Segmentation of Portal Vein in Multiphase CTA Image Based on Unsupervised Domain Transfer and Pseudo Label
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Description
2.2. Image Preprocessing
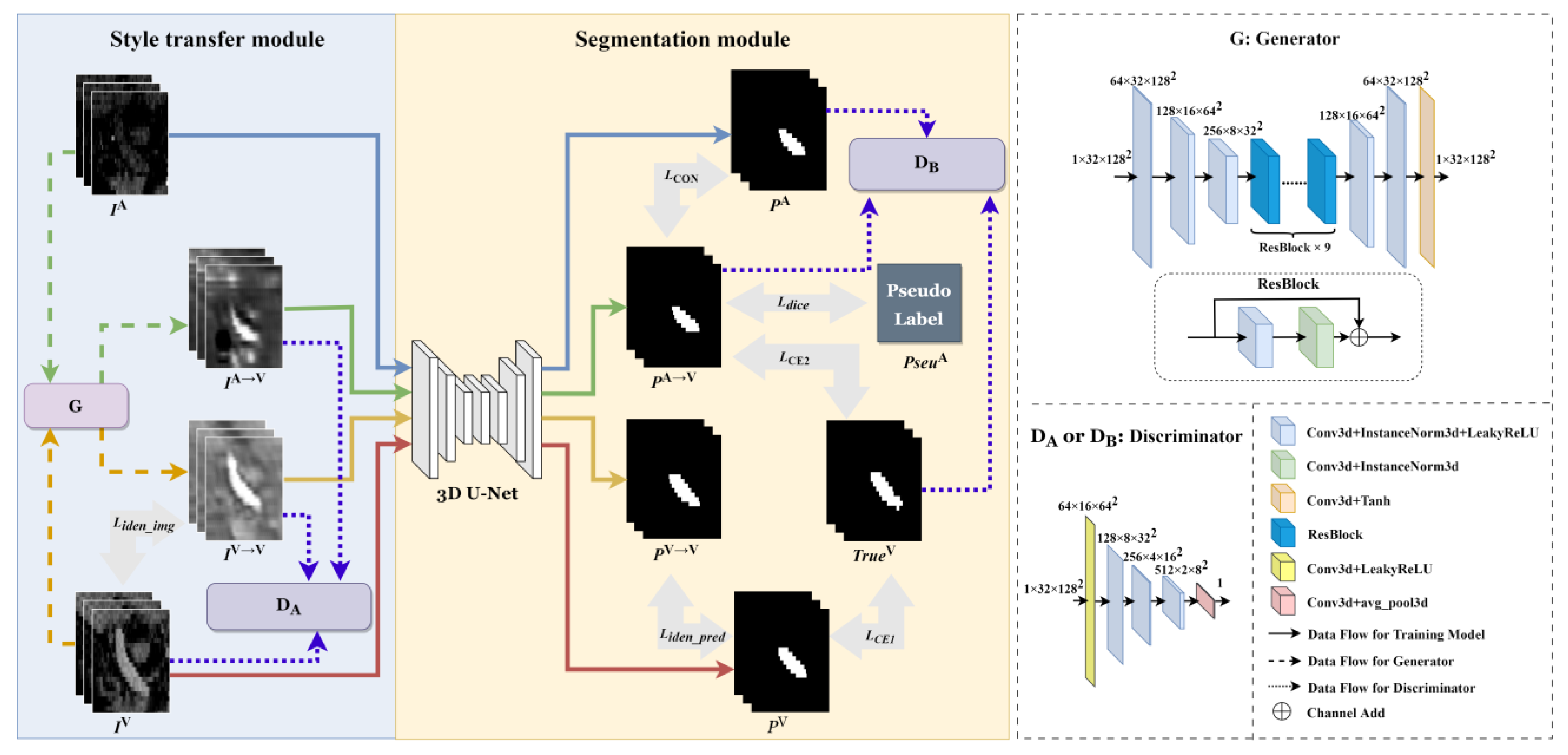
2.3. Multiphase Segmentation Network
2.3.1. Style Transfer Module
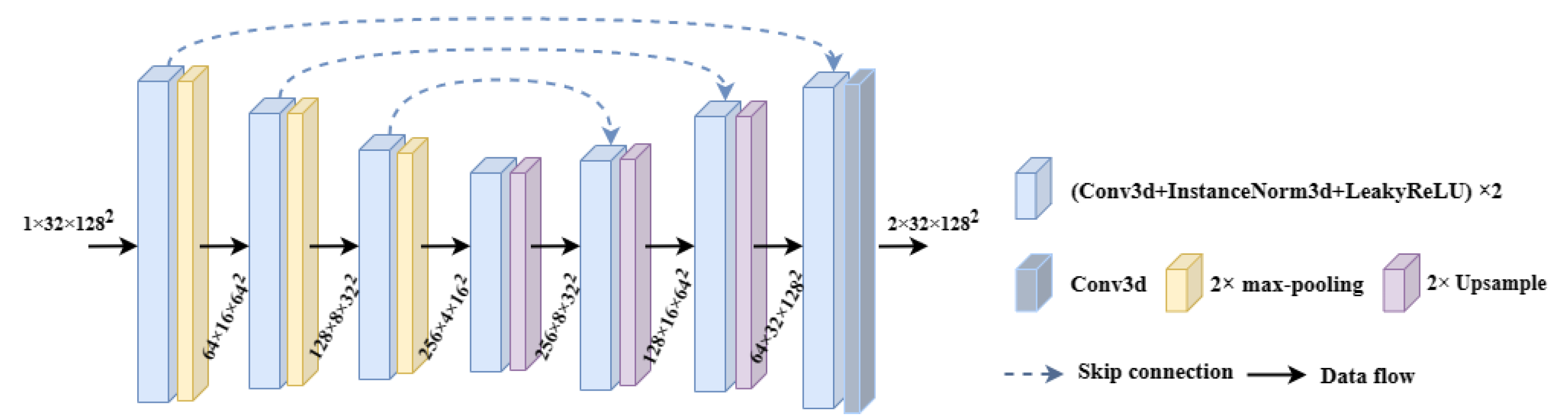
2.3.2. Segmentation Module
2.3.3. Construction of Pseudo Label
2.3.4. Loss Function
3. Experiments
3.1. Experimental Setup and Evaluation Index
3.2. Comparison of Experimental Results
3.3. Ablation Experimental Results
- (1)
- Effect of each module on experimental results
- (2)
- Effect of pseudo label weights on experimental results
3.4. Visualization Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Methods (vs. PVSegNet) | IA | IV | TrueA | TrueV | H-Phase | E-Phase | ||
|---|---|---|---|---|---|---|---|---|
| DSC | Jaccard | DSC | Jaccard | |||||
| nnUNet (Supervised) | ✔ | ✔ | ** 4.91 × 10−22 | ** 9.09× 10−46 | ** 1.15 × 10−26 | 2.73 × 10−1 | ||
| 3D U-Net (Supervised) | ✔ | ✔ | ** 4.02 × 10−3 | ** 4.98 × 10−3 | 2.40 × 10−1 | 1.50 × 10−1 | ||
