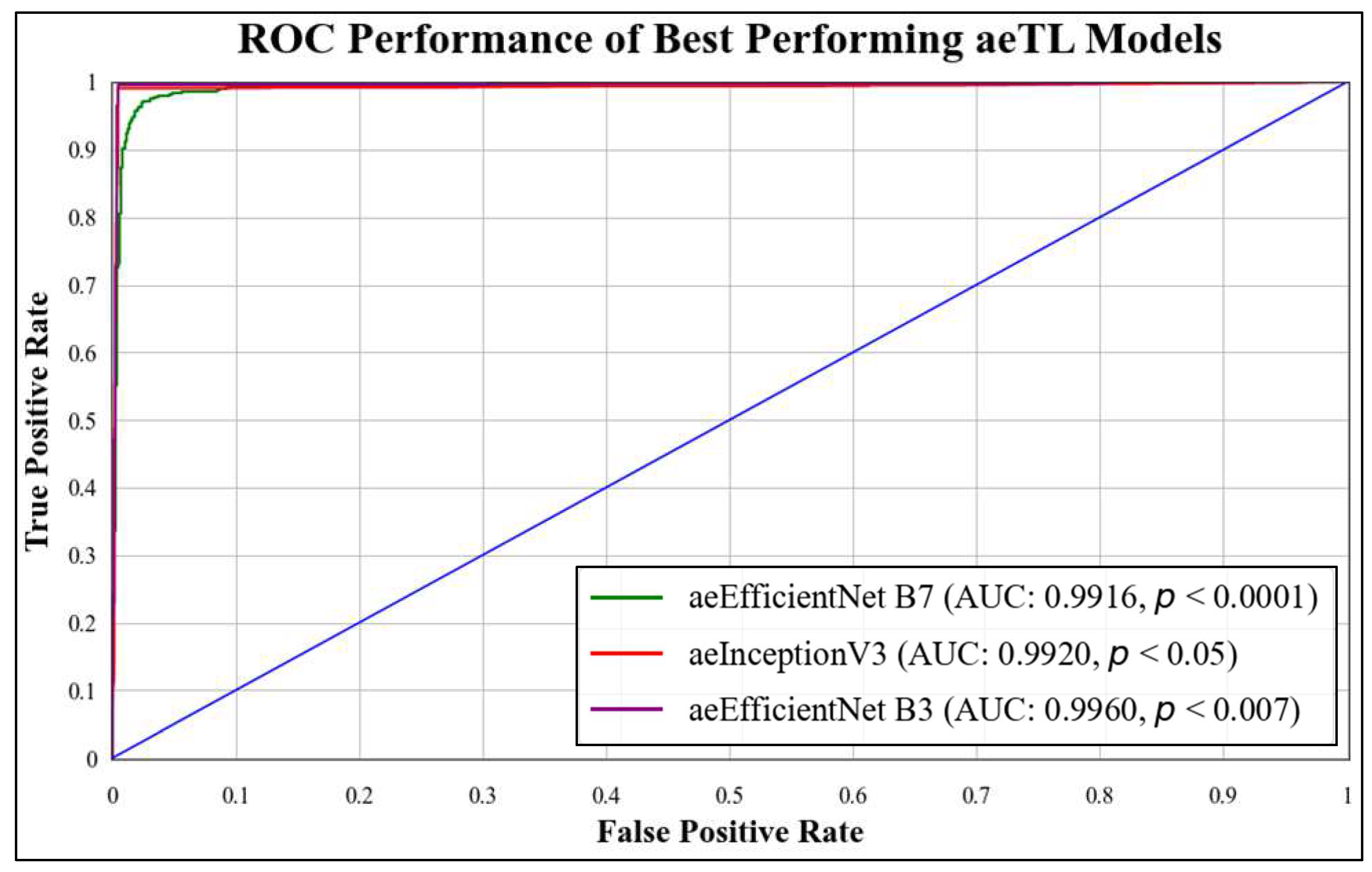
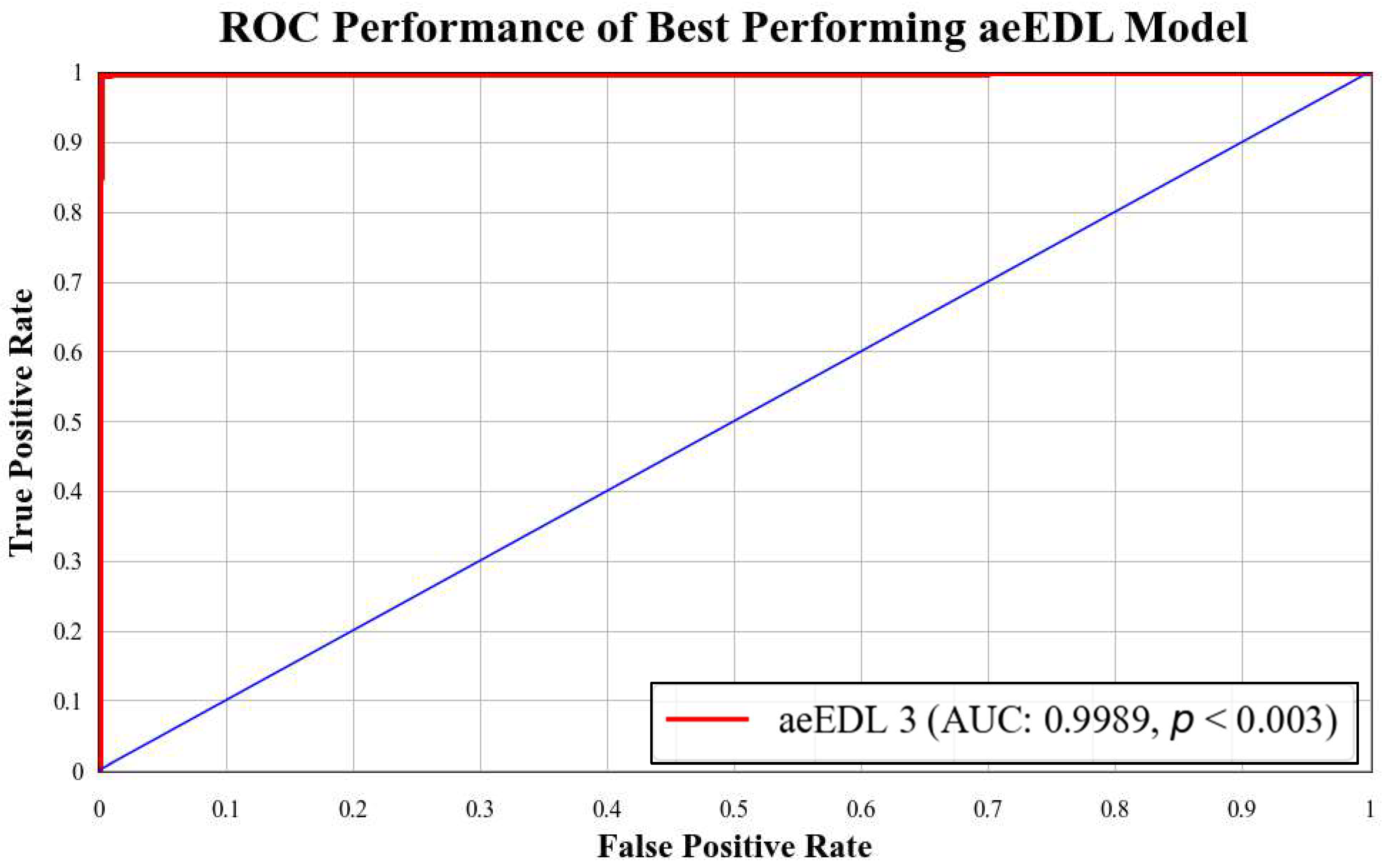
5.1. Receiver Operating Characteristics
5.2. Stability Validation Using Statistical Tests
5.3. Reliability Analysis through Misclassification Results
6. Discussion
6.1. Principal Findings
6.2. Benchmarking
6.3. Special Note on the Use of Attention in Skin Lesion Classification
6.4. Special Note on the Use of Segmentation of Lesion
6.5. Special Strengths, Weakness, and Extension
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Acronym Table
| SN | Abb * | Definition | SN | Abb * | Definition |
| 1 | ACT | Active contour technique | 12 | GLCM | Gray Level Co-occurrence Matrix |
| 2 | aeEBDL | Attention-enabled EBDL | 13 | ISU | Idaho State University |
| 3 | aeTL | Attention-enabled transfer learning | 14 | KNN | k-Nearest Neighbors |
| 4 | AUC | Area-under-the-curve | 15 | ML | Machine Learning |
| 5 | CNN | Convolutional Neural Network | 16 | NASNet | Neural Architecture Search Network |
| 6 | DCNN | Deep CNN | 17 | ROI | Region of interest |
| 7 | DL | Deep Learning | 18 | ROC | Receiver operating characteristics |
| 8 | DT | Decision Trees | 19 | SVM | Support Vector Machine |
| 9 | EBDL | Ensemble-based Deep Learning | 20 | TL | Transfer Learning |
| 10 | FP | False Positive | 21 | TP | True Positive |
| 11 | FN | False Negative | 22 | TN | True Negative |
| Abb * = abbreviation. |
Symbol Table
| SN | Symbols | Explanation |
| 1 | akiec | Actinic keratoses and intraepithelial carcinoma |
| 2 | bcc | Basal cell carcinoma |
| 3 | bkl | Benign keratosis-like lesions |
| 4 | df | Dermatofibroma |
| 5 | mel | Melanoma |
| 6 | nv | Melanocytic nevi |
| 7 | vasc | Vascular lesions |
| 8 | Accuracy | |
| 9 | R | Recall |
| 10 | P | Precision |
| 11 | F1-Score | |
| 12 | M | Total models that were used in the system |
| 13 | D | Total datasets that were used in the system |
| 14 | m | Current model that is being studied |
| 15 | d | Current dataset that is being studied |
| 16 | Denotes the mth model’s prediction score containing the prediction of each class | |
| 17 | Denotes the mth model’s attention output score containing the prediction of each class | |
| 18 | Denotes the mth model’s attention weight | |
| 19 | C | Denotes the number of classes in the multiclass framework |
| 20 | Is the final attention-enabled and ensemble-based model’s output | |
| 21 | Accuracy of model “m” over all D datasets over the K10 protocol | |
| 22 | Accuracy achieved over the dataset “d” over all M Models over the K10 protocol | |
| 23 | Overall system accuracy over M models and D datasets | |
| 24 | AUC of model m summarized over all D datasets | |
| 25 | AUC achieved over dataset d over all M Models | |
| 26 | Overall system AUC over M models and D datasets | |
| 27 | Standard Deviation of the system | |
| 28 | Mean Reliability Index of the system | |
| 29 | Mean Misclassification value | |
| 30 | Mean of Misclassification of ith image over all AI models | |
| 31 | Total number of images for misclassification probability |
References
- Clark, W.H., Jr.; Elder, D.E.; Guerry, D.; Epstein, M.N.; Greene, M.H.; Van Horn, M. A study of tumor progression: The precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol. 1984, 15, 1147–1165. [Google Scholar] [CrossRef] [PubMed]
- Jones, O.T.; Ranmuthu, C.K.I.; Hall, P.N.; Funston, G.; Walter, F.M. Recognising skin cancer in primary care. Adv. Ther. 2020, 37, 603–616. [Google Scholar] [CrossRef]
- D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. UV radiation and the skin. Int. J. Mol. Sci. 2013, 14, 12222–12248. [Google Scholar] [CrossRef] [PubMed]
- Qadir, M.I. Skin cancer: Etiology and management. Pak. J. Pharm. Sci. 2016, 29, 999–1003. [Google Scholar]
- Gordon, R. Skin cancer: An overview of epidemiology and risk factors. Semin. Oncol. Nurs. 2013, 29, 160–169. [Google Scholar] [CrossRef]
- Foote, M.; Harvey, J.; Porceddu, S.; Dickie, G.; Hewitt, S.; Colquist, S.; Zarate, D.; Poulsen, M. Effect of radiotherapy dose and volume on relapse in Merkel cell cancer of the skin. Int. J. Radiat. Oncol. Biol. Phys. 2010, 77, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Jerant, A.F.; Johnson, J.T.; Sheridan, C.D.; Caffrey, T.J. Early detection and treatment of skin cancer. Am. Fam. Physician 2000, 62, 357–368. [Google Scholar]
- Narayanan, D.L.; Saladi, R.N.; Fox, J.L. Ultraviolet radiation and skin cancer. Int. J. Dermatol. 2010, 49, 978–986. [Google Scholar] [CrossRef]
- Rodrigues, D.D.A.; Ivo, R.F.; Satapathy, S.C.; Wang, S.; Hemanth, J.; Reboucas Filho, P.P. A new approach for classification skin lesion based on transfer learning, deep learning, and IoT system. Pattern Recognit. Lett. 2020, 136, 8–15. [Google Scholar] [CrossRef]
