EML4-ALK Gene Mutation Detected with New NGS Lung Cancer Panel CDx Using Sputum Cytology in a Case of Advanced NSCLC
Abstract
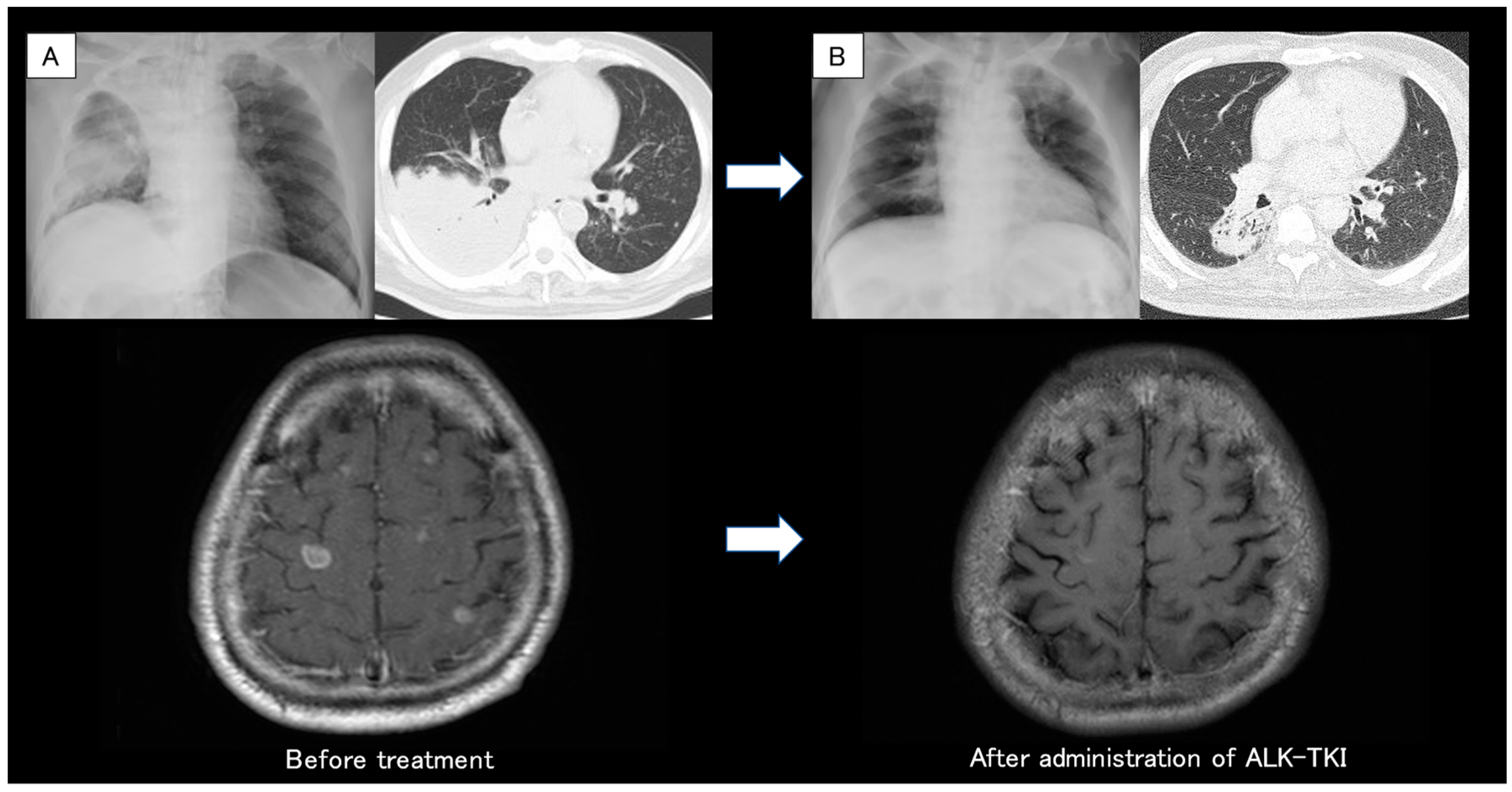
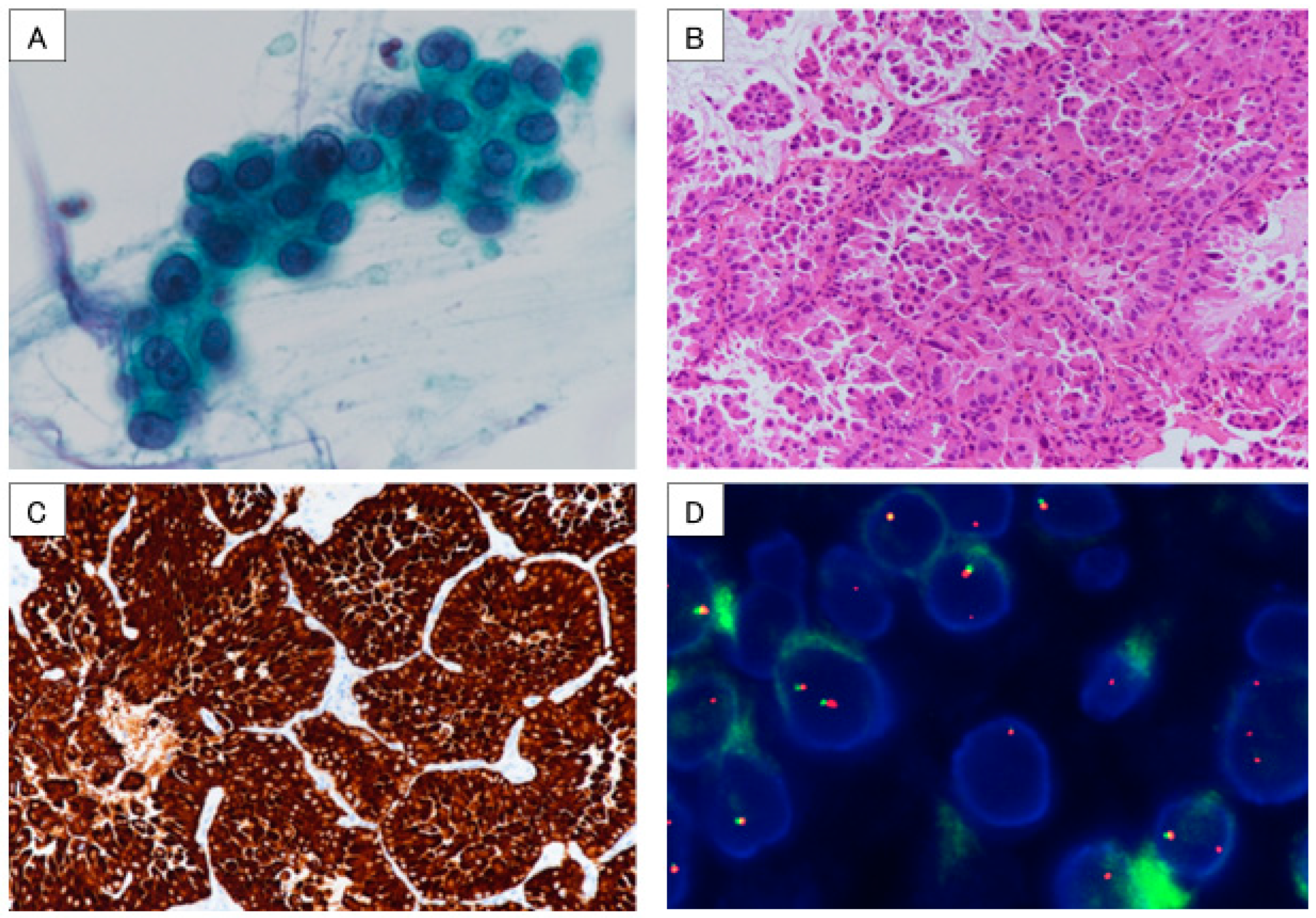
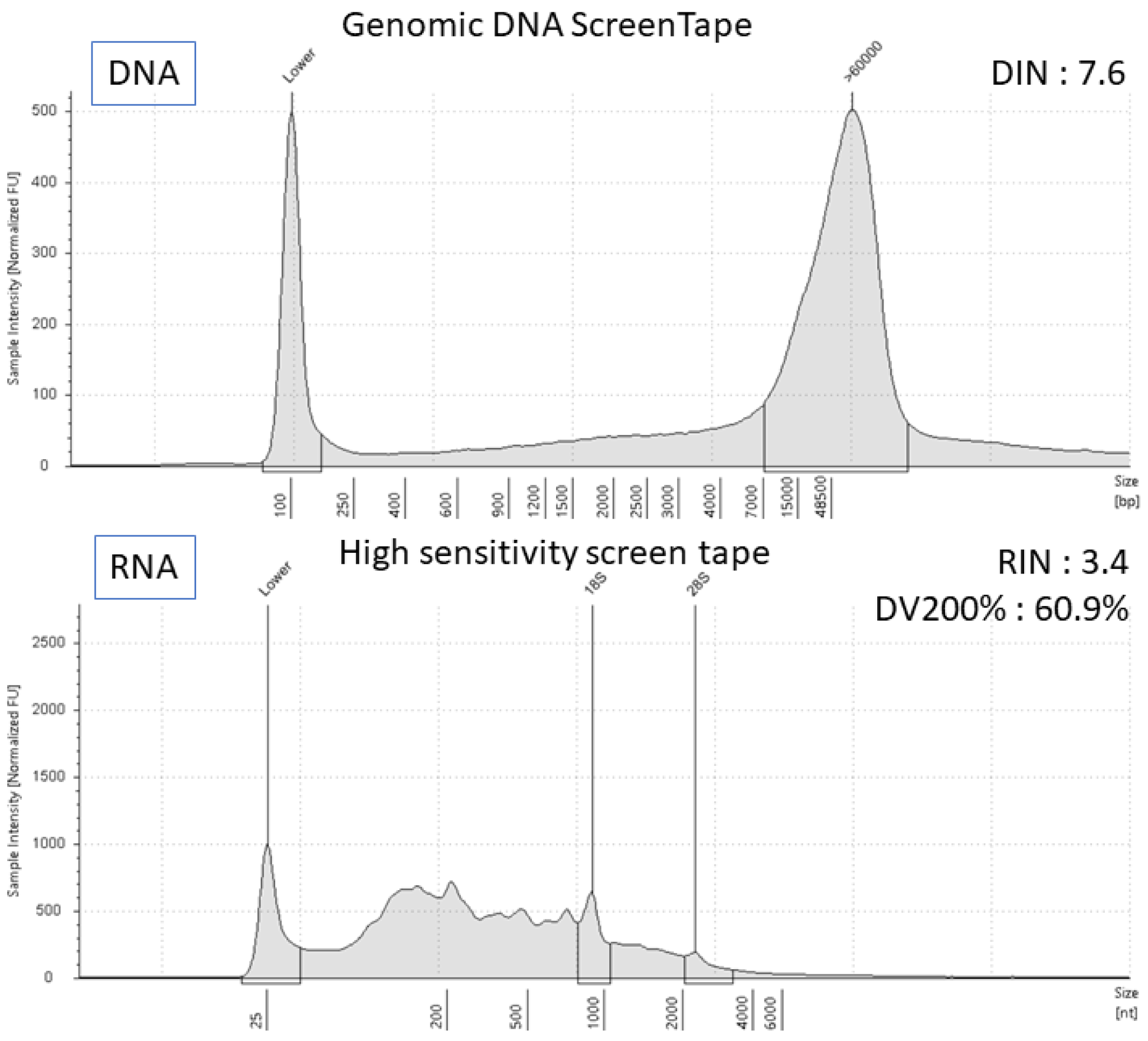
:
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanna, N.; Johnson, D.; Temin, S.; Baker, S.; Brahmer, J.; Ellis, P.M.; Giaccone, G.; Hesketh, P.J.; Jaiyesimi, I.; Leighl, N.B.; et al. Systemic therapy for stage IV non-small cell lung cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J. Clin. Oncol. 2017, 35, 3484–3515. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, K.; Kida, H.; Handa, H.; Inoue, T.; Saji, H.; Koike, J.; Nakamura, S.; Sato, Y.; Ueda, Y.; Suzuki, F.; et al. A Prospective Validation Study of Lung Cancer Gene Panel Testing Using Cytological Specimens. Cancers 2022, 14, 3784. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, K.; Kinoshita, K.; Kida, H.; Inoue, T.; Mineshita, M. Preliminary Results of NGS Gene Panel Test Using NSCLC Sputum Cytology and Therapeutic Effect Using Corresponding Molecular-Targeted Drugs. Genes 2022, 13, 812. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Okami, J.; Nakamura, H.; Honma, K.; Sato, Y.; Nakamura, S.; Kukita, Y.; Nakatsuka, S.-I.; Higashiyama, M. Analytical performance of a highly sensitive system to detect gene variants using next-generation sequencing for lung cancer companion diagnostics. Diagnostics 2023, 13, 1476. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.P.; Mehta, A.C.; Wahidi, M.M. Establishing the Diagnosis of Lung Cancer. Chest 2013, 143, e142S–e165S. [Google Scholar] [CrossRef] [PubMed]
- Petty, T.L. Sputum cytology for the detection of early lung cancer. Curr. Opin. Pulm. Med. 2003, 9, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Petty, T.L.; Tockman, M.S.; Palcic, B. Diagnosis of roentgenographically occult lung cancer by sputum cytology. Clin. Chest Med. 2002, 23, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Isaka, T.; Yokose, T.; Ito, H.; Nakayama, H.; Miyagi, Y.; Saito, H.; Masuda, M. Detection of EGFR mutation of pulmonary adenocarcinoma in sputum using droplet digital PCR. BMC Pulm. Med. 2021, 21, 100. [Google Scholar] [CrossRef] [PubMed]
- Hackner, K.; Buder, A.; Hochmair, M.J.; Strieder, M.; Grech, C.; Fabikan, H.; Burghuber, O.C.; Errhalt, P.; Filipits, M. Detection of EGFR Activating and Resistance Mutations by Droplet Digital PCR in Sputum of EGFR-Mutated NSCLC Patients. Clin. Med. Insights Oncol. 2021, 15, 1179554921993072. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, Z.; Li, C.S.; Zhao, W.; Liang, Z.X.; Liang, Z.X.; Dai, Y.; Zhu, Q.; Miao, K.L.; Cui, D.H.; et al. Differences in the genomic profiles of cell-free DNA between plasma sputum urine tumor tissue in advanced NSCLC. Cancer Med. 2019, 8, 910–919. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wei, C.; Wen, J.; Chen, S.; Chen, L.; Wu, Y.; Shen, Y.; Bai, H.; Zhang, Y.; Chen, X.; et al. Comprehensive analysis of NGS and ARMS-PCR for detecting EGFR mutations based on 4467 cases of NSCLC patients. J. Cancer Res. Clin. Oncol. 2021, 148, 321–330. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morikawa, K.; Kinoshita, K.; Matsuzawa, S.; Kida, H.; Handa, H.; Inoue, T.; Nakamura, S.; Sato, Y.; Mineshita, M. EML4-ALK Gene Mutation Detected with New NGS Lung Cancer Panel CDx Using Sputum Cytology in a Case of Advanced NSCLC. Diagnostics 2023, 13, 2327. https://doi.org/10.3390/diagnostics13142327
