Identification of the Immune Landscapes and Follicular Helper T Cell-Related Genes for the Diagnosis of Age-Related Macular Degeneration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Filtering
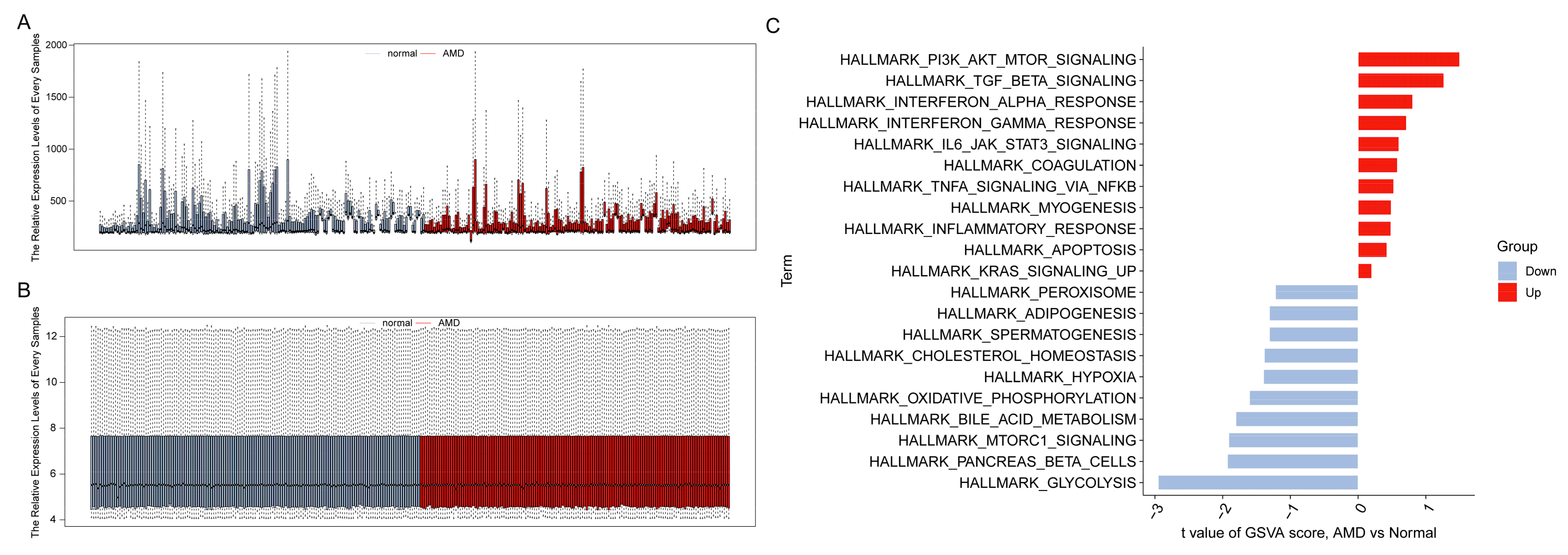
2.2. Gene Set Variation Analysis
2.3. CIBERSORT Analysis
2.4. Weighted Gene Correlation Network Analysis
2.5. Differently Expressed Genes Analysis
2.6. Identified the Potential Immune-Related Biomarkers of AMD
3. Results
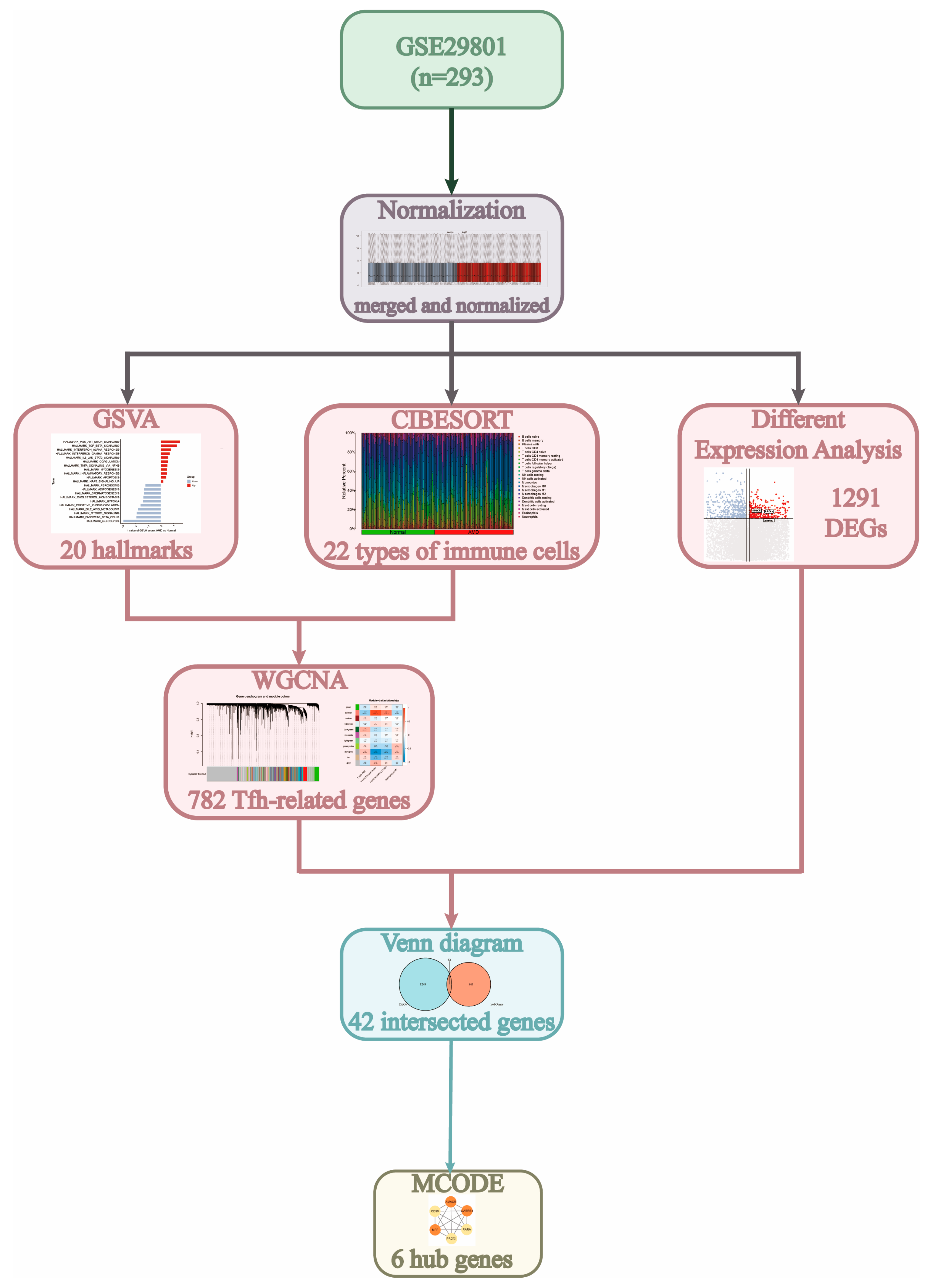
3.1. Study Process and Data Normalization
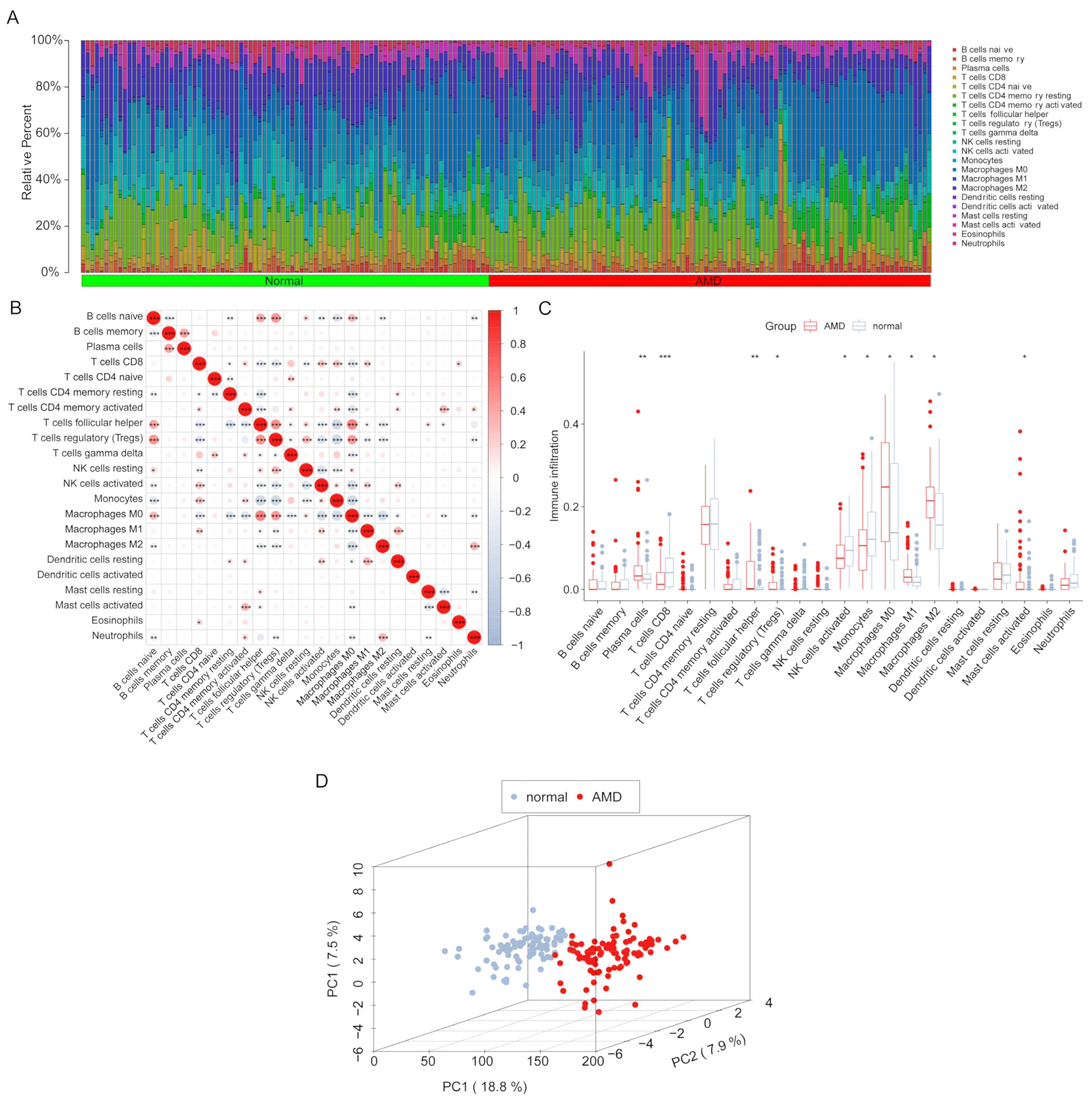
3.2. Infiltration Levels of Immune Cells
3.3. Filtration of Immune Cell-Related Modules
3.4. Identified the Tfh-Related Hub Genes
3.5. Development of Risk-Model and Construction of the Diagnostic Nomogram
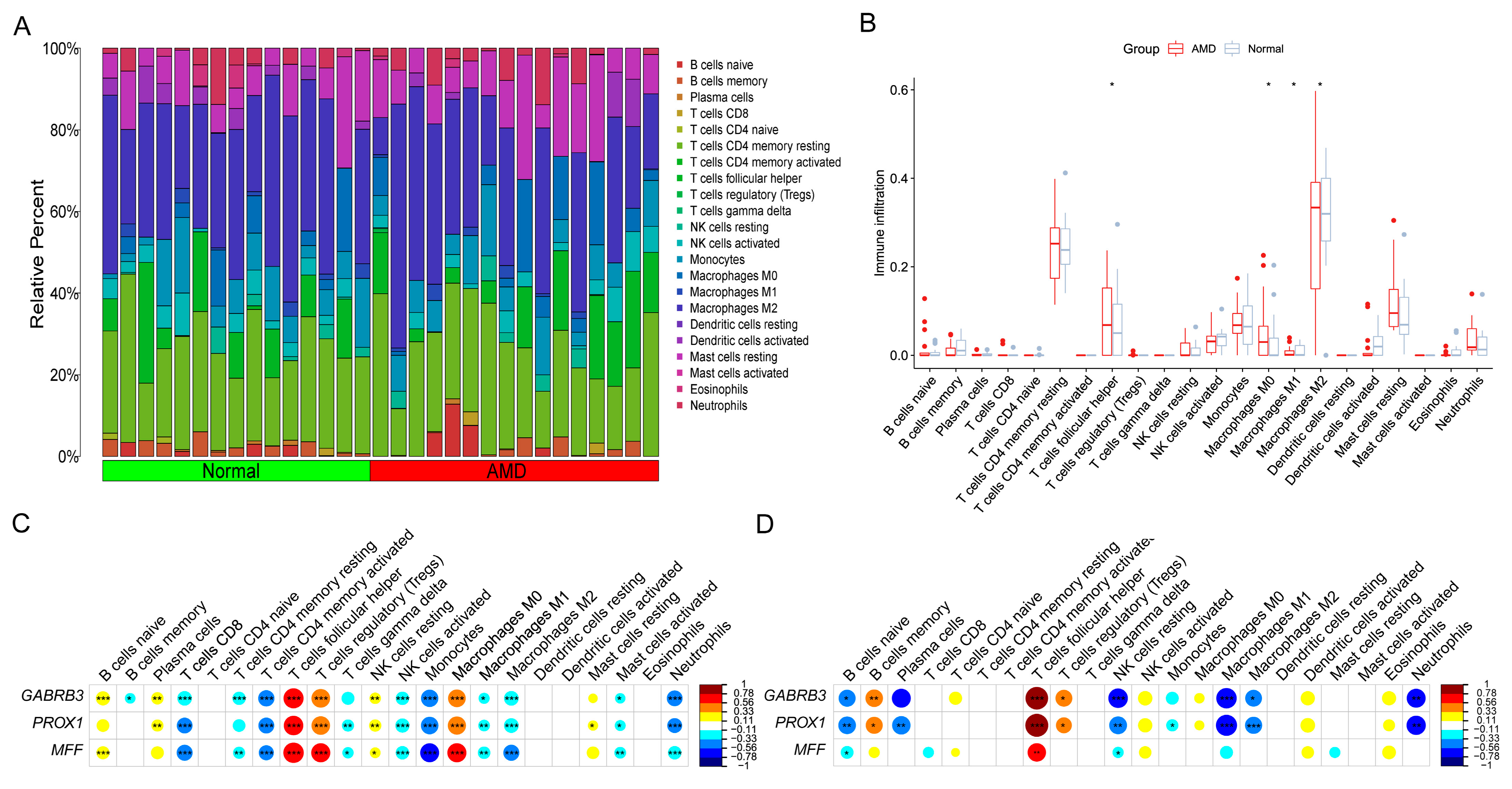
3.6. Correlation Analysis between Model-Related Risk Score and Tfh Cells Infiltration Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, J.; Huang, X.; Dou, L.; Yan, M.; Shen, T.; Tang, W.; Li, J. Aging and Aging-Related Diseases: From Molecular Mechanisms to Interventions and Treatments. Signal Transduct. Target Ther. 2022, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Qiao, L.; Du, M.; Qu, C.; Wan, L.; Li, J.; Huang, L. Age-Related Macular Degeneration: Epidemiology, Genetics, Pathophysiology, Diagnosis, and Targeted Therapy. Genes Dis. 2022, 9, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Kevany, B.M.; Palczewski, K. Phagocytosis of Retinal Rod and Cone Photoreceptors. Physiology 2010, 25, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Molins, B.; Romero-Vázquez, S.; Fuentes-Prior, P.; Adan, A.; Dick, A.D. C-Reactive Protein as a Therapeutic Target in Age-Related Macular Degeneration. Front. Immunol. 2018, 9, 808. [Google Scholar] [CrossRef]
- Hernández-Zimbrón, L.F.; Zamora-Alvarado, R.; Ochoa-De la Paz, L.; Velez-Montoya, R.; Zenteno, E.; Gulias-Cañizo, R.; Quiroz-Mercado, H.; Gonzalez-Salinas, R. Age-Related Macular Degeneration: New Paradigms for Treatment and Management of AMD. Oxidative Med. Cell. Longev. 2018, 2018, 8374647. [Google Scholar] [CrossRef]
- Kijlstra, A.; La Heij, E.; Hendrikse, F. Immunological Factors in the Pathogenesis and Treatment of Age-Related Macular Degeneration. Ocul. Immunol. Inflamm. 2005, 13, 3–11. [Google Scholar] [CrossRef]
- Rosenberg, D.; Deonarain, D.M.; Gould, J.; Sothivannan, A.; Phillips, M.R.; Sarohia, G.S.; Sivaprasad, S.; Wykoff, C.C.; Cheung, C.M.G.; Sarraf, D.; et al. Efficacy, Safety, and Treatment Burden of Treat-and-Extend versus Alternative Anti-VEGF Regimens for NAMD: A Systematic Review and Meta-Analysis. Eye 2023, 37, 6–16. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Li, Q. Increased Th1/Th17 Responses Contribute to Low-Grade Inflammation in Age-Related Macular Degeneration. Cell Physiol. Biochem. 2017, 44, 357–367. [Google Scholar] [CrossRef]