- Cai, C.J.; Winter, S.; Steiner, D.; Wilcox, L.; Terry, M. “Hello AI”: Uncovering the onboarding needs of medical practitioners for human-AI collaborative decision-making. Proc. ACM Hum. Comput. Interact. 2019, 3, 1–24. [Google Scholar] [CrossRef]
- Soenksen, L.R.; Kassis, T.; Conover, S.T.; Marti-Fuster, B.; Birkenfeld, J.S.; Tucker-Schwartz, J.; Naseem, A.; Stavert, R.R.; Kim, C.C.; Senna, M.M.; et al. Using deep learning for dermatologist-level detection of suspicious pigmented skin lesions from wide-field images. Sci. Transl. Med. 2021, 13, eabb3652. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Allugunti, V.R. A machine learning model for skin disease classification using convolution neural network. Int. J. Comput. Program. Database Manag. 2022, 3, 141–147. [Google Scholar] [CrossRef]
- Huang, S.-F.; Ruey-Feng, C.; Moon, W.K.; Lee, Y.-H.; Dar-Ren, C.; Suri, J.S. Analysis of tumor vascularity using three-dimensional power Doppler ultrasound images. IEEE Trans. Med. Imaging 2008, 27, 320–330. [Google Scholar] [CrossRef]
- Niu, S.; Liu, Y.; Wang, J.; Song, H. A decade survey of transfer learning (2010–2020). IEEE Trans. Artif. Intell. 2020, 1, 151–166. [Google Scholar] [CrossRef]
- Torrey, L.; Shavlik, J. Transfer learning. In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques; IGI Global: Hershey, PA, USA, 2010; pp. 242–264. [Google Scholar]
- Shaha, M.; Pawar, M. Transfer learning for image classification. In Proceedings of the 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 29–31 March 2018; pp. 656–660. [Google Scholar]
- Krishna, S.T.; Kalluri, H.K. Deep learning and transfer learning approaches for image classification. Int. J. Recent Technol. Eng. IJRTE 2019, 7, 427–432. [Google Scholar]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE 2019, 14, e0217293. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, R.; Afzal, S.; Rehman, A.U.; Gul, S.; Baber, J.; Bakhtyar, M.; Mehmood, I.; Song, O.-Y.; Maqsood, M. Region-of-interest based transfer learning assisted framework for skin cancer detection. IEEE Access 2020, 8, 147858–147871. [Google Scholar] [CrossRef]
- Fraiwan, M.; Faouri, E. On the automatic detection and classification of skin cancer using deep transfer learning. Sensors 2022, 22, 4963. [Google Scholar] [CrossRef]
- Weatheritt, J.; Rueckert, D.; Wolz, R. Transfer learning for brain segmentation: Pre-task selection and data limitations. In Proceedings of the Medical Image Understanding and Analysis: 24th Annual Conference, MIUA 2020, Oxford, UK, 15–17 July 2020; Volume 24, pp. 118–130. [Google Scholar]
- Lehnert, L.; Tellex, S.; Littman, M.L. Advantages and limitations of using successor features for transfer in reinforcement learning. arXiv 2017, arXiv:1708.00102. [Google Scholar]
- Ganaie, M.A.; Hu, M.; Malik, A.; Tanveer, M.; Suganthan, P. Ensemble deep learning: A review. Eng. Appl. Artif. Intell. 2022, 115, 105151. [Google Scholar] [CrossRef]
- Yang, Y.; Lv, H.; Chen, N. A survey on ensemble learning under the era of deep learning. Artif. Intell. Rev. 2023, 56, 5545–5589. [Google Scholar] [CrossRef]
- Ozkan, I.A.; Koklu, M. Skin lesion classification using machine learning algorithms. Int. J. Intell. Syst. Appl. Eng. 2017, 5, 285–289. [Google Scholar] [CrossRef]
- Singh, A.; Thakur, N.; Sharma, A. A review of supervised machine learning algorithms. In Proceedings of the 2016 3rd International Conference on Computing for Sustainable Global Development (INDIACom), New Delhi, India, 16–18 March 2016; pp. 1310–1315. [Google Scholar]
- Sajid, P.; Rajesh, D. Performance evaluation of classifiers for automatic early detection of skin cancer. J. Adv. Res. Dyn. Control. Syst. 2018, 10, 454–461. [Google Scholar]
- Senan, E.M.; Jadhav, M.E. Diagnosis of dermoscopy images for the detection of skin lesions using SVM and KNN. In Proceedings of the Third International Conference on Sustainable Computing: SUSCOM 2021, Jaipur, India, 19–20 March 2021; Springer: Singapore, 2022; pp. 125–134. [Google Scholar]
- Karamizadeh, S.; Abdullah, S.M.; Halimi, M.; Shayan, J.; Rajabi, M.J. Advantage and drawback of support vector machine functionality. In Proceedings of the 2014 International Conference on Computer, Communications, and Control Technology (I4CT), Langkawi, Malaysia, 2–4 September 2014; pp. 63–65. [Google Scholar]
- Lopez, A.R.; Giro-i-Nieto, X.; Burdick, J.; Marques, O. Skin lesion classification from dermoscopic images using deep learning techniques. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–21 February 2017; pp. 49–54. [Google Scholar]
- Serte, S.; Demirel, H. Gabor wavelet-based deep learning for skin lesion classification. Comput. Biol. Med. 2019, 113, 103423. [Google Scholar] [CrossRef]
- Mirunalini, P.; Chandrabose, A.; Gokul, V.; Jaisakthi, S. Deep learning for skin lesion classification. arXiv 2017, arXiv:1703.04364. [Google Scholar]
