Distribution of Human Papillomavirus Genotypes among the Women of South Andaman Island, India
Abstract
:1. Background
2. Methodology
2.1. Study Population
2.2. Exclusion Criteria
2.3. Ethical Approval
2.4. Sampling and Sample Size
2.5. Awareness Programmes
2.6. Sample Collection and Storage
2.7. Sample Processing
2.8. DNA Extraction
2.9. PCR Assays
2.10. Detection of HPV 16 & 18
2.11. PCR Sequencing
2.12. Phylogenetic Analysis
3. Results
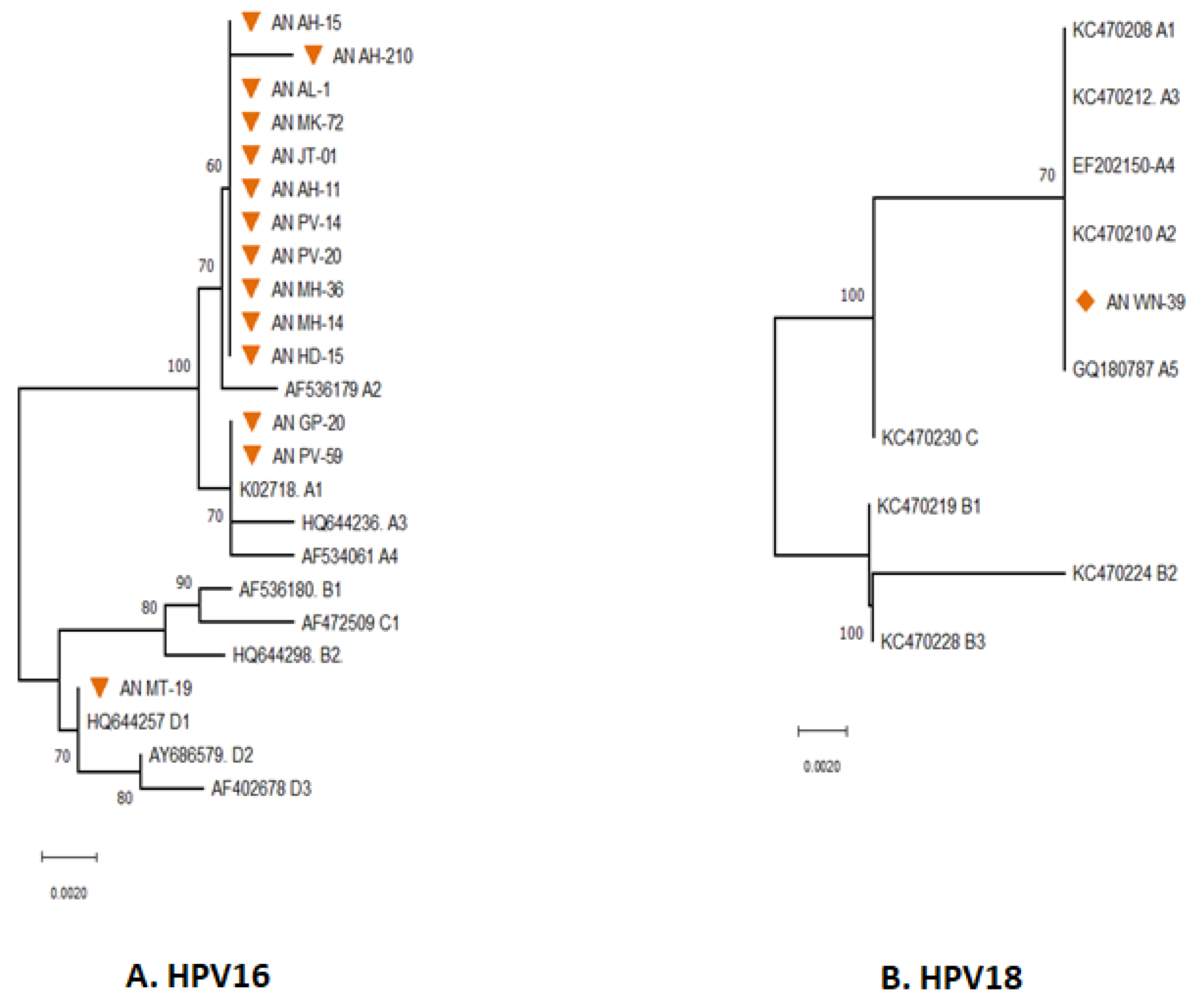
3.1. Phylogenetic Analysis of E6 Gene
3.2. Phylogenetic Analysis of E7 Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organisation Team. Cervical Cancer. World Health Organisation (WHO). 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/cervical-cancer (accessed on 20 April 2023).