- Ezzat, M.-K.; Hann, C.R.; Vuk-Pavlovic, S.; Pulido, J.S. Immune Cells in the Human Choroid. Br. J. Ophthalmol. 2008, 92, 976–980. [Google Scholar] [CrossRef]
- Morita, R.; Schmitt, N.; Bentebibel, S.-E.; Ranganathan, R.; Bourdery, L.; Zurawski, G.; Foucat, E.; Dullaers, M.; Oh, S.; Sabzghabaei, N.; et al. Human Blood CXCR5(+)CD4(+) T Cells Are Counterparts of T Follicular Cells and Contain Specific Subsets That Differentially Support Antibody Secretion. Immunity 2011, 34, 108–121. [Google Scholar] [CrossRef]
- Choi, Y.S.; Yang, J.A.; Yusuf, I.; Johnston, R.J.; Greenbaum, J.; Peters, B.; Crotty, S. Bcl6 Expressing Follicular Helper CD4 T Cells Are Fate Committed Early and Have the Capacity to Form Memory. J. Immunol. 2013, 190, 4014–4026. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Deenick, E.K. Human T Follicular Helper (Tfh) Cells and Disease. Immunol. Cell Biol. 2014, 92, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef]
- Li, Y.; Fu, J.; Liu, J.; Feng, H.; Chen, X. Diagnostic Markers and Molecular Dysregulation Mechanisms in the Retinal Pigmented Epithelium and Retina of Age-Related Macular Degeneration. J. Healthc. Eng. 2022, 2022, 3787567. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Grant, G.R.; Bowman, A.S.; Haider, N.; Gudiseva, H.V.; Chavali, V.R.M. Complete Transcriptome Profiling of Normal and Age-Related Macular Degeneration Eye Tissues Reveals Dysregulation of Anti-Sense Transcription. Sci. Rep. 2018, 8, 3040. [Google Scholar] [CrossRef]