- Mahbod, A.; Schaefer, G.; Wang, C.; Ecker, R.; Ellinge, I. Skin lesion classification using hybrid deep neural networks. In Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1229–1233. [Google Scholar]
- Younis, H.; Bhatti, M.H.; Azeem, M. Classification of skin cancer dermoscopy images using transfer learning. In Proceedings of the in 2019 15th International Conference on Emerging Technologies (ICET), Peshawar, Pakistan, 2–3 December 2019; pp. 1–4. [Google Scholar]
- Harangi, B.; Baran, A.; Hajdu, A. Classification of skin lesions using an ensemble of deep neural networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 2575–2578. [Google Scholar]
- Shehzad, K.; Zhenhua, T.; Shoukat, S.; Saeed, A.; Ahmad, I.; Bhatti, S.S.; Chelloug, S.A. A Deep-Ensemble-Learning-Based Approach for Skin Cancer Diagnosis. Electronics 2023, 12, 1342. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Ali, M.S.; Miah, M.S.; Haque, J.; Rahman, M.M.; Islam, M.K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef]
- Salehi, A.W.; Khan, S.; Gupta, G.; Alabduallah, B.I.; Almjally, A.; Alsolai, H.; Siddiqui, T.; Mellit, A. A Study of CNN and Transfer Learning in Medical Imaging: Advantages, Challenges, Future Scope. Sustainability 2023, 15, 5930. [Google Scholar] [CrossRef]
- Ghosal, P.; Nandanwar, L.; Kanchan, S.; Bhadra, A.; Chakraborty, J.; Nandi, D. Brain tumor classification using ResNet-101 based squeeze and excitation deep neural network. In Proceedings of the 2019 Second International Conference on Advanced Computational and Communication Paradigms (ICACCP), Gangtok, India, 25–28 February 2019; pp. 1–6. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8697–8710. [Google Scholar]
- Kausar, N.; Hameed, A.; Sattar, M.; Ashraf, R.; Imran, A.S.; Abidin, M.Z.U.; Ali, A. Multiclass skin cancer classification using ensemble of fine-tuned deep learning models. Appl. Sci. 2021, 11, 10593. [Google Scholar] [CrossRef]
- Hoang, L.; Lee, S.-H.; Lee, E.-J.; Kwon, K.-R. Multiclass skin lesion classification using a novel lightweight deep learning framework for smart healthcare. Appl. Sci. 2022, 12, 2677. [Google Scholar] [CrossRef]
- Chaturvedi, S.S.; Tembhurne, J.V.; Diwan, T. A multi-class skin Cancer classification using deep convolutional neural networks. Multimed. Tools Appl. 2020, 79, 28477–28498. [Google Scholar] [CrossRef]
- Rahi, M.M.I.; Khan, F.T.; Mahtab, M.T.; Ullah, A.A.; Alam, M.G.R.; Alam, M.A. Detection of skin cancer using deep neural networks. In Proceedings of the 2019 IEEE Asia-Pacific Conference on Computer Science and Data Engineering (CSDE), Melbourne, VIC, Australia, 9–11 December 2019; pp. 1–7. [Google Scholar]
- Sae-Lim, W.; Wettayaprasit, W.; Aiyarak, P. Convolutional neural networks using MobileNet for skin lesion classification. In Proceedings of the 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), Chonburi, Thailand, 10–12 July 2019; pp. 242–247. [Google Scholar]
- Moldovan, D. Transfer learning based method for two-step skin cancer images classification. In Proceedings of the 2019 E-Health and Bioengineering Conference (EHB), Iasi, Romania, 21–23 November 2019; pp. 1–4. [Google Scholar]
- Iqbal, I.; Younus, M.; Walayat, K.; Kakar, M.U.; Ma, J. Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images. Comput. Med. Imaging Graph. 2021, 88, 101843. [Google Scholar] [CrossRef]
- Sai Charan, D.; Nadipineni, H.; Sahayam, S.; Jayaraman, U. Method to Classify Skin Lesions using Dermoscopic images. arXiv 2020, arXiv:2008.09418. [Google Scholar]
- Phung, S.L.; Bouzerdoum, A.; Chai, D. Skin segmentation using color pixel classification: Analysis and comparison. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, V.K.; Londhe, N.D.; Sonawane, R.S.; Suri, J.S. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput. Methods Programs Biomed. 2017, 150, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.B.; Mercedes Filho, E.; Ma, Z.; Papa, J.P.; Pereira, A.S.; Tavares, J.M.R. Computational methods for the image segmentation of pigmented skin lesions: A review. Comput. Methods Programs Biomed. 2016, 131, 127–141. [Google Scholar] [CrossRef]
- Sumithra, R.; Suhil, M.; Guru, D. Segmentation and classification of skin lesions for disease diagnosis. Procedia Comput. Sci. 2015, 45, 76–85. [Google Scholar] [CrossRef]
- Liu, K.; Suri, J.S. Automatic Vessel Indentification for Angiographic Screening. U.S. Patent 6,845,260, 18 January 2005. [Google Scholar]