- International Agency for Research on Cancer (IARC). Global Cancer Observatory (GCO). World Health Organisation-International Agency for Research on Cancer. 2020. Available online: http://gco.iarcfr/today/online-analysis (accessed on 20 April 2023).
- International Institute for Population Sciences (IIPS). National Family Health Survey (NFHS-4), India, 2015–2016. IIPS. 2017. Available online: http://rchiips.org/nfhs/nfhs-4Reports/India.pdf (accessed on 12 November 2022).
- Thobias, A.R.; Patel, K.A.; Gokani, R.; Parekh, C.; Desai, A.; Patel, J.B. Prevalence of Human Papilloma Virus Infection in Cervical Cancer Patients from Western Region of India. Indian J. Gynecol. Oncol. 2019, 17, 41. [Google Scholar] [CrossRef]
- Press, D. Epidemiology of cervical cancer with special focus on India. Int. J. Womens Health 2015, 7, 405–414. [Google Scholar]
- Graham, S.V. Human papillomavirus: Gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010, 5, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation—International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. World Health Organisation. Volume 90 Human Papillomaviruses. 2007. Available online: https://monographs.iarc.who.int/wp-content/uploads/2018/06/mono90.pdf (accessed on 21 May 2023).
- Muñoz, N.; Bosch, F.X.; De Sanjosé, S.; Herrero, R.; Castellsagué, X.; Shah, K.V.; Snijders, P.J.F.; Meijer, C.J.L.M. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Tommasino, M.; Depuydt, C.; Dillner, J. Are 20 human papillomavirus types causing cervical cancer? J. Pathol. 2014, 234, 431–435. [Google Scholar] [CrossRef]
- Office of the Registrar General & Census Commissioner, India. Census 2011. Ministry of Home Affairs, Government of India. 2011. Available online: https://censusindia.gov.in/census.website/data/data-visualizations/PopulationSearch_PCA_Indicators (accessed on 20 April 2023).
- Parvez, R.; Hedau, S.; Bhattacharya, D.; Bhattacharjee, H.; Muruganandam, N.; Das, B.; Saha, M.; Sugunan, A.; Vijayachari, P. High-risk HPV infection among the tribal and non-tribal women of the Andaman and Nicobar Islands, India. Public Health 2012, 126, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Gopalkrishna, V.; Aggarwal, N.; Malhotra, V.; Koranne, R.; Mohan, V.; Mittal, A.; Das, B. Chlamydia trachomatis and human papillomavirus infection in Indian women with sexually transmitted diseases and cervical precancerous and cancerous lesions. Clin. Microbiol. Infect. 2000, 6, 88–93. [Google Scholar] [CrossRef]
- Das, B.C.; Sharma, J.K.; Gopalkrishna, V.; Das, D.K.; Singh, V.; Gissmann, L.; Hausen, H.Z.; Luthra, U.K. A high frequency of human papillomavirus DNA sequences in cervical carcinomas of Indian women as revealed by southern blot hybridization and polymerase chain reaction. J. Med. Virol. 1992, 36, 239–245. [Google Scholar] [CrossRef]
- Pande, S.; Jain, N.; Prusty, B.K.; Bhambhani, S.; Gupta, S.; Sharma, R.; Batra, S.; Das, B.C. Human papillomavirus type 16 variant analysis of E6, E7, and L1 genes and long control region in biopsy samples from cervical cancer patients in north India. J. Clin. Microbiol. 2008, 46, 1060–1066. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Du, X.; Lu, M.; Zhang, W.; Sun, Z.; Li, L.; Ye, M.; Fan, W.; Jiang, S.; Liu, A.; et al. Prevalence characteristics of single and multiple HPV infections in women with cervical cancer and precancerous lesions in Beijing, China. J. Med. Virol. 2019, 91, 473–481. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. HPV and Cancer. National Institute of Health. 2023. Available online: https://www.cancer.gov/about-cancer/causes-prevention/risk/infectious-agents/hpv-and-cancer (accessed on 20 April 2023).
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Collado, J.J.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in Asia. Summary Report 10 March 2023. Available online: https://hpvcentre.net/statistics/reports/XWX.pdf (accessed on 20 April 2023).
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Collado, J.J.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in India. Summary Report 10 March 2023. Available online: https://hpvcentre.net/statistics/reports/IND.pdf (accessed on 20 April 2023).