- Nagy, Á.; Győrffy, B. MuTarget: A Platform Linking Gene Expression Changes and Mutation Status in Solid Tumors. Int. J. Cancer 2021, 148, 502–511. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Zhou, S.; Lu, H.; Xiong, M. Identifying Immune Cell Infiltration and Effective Diagnostic Biomarkers in Rheumatoid Arthritis by Bioinformatics Analysis. Front. Immunol. 2021, 12, 726747. [Google Scholar] [CrossRef]
- Jia, J.; Qiu, D.; Lu, C.; Wang, W.; Li, N.; Han, Y.; Tong, P.; Sun, X.; Wu, M.; Dai, J. Transcriptome Analysis of Choroid and Retina From Tree Shrew With Choroidal Neovascularization Reveals Key Signaling Moieties. Front. Genet. 2021, 12, 654955. [Google Scholar] [CrossRef]
- Li, Y.; Masse-Ranson, G.; Garcia, Z.; Bruel, T.; Kök, A.; Strick-Marchand, H.; Jouvion, G.; Serafini, N.; Lim, A.I.; Dusseaux, M.; et al. A Human Immune System Mouse Model with Robust Lymph Node Development. Nat. Methods 2018, 15, 623–630. [Google Scholar] [CrossRef]
- Caceres, P.S.; Rodriguez-Boulan, E. Retinal Pigment Epithelium Polarity in Health and Blinding Diseases. Curr. Opin. Cell Biol. 2020, 62, 37–45. [Google Scholar] [CrossRef]