Morikawa K, Kinoshita K, Matsuzawa S, Kida H, Handa H, Inoue T, Nakamura S, Sato Y, Mineshita M. EML4-ALK Gene Mutation Detected with New NGS Lung Cancer Panel CDx Using Sputum Cytology in a Case of Advanced NSCLC. Diagnostics. 2023; 13(14):2327. https://doi.org/10.3390/diagnostics13142327
Chicago/Turabian StyleMorikawa, Kei, Kohei Kinoshita, Shin Matsuzawa, Hirotaka Kida, Hiroshi Handa, Takeo Inoue, Seiji Nakamura, Yoshiharu Sato, and Masamichi Mineshita. 2023. "EML4-ALK Gene Mutation Detected with New NGS Lung Cancer Panel CDx Using Sputum Cytology in a Case of Advanced NSCLC" Diagnostics 13, no. 14: 2327. https://doi.org/10.3390/diagnostics13142327
APA StyleMorikawa, K., Kinoshita, K., Matsuzawa, S., Kida, H., Handa, H., Inoue, T., Nakamura, S., Sato, Y., & Mineshita, M. (2023). EML4-ALK Gene Mutation Detected with New NGS Lung Cancer Panel CDx Using Sputum Cytology in a Case of Advanced NSCLC. Diagnostics, 13(14), 2327. https://doi.org/10.3390/diagnostics13142327