- Li, H.; He, X.; Zhou, F.; Yu, Z.; Ni, D.; Chen, S.; Wang, T.; Lei, B. Dense deconvolutional network for skin lesion segmentation. IEEE J. Biomed. Health Inform. 2018, 23, 527–537. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.; Simpson, J.; Kane, A.; Menon, D.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- El-Baz, A.; Gimel’farb, G.; Suri, J.S. Stochastic Modeling for Medical Image Analysis; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Suri, J.S. Two-dimensional fast magnetic resonance brain segmentation. IEEE Eng. Med. Biol. Mag. 2001, 20, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Faust, O.; Alvin, A.P.; Krishnamurthi, G.; Seabra, J.C.; Sanches, J.; Suri, J.S. Understanding symptomatology of atherosclerotic plaque by image-based tissue characterization. Comput. Methods Programs Biomed. 2013, 110, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Sree, S.V.; Krishnan, M.M.R.; Krishnananda, N.; Ranjan, S.; Umesh, P.; Suri, J.S. Automated classification of patients with coronary artery disease using grayscale features from left ventricle echocardiographic images. Comput. Methods Programs Biomed. 2013, 112, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Faust, O.; Sree, S.V.; Molinari, F.; Saba, L.; Nicolaides, A.; Suri, J.S. An accurate and generalized approach to plaque characterization in 346 carotid ultrasound scans. IEEE Trans. Instrum. Meas. 2011, 61, 1045–1053. [Google Scholar] [CrossRef]
- Singh, J.; Singh, N.; Fouda, M.M.; Saba, L.; Suri, J.S. Attention-Enabled Ensemble Deep Learning Models and Their Validation for Depression Detection: A Domain Adoption Paradigm. Diagnostics 2023, 13, 2092. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Pathak, R.; Ketireddy, V.; Columbu, M.; Saba, L.; Gupta, S.K.; Faa, G.; Singh, I.M.; Turk, M.; et al. COVLIAS 1.0: Lung segmentation in COVID-19 computed tomography scans using hybrid deep learning artificial intelligence models. Diagnostics 2021, 11, 1405. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.S.C.; Saba, L.; Mehmedović, A.; Faa, G.; Singh, I.M.; et al. COVLIAS 2.0-cXAI: Cloud-based explainable deep learning system for COVID-19 lesion localization in computed tomography scans. Diagnostics 2022, 12, 1482. [Google Scholar] [CrossRef]
- Saba, L.; Banchhor, S.K.; Suri, H.S.; Londhe, N.D.; Araki, T.; Ikeda, N.; Viskovic, K.; Shafique, S.; Laird, J.R.; Gupta, A.; et al. Accurate cloud-based smart IMT measurement, its validation and stroke risk stratification in carotid ultrasound: A web-based point-of-care tool for multicenter clinical trial. Comput. Biol. Med. 2016, 75, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, V.K.; Londhe, N.D.; Sonawane, R.S.; Suri, J.S. Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: A first comparative study of its kind. Comput. Methods Programs Biomed. 2016, 126, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Molinari, F.; Mantovani, A.; Deandrea, M.; Limone, P.; Garberoglio, R.; Suri, J.S. Characterization of single thyroid nodules by contrast-enhanced 3-D ultrasound. Ultrasound Med. Biol. 2010, 36, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Banchhor, S.K.; Araki, T.; Viskovic, K.; Londhe, N.D.; Laird, J.R.; Suri, H.S.; Suri, J.S. Intra-and inter-operator reproducibility of automated cloud-based carotid lumen diameter ultrasound measurement. Indian Heart J. 2018, 70, 649–664. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Mookiah, M.R.K.; Sree, S.V.; Yanti, R.; Martis, R.J.; Saba, L.; Molinari, F.; Guerriero, S.; Suri, J.S. Evolutionary algorithm-based classifier parameter tuning for automatic ovarian cancer tissue characterization and classification. Ultraschall Med. Eur. J. Ultrasound 2014, 35, 237–245. [Google Scholar]
- Agarwal, M.; Agarwal, S.; Saba, L.; Chabert, G.L.; Gupta, S.; Carriero, A.; Pasche, A.; Danna, P.; Mehmedovic, A.; Faa, G.; et al. Eight pruning deep learning models for low storage and high-speed COVID-19 computed tomography lung segmentation and heatmap-based lesion localization: A multicenter study using COVLIAS 2.0. Comput. Biol. Med. 2022, 146, 105571. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Viskovic, K.; Suri, N.; Alizad, A.; El-Baz, A.; Saba, L.; Fatemi, M.; et al. Systematic review of artificial intelligence in acute respiratory distress syndrome for COVID-19 lung patients: A biomedical imaging perspective. IEEE J. Biomed. Health Inform. 2021, 25, 4128–4139. [Google Scholar] [CrossRef]
- Suri, J.S.; Bhagawati, M.; Agarwal, S.; Paul, S.; Pandey, A.; Gupta, S.K.; Saba, L.; Paraskevas, K.I.; Khanna, N.N.; Laird, J.R.; et al. UNet Deep Learning Architecture for Segmentation of Vascular and Non-Vascular Images: A Microscopic Look at UNet Components Buffered With Pruning, Explainable Artificial Intelligence, and Bias. IEEE Access 2022, 11, 595–645. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Jena, B.; Saxena, S.; El-Baz, A.; Agarwal, V.; Kalra, M.K.; Saba, L.; Viskovic, K.; Fatemi, M.; et al. Five strategies for bias estimation in artificial intelligence-based hybrid deep learning for acute respiratory distress syndrome COVID-19 lung infected patients using AP (ai) Bias 2.0: A systematic review. IEEE Trans. Instrum. Meas. 2022. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017).