- Chen, M.; Xu, H. Parainflammation, Chronic Inflammation, and Age-Related Macular Degeneration. J. Leukoc. Biol. 2015, 98, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Olatunde, A.C.; Hale, J.S.; Lamb, T.J. Cytokine-Skewed Tfh Cells: Functional Consequences for B Cell Help. Trends Immunol. 2021, 42, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Behnke, V.; Wolf, A.; Langmann, T. The Role of Lymphocytes and Phagocytes in Age-Related Macular Degeneration (AMD). Cell Mol. Life Sci. 2020, 77, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Z.; Lai, P.; Huang, Z.; Sun, X.; Zhou, T.; He, C.; Liu, X. Bcl-6-Directed Follicular Helper T Cells Promote Vascular Inflammatory Injury in Diabetic Retinopathy. Theranostics 2020, 10, 4250–4264. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, B.; Yuan, L.; Peng, Q.; Cheng, L.; Zhong, P.; Yang, X.; Yu, H. Dysregulations of Follicular Helper T Cells through IL-21 Pathway in Age-Related Macular Degeneration. Mol. Immunol. 2019, 114, 243–250. [Google Scholar] [CrossRef]
- Yang, Y.; Zeng, Q.; Cheng, M.; Niu, X.; Xiangwei, W.; Gong, P.; Li, W.; Ma, J.; Zhang, X.; Yang, X.; et al. GABRB3-Related Epilepsy: Novel Variants, Clinical Features and Therapeutic Implications. J. Neurol. 2022, 269, 2649–2665. [Google Scholar] [CrossRef]
- Pangou, E.; Bielska, O.; Guerber, L.; Schmucker, S.; Agote-Arán, A.; Ye, T.; Liao, Y.; Puig-Gamez, M.; Grandgirard, E.; Kleiss, C.; et al. A PKD-MFF Signaling Axis Couples Mitochondrial Fission to Mitotic Progression. Cell Rep. 2021, 35, 109129. [Google Scholar] [CrossRef]