- Gheit, T.; Vaccarella, S.; Schmitt, M.; Pawlita, M.; Franceschi, S.; Sankaranarayanan, R.; Sylla, B.S.; Tommasino, M.; Gangane, N. Prevalence of human papillomavirus types in cervical and oral cancers in central India. Vaccine 2009, 27, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Munjal, K.; Adamson, C.S.; Rajendran, V.; Nandedkar, S.; Cooper, K.; Evans, M.F. Human papillomavirus type distribution in invasive cervical cancers from Madhya Pradesh: Implications for vaccination programs in central India. Int. J. Gynecol. Pathol. 2014, 33, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.M.; Tenet, V.; Georges, D.; Alemany, L.; Pavón, M.A.; Chen, Z.; Yeager, M.; Cullen, M.; Boland, J.F.; Bass, S.; et al. Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: Whole viral genome sequences from 7116 HPV 16-positive women. Papillomavirus Res. 2019, 7, 67–74. [Google Scholar] [CrossRef]
- Pérez, S.; Cid, A.; Iñarrea, A.; Pato, M.; Lamas, M.J.; Couso, B.; Gil, M.; Álvarez, M.J.; Rey, S.; López-Miragaya, I.; et al. Prevalence of HPV 16 and HPV 18 Lineages in Galicia, Spain. PLoS ONE 2014, 9, e104678. [Google Scholar] [CrossRef]
- Mandal, P.; Bhattacharjee, B.; Sen, S.; Bhattacharya, A.; Saha, S.S.; Chowdhury, R.R.; Mondal, N.R.; Chakrabarty, B.; Chatterjee, T.; Roy, S.; et al. Predominance of genomically defined A lineage of HPV 16 over D lineage in Indian patients from eastern India with squamous cell carcinoma of the cervix in association with distinct oncogenic phenotypes. Transl. Oncol. 2022, 15, 101256. [Google Scholar] [CrossRef]
- Chen, A.A.; Gheit, T.; Franceschi, S.; Tommasino, M.; Clifford, G.M. IARC HPV Variant Study Group. Human Papillomavirus 18 Genetic Variation and Cervical Cancer Risk Worldwide. J. Virol. 2015, 89, 10680–10687. [Google Scholar] [CrossRef]
- Zu, Y.; Ou, Z.; Wu, D.; Liu, W.; Liu, L.; Wu, D.; Zhao, Y.; Ren, P.; Zhang, Y.; Li, W.; et al. Genetic characteristics of human papillomavirus type 16, 18, 52 and 58 in southern China. Genomics 2021, 113, 3895–3906. [Google Scholar] [CrossRef]
- Wang, X.; Han, S.; Li, X.; Wang, X.; Wang, S.; Ma, L. Prevalence and distribution of humanpapillomavirus (HPV) in Luoyang city of Henan province during 2015–2021 and the genetic variability of HPV 16 and 52. Virol. J. 2022, 19, 37. [Google Scholar] [CrossRef]
- Salavatiha, Z.; Shoja, Z.; Heydari, N.; Marashi, S.M.; Younesi, S.; Nozarian, Z.; Jalilvand, S. Lineage analysis of human papillomavirus type 18 based on E6 region in cervical samples of Iranian women. J. Med. Virol. 2020, 92, 3815–3820. [Google Scholar] [CrossRef] [PubMed]


| S.No | Lab. ID | Urban/Rural | HPV Genotype | Genetically Close Related Worldwide Reference Sequences of HPV Types | K2P Distance Value | |
|---|---|---|---|---|---|---|
| Accession Number | Country Origin | |||||
| HR-HPV Types | ||||||
| 1 | AN-MT-19 | Rural | HPV 16 | AF472509 | NY, USA | 0.035 |
| 2 | AN-MK-72 | Rural | HPV 16 | AF534061 | NY, USA | 0.002 |
| 3 | AN-AH-11(i) | Rural | HPV 16 | AF534061 | NY, USA | 0.000 |
| 4 | AN-AL1 | Rural | HPV 16 | AF534061 | NY, USA | 0.000 |
| 5 | AN-PV-40(i) | Urban | HPV 16 | AF534061 | NY, USA | 0.000 |
| 6 | AN-AH-210 | Urban | HPV 16 | AF534061 | NY, USA | 0.000 |
| 7 | AN-MH-36 | Rural | HPV 16 | K02718 | Britain | 0.022 |