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. CSUR 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Suri, J.; Liu, K.; Singh, S.; Laxminarayan, S.; Zeng, X.; Reden, L. Shape recovery algorithms using level sets in 2-D/3-D medical imagery: A state-of-the-art review. IEEE Trans. Inf. Technol. Biomed. 2002, 6, 8–28. [Google Scholar] [CrossRef] [PubMed]



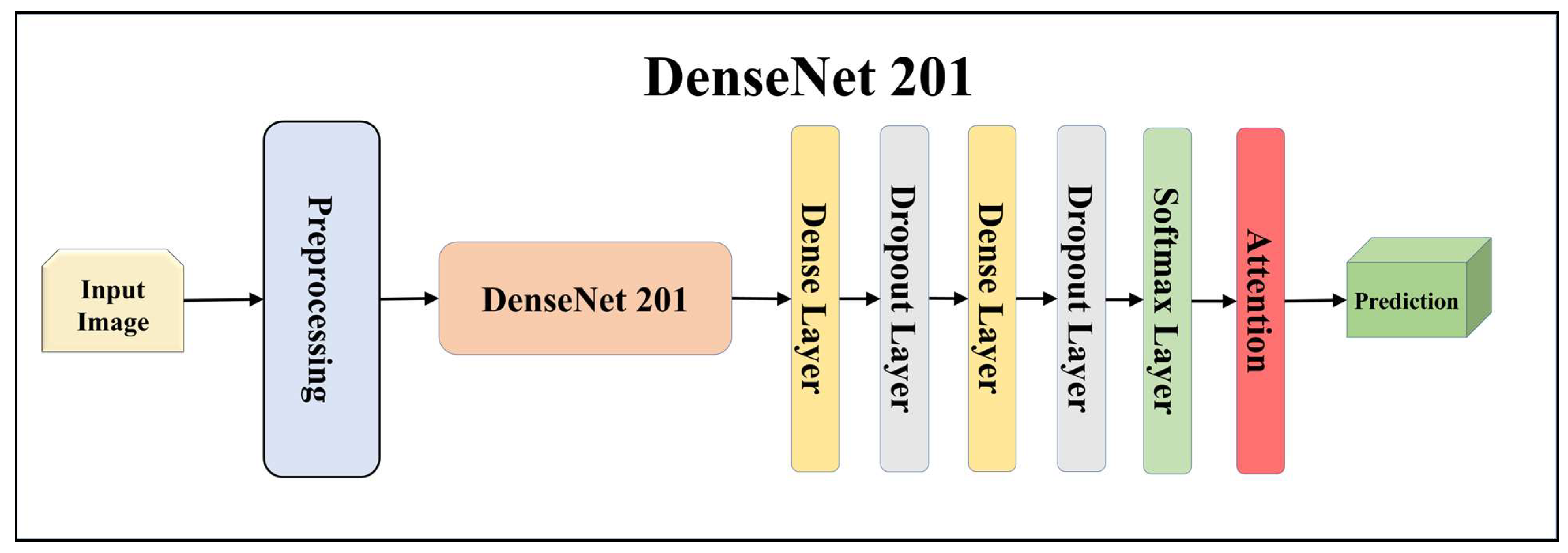

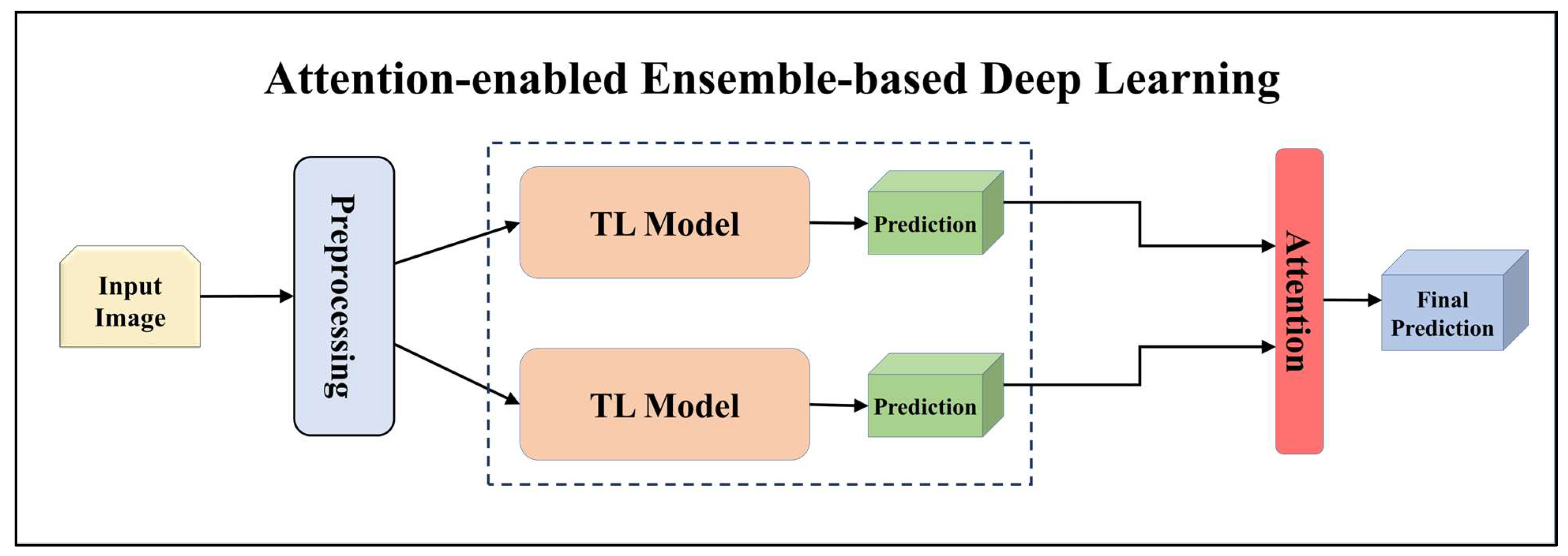


| Performance Metrics of seven TL Models | |||||||
|---|---|---|---|---|---|---|---|
| TL | TL Model | Accuracy (%) | AUC [0–1] | Precision [0–1] | Recall [0–1] | F1-Score [0–1] | S.D. (%) |
| TL 1 | ResNet 101 | 94.10% | 0.9697 | 0.9428 | 0.9407 | 0.9406 | 1.51 |
| TL 2 | MobileNet | 94.73% | 0.9748 | 0.9501 | 0.9484 | 0.9482 | 1.5 |
| TL 3 | NASNet Mobile | 94.93% | 0.9755 | 0.9508 | 0.9488 | 0.9485 | 1.48 |
| TL 4 | DenseNet 201 | 95.18% | 0.9780 | 0.9535 | 0.9523 | 0.9518 | 1.51 |
| TL 5 | Inception V3 | 95.41% | 0.9772 | 0.9550 | 0.9542 | 0.9539 | 1.49 |
| TL 6 | Efficient Net B7 | 95.76% | 0.9813 | 0.9589 | 0.9565 | 0.9558 | 1.49 |
| TL 7 | Efficient Net B3 | 97.01% | 0.9870 | 0.9701 | 0.9700 | 0.9698 | 1.51 |
| Mean accuracy of all models = 95.30% | |||||||
| Performance Metrics of six EBDL Models | ||||||||
|---|---|---|---|---|---|---|---|---|
| EBDL | Ensemble Technique | Base Models | Accuracy (%) | AUC [0–1] | Precision [0–1] | Recall [0–1] | F1-Score [0–1] | S.D. (%) |
| EBDL 1 | Weighted Average | MobileNet + EfficientNet B3 + InceptionV3 | 99.11% | 0.9984 | 0.9960 | 0.9960 | 0.9960 | 1.51 |
| EBDL 2 | Max Voting | NASNet Mobile + ResNet101 + EfficientNet B7 | 99.34% | 0.9972 | 0.9934 | 0.9934 | 0.9933 | 1.50 |
| EBDL 3 | Max Voting | ResNet101 + MobileNet + InceptionV3 | 99.58% | 0.9981 | 0.9958 | 0.9958 | 0.9958 | 1.51 |
| EBDL 4 | Weighted Average | MobileNet + EfficientNet B7 + ResNet101 + DenseNet201 | 99.68% | 0.9987 | 0.9968 | 0.9968 | 0.9967 | 1.51 |
| EBDL 5 | Stack | ResNet101 + MobileNet + InceptionV3 | 99.69% | 0.9986 | 0.9969 | 0.9969 | 0.9969 | 1.51 |
| EBDL 6 | Stack | DenseNet201 + ResNet101 + NASNet Mobile | 99.69% | 0.9987 | 0.9969 | 0.9969 | 0.9969 | 1.51 |
| Mean accuracy of all models = 99.52% | ||||||||
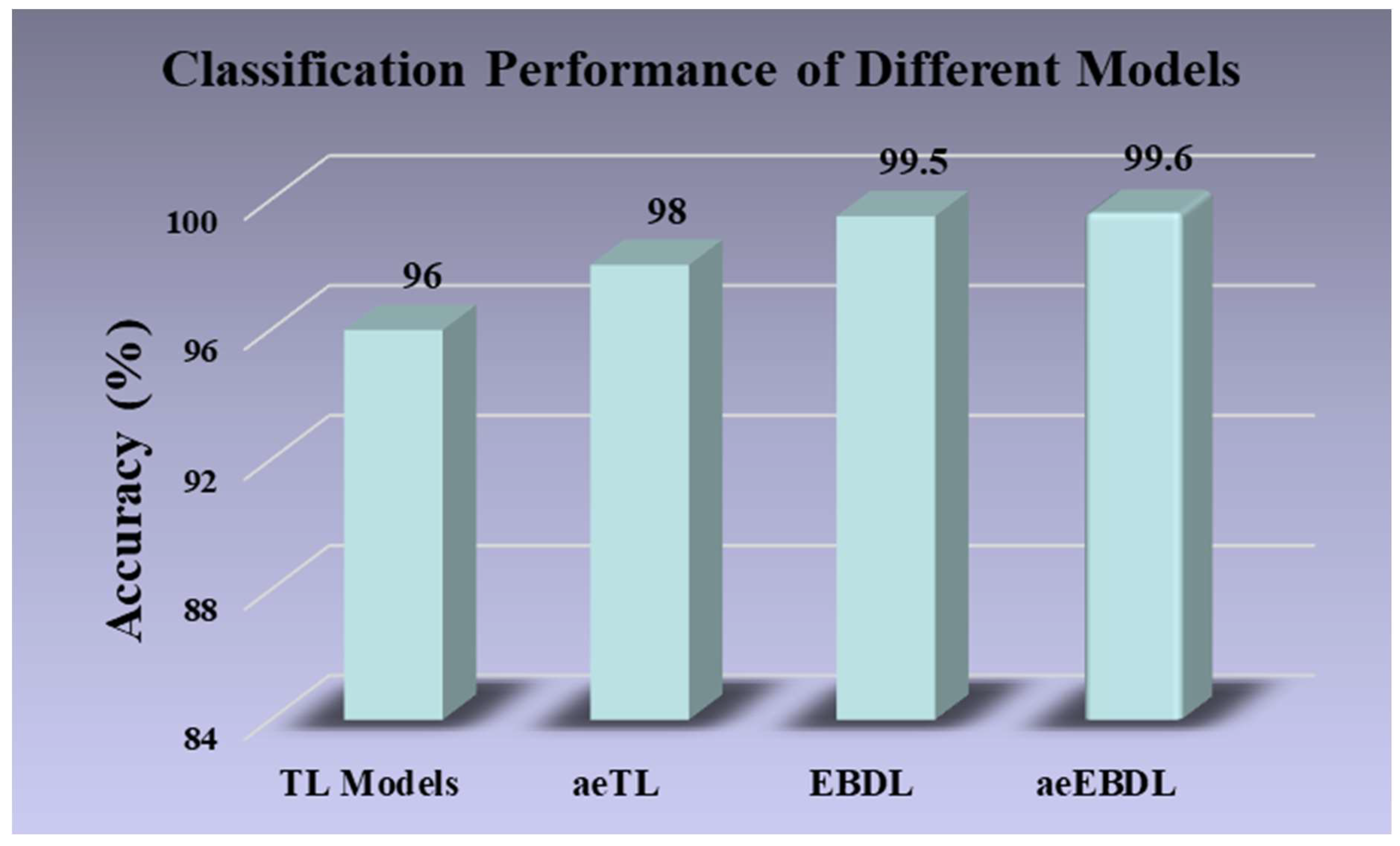
| Comparison of EBDL Models vs. TL Models | |||||
|---|---|---|---|---|---|
| Model Type | Mean Accuracy (%) | Mean AUC [0–1] | Mean Precision [0–1] | Mean Recall [0–1] | Mean F1-Score [0–1] |
| TL Models | 95.30% | 0.9776 | 0.9544 | 0.9529 | 0.9526 |
| EBDL Models | 99.52% | 0.9982 | 0.9959 | 0.9959 | 0.9959 |