- Doğanlar, Z.B.; Doğanlar, O.; Kurtdere, K.; Güçlü, H.; Chasan, T.; Turgut, E. Melatonin Prevents Blood-Retinal Barrier Breakdown and Mitochondrial Dysfunction in High Glucose and Hypoxia-Induced in Vitro Diabetic Macular Edema Model. Toxicol. Vitr. 2021, 75, 105191. [Google Scholar] [CrossRef]
- Peng, F.; Jiang, D.; Xu, W.; Sun, Y.; Zha, Z.; Tan, X.; Yu, J.; Pan, C.; Zheng, Q.; Chen, W. AMPK/MFF Activation: Role in Mitochondrial Fission and Mitophagy in Dry Eye. Investig. Ophthalmol. Vis. Sci. 2022, 63, 18. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2020, 15, 235–259. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, R.; Higgs, H.N. Revolutionary View of Two Ways to Split a Mitochondrion. Nature 2021, 593, 346–347. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Tian, Q.; Gao, H.; Wu, K.; Wang, B.; Ge, G.; Jiang, S.; Wang, K.; Zhou, C.; He, J.; et al. PROX1 Promotes Breast Cancer Invasion and Metastasis through WNT/β-Catenin Pathway via Interacting with HnRNPK. Int. J. Biol. Sci. 2022, 18, 2032–2046. [Google Scholar] [CrossRef]
- Huang, X.; Hao, S.; Liu, J.; Huang, Y.; Liu, M.; Xiao, C.; Wang, Y.; Pei, S.; Yu, T.; Xu, J.; et al. The Ubiquitin Ligase Peli1 Inhibits ICOS and Thereby Tfh-Mediated Immunity. Cell Mol. Immunol. 2021, 18, 969–978. [Google Scholar] [CrossRef] [PubMed]



| Variable | Coefficient | Odds Ratio (95% CI) | p-Value |
|---|---|---|---|
| (Intercept) | −36.214 | 0 (0~0.004) | 0.029 |
| AMACR | 0.666 | 1.946 (0.742~5.627) | 0.192 |