| 8 | AN-PV-03 | Urban | HPV 33 | M12732 | NY, USA | 0.007 |
| 9 | AN-PV-06 | Urban | HPV 52 | AB819273 | Japan | 0.000 |
| 10 | AN-WN-08 | Rural | HPV 58 | D90400 | Osaka, Japan | 0.004 |
| 11 | AN-WN-34 | Rural | HPV 58 | D90400 | Osaka, Japan | 0.004 |
| 12 | AN-AH-53 | Urban | HPV 66 | EF177188 | NY, USA | 0.017 |
| 13 | AN-AH-88 | Urban | HPV 66 | EF177188 | NY, USA | 0.022 |
| 14 | AN-AH-09 | Urban | HPV 73 | X94165 | Germany | 0.000 |
| 15 | AN-TM-08 | Rural | HPV 53 | EF546482 | NY, USA | 0.000 |
| LR-HPV types | ||||||
| 16 | AN-MH-45 | Rural | HPV 6 | FR751328 | Slovania | 0.009 |
| 17 | AN-NS-28 | Rural | HPV 6 | FR751328 | Slovania | 0.002 |
| 18 | AN-AH-32 | Urban | HPV 30 | KF436837 | NY, USA | 0.028 |
| 19 | AN-AH-11(ii) | Rural | HPV 61 | U31793 | Los Alamos, NM, USA | 0.00 |
| 20 | AN-PV-40(ii) | Urban | HPV 71 | NC039089 | Tokyo, Japan | 0.002 |
| 21 | AN-MT-04 | Rural | HPV 81 | KU298939 | Brazil | 0.039 |
| 22 | AN-FG-19 | Rural | HPV 84 | AJ621384 | Cyprus | 0.053 |
| 23 | AN-AH-15 | Rural | HPV 84 | AJ621384 | Cyprus | 0.00 |
| 24 | AN-AH-41(ii) | Rural | HPV 87 | KU298942 | Brazil | 0.00 |
| Genus-Species | Genotype | Risk Group | Frequency (n) | Percentage (%) |
|---|---|---|---|---|
| Alpha-PV-9 | HPV 16 | High-Risk | 32 | 57.2 |
| Alpha-PV-9 | HPV 52 | High-Risk | 1 | 1.8 |
| Alpha-PV-9 | HPV 58 | High-Risk | 2 | 3.6 |
| Alpha-PV-6 | HPV 66 | High-Risk | 3 | 5.3 |
| Alpha-PV-9 | HPV 33 | High-Risk | 1 | 1.8 |
| Alpha-PV-7 | HPV 18 | High-Risk | 3 | 5.3 |
| Alpha-PV-11 | HPV 73 | High-Risk | 2 | 3.5 |
| Alpha-PV-6 | HPV 53 | High-Risk | 1 | 1.8 |
| Alpha-PV-6 | HPV 30 | Low-Risk | 1 | 1.8 |
| Alpha-PV-10 | HPV 6 | Low-Risk | 2 | 3.6 |
| Alpha-PV-3 | HPV 61 | Low-Risk | 1 | 1.8 |
| Alpha-PV-14 | HPV 71 | Low-Risk | 1 | 1.8 |
| Alpha-PV-3 | HPV 81 | Low-Risk | 1 | 1.8 |
| Alpha-PV-3 | HPV 84 | Low-Risk | 2 | 3.6 |
| Alpha-PV-3 | HPV 87 | Low-Risk | 1 | 1.8 |
| Un-typed | - | - | 2 | 3.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parvez, R.; Vijayachari, P.; Saha, M.K.; Biswas, L.; Ramasamy, J.; Vins, A.; Beniwal, N.; Vasanthi, S.; Ramadoss, S.; Kaur, H.; et al. Distribution of Human Papillomavirus Genotypes among the Women of South Andaman Island, India. Diagnostics 2023, 13, 2765. https://doi.org/10.3390/diagnostics13172765
Parvez R, Vijayachari P, Saha MK, Biswas L, Ramasamy J, Vins A, Beniwal N, Vasanthi S, Ramadoss S, Kaur H, et al. Distribution of Human Papillomavirus Genotypes among the Women of South Andaman Island, India. Diagnostics. 2023; 13(17):2765. https://doi.org/10.3390/diagnostics13172765
Chicago/Turabian StyleParvez, Rehnuma, Paluru Vijayachari, Mrinmoy Kumar Saha, Lipika Biswas, Jawahar Ramasamy, Alwin Vins, Nisha Beniwal, S. Vasanthi, Sasikala Ramadoss, Harpreet Kaur, and et al. 2023. "Distribution of Human Papillomavirus Genotypes among the Women of South Andaman Island, India" Diagnostics 13, no. 17: 2765. https://doi.org/10.3390/diagnostics13172765
APA StyleParvez, R., Vijayachari, P., Saha, M. K., Biswas, L., Ramasamy, J., Vins, A., Beniwal, N., Vasanthi, S., Ramadoss, S., Kaur, H., & Nagarajan, M. (2023). Distribution of Human Papillomavirus Genotypes among the Women of South Andaman Island, India. Diagnostics, 13(17), 2765. https://doi.org/10.3390/diagnostics13172765