| Mean Increase (%) | 4.22% | 2.06% | 4.15% | 4.30% | 4.33% |
| Performance Metrics of Seven aeTL Models | |||||||
|---|---|---|---|---|---|---|---|
| aeTL | TL Model | Accuracy (%) | AUC [0–1] | Precision [0–1] | Recall [0–1] | F1-Score [0–1] | S.D. (%) |
| aeTL 1 | ResNet 101 | 98.17% | 0.9908 | 0.9823 | 0.9817 | 0.9818 | 1.52 |
| aeTL 2 | MobileNet | 98.21% | 0.9919 | 0.9824 | 0.9821 | 0.9821 | 1.51 |
| aeTL 3 | NASNet Mobile | 97.45% | 0.9871 | 0.9750 | 0.9745 | 0.9745 | 1.49 |
| aeTL 4 | DenseNet 201 | 97.91% | 0.9906 | 0.9792 | 0.9791 | 0.9790 | 1.51 |
| aeTL 5 | Inception V3 | 98.56% | 0.9920 | 0.9858 | 0.9856 | 0.9856 | 1.5 |
| eTL 6 | Efficient Net B7 | 98.96% | 0.9916 | 0.9816 | 0.9804 | 0.9805 | 1.52 |
| aeTL 7 | Efficient Net B3 | 98.96% | 0.996 | 0.9897 | 0.9896 | 0.9896 | 1.52 |
| Mean accuracy of all models = 98.31% | |||||||
| Comparison of TL Models vs. aeTL Models | |||||
|---|---|---|---|---|---|
| Model Type | Mean Accuracy (%) | Mean AUC [0–1] | Mean Precision [0–1] | Mean Recall [0–1] | Mean F1-Score [0–1] |
| TL Models | 95.30% | 0.9776 | 0.9544 | 0.9529 | 0.9526 |
| aeTL Models | 98.31% | 0.9914 | 0.9822 | 0.9818 | 0.9818 |
| Mean Increase (%) | 3.01% | 1.38% | 2.78% | 2.89% | 2.92% |
| Performance Metrics of aeEBDL Models | ||||||||
|---|---|---|---|---|---|---|---|---|
| EBDL | Ensemble Technique | Base Models | Accuracy (%) | AUC [0–1] | Precision [0–1] | Recall [0–1] | F1-Score [0–1] | S.D. (%) |
| aeEBDL 1 | Attention-enabled | ResNet101+ MobileNet + InceptionV3 | 99.58% | 0.9981 | 0.9958 | 0.9958 | 0.9958 | 1.51 |
| aeEBDL 2 | Attention-enabled | MobileNet + InceptionV3 + NASNet Mobile | 99.49% | 0.9977 | 0.9949 | 0.9949 | 0.9949 | 1.50 |
| aeEBDL 3 | Attention-enabled | ResNet101 + MobileNet + InceptionV3 + EfficientNet B3 + EfficientNet B7 + DenseNet201 | 99.73% | 0.9989 | 0.9973 | 0.9973 | 0.9973 | 1.51 |
| Mean accuracy of all models = 99.60% | ||||||||
| Comparison of aeTL Models vs. aeEBDL Models | |||||
|---|---|---|---|---|---|
| Model Type | Mean Accuracy (%) | Mean AUC [0–1] | Mean Precision [0–1] | Mean Recall [0–1] | Mean F1-Score [0–1] |
| aeTL Models | 98.31% | 0.9914 | 0.9822 | 0.9818 | 0.9818 |
| aeEBDL Models | 99.60% | 0.9982 | 0.996 | 0.996 | 0.996 |
| Mean Increase | 1.29% | 0.68% | 1.38% | 1.42% | 1.42% |
| Effect of Training Data on Performance for EfficientNet B3 Model | |||||||
|---|---|---|---|---|---|---|---|
| Model Type | K2 | K4 | K5 | K10 | D1 (%) | D2 (%) | D3 (%) |
| Accuracy (%) | 91.39% | 94.85% | 95.69% | 97.01% | 1.32% | 2.16% | 5.62% |
| AUC [0–1] | 0.935 | 0.9592 | 0.9744 | 0.987 | 1.26% | 2.78% | 5.20% |