| GABRB3 | −1.198 | 0.302 (0.088~0.930) | 0.045 |
| PROX1 | 1.228 | 3.416 (1.210~10.853) | 0.027 |
| MFF | 2.618 | 13.712 (1.555~162.893) | 0.025 |
| CD99 | 0.848 | 2.335 (0.554~10.779) | 0.258 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Sun, Z.; Li, Z.; Wang, Q.; Yan, M.; Li, W.; Xu, K.; Shen, T. Identification of the Immune Landscapes and Follicular Helper T Cell-Related Genes for the Diagnosis of Age-Related Macular Degeneration. Diagnostics 2023, 13, 2732. https://doi.org/10.3390/diagnostics13172732
Yang Y, Sun Z, Li Z, Wang Q, Yan M, Li W, Xu K, Shen T. Identification of the Immune Landscapes and Follicular Helper T Cell-Related Genes for the Diagnosis of Age-Related Macular Degeneration. Diagnostics. 2023; 13(17):2732. https://doi.org/10.3390/diagnostics13172732
Chicago/Turabian StyleYang, Yao, Zhiqiang Sun, Zhenping Li, Que Wang, Mingjing Yan, Wenlin Li, Kun Xu, and Tao Shen. 2023. "Identification of the Immune Landscapes and Follicular Helper T Cell-Related Genes for the Diagnosis of Age-Related Macular Degeneration" Diagnostics 13, no. 17: 2732. https://doi.org/10.3390/diagnostics13172732
APA StyleYang, Y., Sun, Z., Li, Z., Wang, Q., Yan, M., Li, W., Xu, K., & Shen, T. (2023). Identification of the Immune Landscapes and Follicular Helper T Cell-Related Genes for the Diagnosis of Age-Related Macular Degeneration. Diagnostics, 13(17), 2732. https://doi.org/10.3390/diagnostics13172732