| Precision [0–1] | 0.9002 | 0.9259 | 0.9611 | 0.9701 | 0.90% | 4.42% | 6.99% |
| Recall [0–1] | 0.8978 | 0.9258 | 0.9614 | 0.97 | 0.86% | 4.42% | 7.22% |
| F1-Score [0–1] | 0.899 | 0.9258 | 0.9612 | 0.9698 | 0.86% | 4.40% | 7.08% |
| Effect of Training Data on Performance for aeEfficientNet B3 Model | |||||||
|---|---|---|---|---|---|---|---|
| Model Type | K2 | K4 | K5 | K10 | D1 (%) | D2 (%) | D3 (%) |
| Accuracy (%) | 92.68% | 94.83% | 98.15% | 98.96% | 0.81% | 4.13% | 6.28% |
| AUC [0–1] | 0.9416 | 0.9561 | 0.9824 | 0.996 | 1.36% | 3.99% | 5.44% |
| Precision [0–1] | 0.9237 | 0.9629 | 0.9805 | 0.9897 | 0.92% | 2.68% | 6.60% |
| Recall [0–1] | 0.9235 | 0.964 | 0.9801 | 0.9896 | 0.95% | 2.56% | 6.61% |
| F1-Score [0–1] | 0.9236 | 0.9634 | 0.9803 | 0.9896 | 0.93% | 2.62% | 6.60% |
| Model Type | Model Name | Wilcoxon p-Test | Cohen’s Kappa |
|---|---|---|---|
| TL Models | ResNet 101 | p < 0.001 | 0.9251 |
| MobileNet | p < 0.001 | 0.9348 | |
| NASNet Mobile | p < 0.001 | 0.9352 | |
| DenseNet 201 | p < 0.001 | 0.9398 | |
| Inception V3 | p < 0.001 | 0.9422 | |
| Efficient Net B7 | p < 0.001 | 0.9451 | |
| Efficient Net B3 | p < 0.001 | 0.9622 | |
| EBDL Models | EBDL 1 | p < 0.004 | 0.995 |
| EBDL 2 | p < 0.001 | 0.9917 | |
| EBDL 3 | p < 0.001 | 0.9948 | |
| EBDL 4 | p < 0.002 | 0.996 | |
| EBDL 5 | p < 0.06 | 0.9962 | |
| EBDL 6 | p < 0.007 | 0.9962 | |
| aeTL Models | aeResNet 101 | p < 0.001 | 0.9769 |
| aeMobileNet | p < 0.001 | 0.9774 | |
| aeNASNet Mobile | p < 0.001 | 0.9679 | |
| aeDenseNet 201 | p < 0.001 | 0.9736 | |
| aeInception V3 | p < 0.05 | 0.9819 | |
| aeEfficient Net B7 | p < 0.001 | 0.9753 | |
| aeEfficient Net B3 | p < 0.007 | 0.9869 | |
| aeEBDL Models | aeEBDL 1 | p < 0.001 | 0.9948 |
| aeEBDL 2 | p < 0.001 | 0.9936 | |
| aeEBDL 3 | p < 0.003 | 0.9967 |
| Year | Author | AI Models | Approach | Methodology | Dataset | Accuracy (%) | Precision (%) | Recall (%) | F1 (%) | AUC [0,1] | Scientific Val. | Clinical Val. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2019 | Moldovan et al. [54] | DenseNet-121 | TL | Dual-step CNN Based on DenseNet121 | HAM 10000 | 80 | 🗴 | 🗴 | 🗴 | 🗴 | 🗴 | 🗴 |
| 2019 | Lim et al. [53] | MobileNet | TL | Augmented Data on Fine-tuned MobileNet | HAM 10000 | 83.23 | 🗴 | 🗴 | 82 | 🗴 | 🗴 | 🗴 |
| 2019 | Rahi et al. [52] | ResNet-50 | TL | Fine-tuned TL Models and a CNN from Scratch | HAM 10000 | 90 | 91 | 89 | 89 | 🗴 | 🗴 | 🗴 |
| 2020 | Chaturvedi et al. [51] | IRv2 | TL | Fine-tuned ResNetXt101 | HAM 10000 | 93.2 | 87 | 88 | 🗴 | 🗴 | 🗴 | 🗴 |
| 2021 | Iqbal et al. [55] | DCNN | CNN | Deep CNN Models | ISIC-19 | 88.75 | 90.66 | 🗴 | 89.75 | 0.95 | 🗴 | 🗴 |
| 2021 | Ali et al. [39] | DCNN | CNN | Normalization and Data Augmentation with DCNN | HAM 10000 | 90.16 | 94.63 | 93.91 | 94.27 | 🗴 | 🗴 | 🗴 |
| 2021 | Kauser et al. [49] | Ensemble-based Model | EBDL | Fine-tuned InceptionResNetV2 with Ensemble | ISIC-19 | 98 | 99 | 99 | 99 | 🗴 | 🗴 | 🗴 |
| 2022 | Charan et al. [56] | Two-path CNN | CNN | Dual-input CNN | HAM 10000 | 88.6 | 🗴 | 🗴 | 🗴 | 🗴 | 🗴 | 🗴 |
| 2022 | Hoang et al. [50] | ShuffleNet | TL | Segmentation-based ShuffleNet | HAM 10000 | 84.8 | 75.15 | 🗴 | 72.61 | 🗴 | 🗴 | 🗴 |
| 2023 | Proposed Study | EfficientNet B3 + Attention | TL | Fine-tuned EfficientNetB3 with Attention | HAM 10000 | 98.96 | 99 | 99 | 99 | 0.996 | p < 0.007 | 🗴 |
| 2023 | Proposed Study | Ensemble-based Model | EBDL | Ensemble Model of TL models with Attention | HAM 10000 | 99.73 | 100 | 100 | 100 | 0.9989 | p < 0.003 | 🗴 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanga, P.; Singh, J.; Dubey, A.K.; Khanna, N.N.; Laird, J.R.; Faa, G.; Singh, I.M.; Tsoulfas, G.; Kalra, M.K.; Teji, J.S.; et al. DermAI 1.0: A Robust, Generalized, and Novel Attention-Enabled Ensemble-Based Transfer Learning Paradigm for Multiclass Classification of Skin Lesion Images. Diagnostics 2023, 13, 3159. https://doi.org/10.3390/diagnostics13193159
Sanga P, Singh J, Dubey AK, Khanna NN, Laird JR, Faa G, Singh IM, Tsoulfas G, Kalra MK, Teji JS, et al. DermAI 1.0: A Robust, Generalized, and Novel Attention-Enabled Ensemble-Based Transfer Learning Paradigm for Multiclass Classification of Skin Lesion Images. Diagnostics. 2023; 13(19):3159. https://doi.org/10.3390/diagnostics13193159
Chicago/Turabian StyleSanga, Prabhav, Jaskaran Singh, Arun Kumar Dubey, Narendra N. Khanna, John R. Laird, Gavino Faa, Inder M. Singh, Georgios Tsoulfas, Mannudeep K. Kalra, Jagjit S. Teji, and et al. 2023. "DermAI 1.0: A Robust, Generalized, and Novel Attention-Enabled Ensemble-Based Transfer Learning Paradigm for Multiclass Classification of Skin Lesion Images" Diagnostics 13, no. 19: 3159. https://doi.org/10.3390/diagnostics13193159
APA StyleSanga, P., Singh, J., Dubey, A. K., Khanna, N. N., Laird, J. R., Faa, G., Singh, I. M., Tsoulfas, G., Kalra, M. K., Teji, J. S., Al-Maini, M., Rathore, V., Agarwal, V., Ahluwalia, P., Fouda, M. M., Saba, L., & Suri, J. S. (2023). DermAI 1.0: A Robust, Generalized, and Novel Attention-Enabled Ensemble-Based Transfer Learning Paradigm for Multiclass Classification of Skin Lesion Images. Diagnostics, 13(19), 3159. https://doi.org/10.3390/diagnostics13193159