| 3D U-Net | ✔ | ✔ | ** 1.24 × 10−9 | ** 1.38 × 10−11 | ** 1.52 × 10−33 | ** 5.89 × 10−40 | ||
| 3D U-Net | ✔ | ✔ | ** 1.75 × 10−75 | ** 1.36 × 10−74 | ** 1.06 × 10−5 | ** 1.37 × 10−5 | ||
| CycleGan | ✔ | ✔ | ✔ | ** 4.15 × 10−4 | ** 3.66 × 10−5 | ** 6.48 × 10−34 | ** 5.15 × 10−41 | |
| # | DB | G&DA | PL | H-Phase | E-Phase | |||
|---|---|---|---|---|---|---|---|---|
| DSC | Jaccard | DSC | Jaccard | |||||
| PVSegNet Based vs. PVSegNet | 1 | ** 1.59 × 10−4 | ** 4.15 × 10−6 | ** 2.98 × 10−20 | * 9.82 × 10−22 | |||
| 2 | ✔ | ✔ | * 1.72 × 10−2 | ** 1.05 × 10−3 | 9.76 × 10−2 | 5.92 × 10−2 | ||
| 3 | ✔ | ✔ | ** 3.47 × 10−4 | ** 2.58 × 10−6 | ** 3.93 × 10−18 | * 1.61 × 10−20 | ||
| 4 | ✔ | ✔ | 1.06 × 10−1 | 1.10 × 10−1 | 4.31 × 10−1 | 6.63 × 10−1 | ||
| H-Phase | E-Phase | |||
|---|---|---|---|---|
| DSC | Jaccard | DSC | Jaccard | |
| 0.1 | * 4.65 × 10−2 | * 2.77 × 10−2 | ||
| 0.3 | ** 8.89 × 10−3 | ** 5.24 × 10−3 | ** 6.81 × 10−3 | ** 8.92 × 10−3 |
| 0.5 | ** 8.79 × 10−3 | ** 1.58 × 10−3 | 8.66 × 10−2 | 9.18 × 10−2 |
| 0.7 | ** 4.60 × 10−10 | ** 3.09 × 10−11 | ||
| 0.9 | ** 8.12 × 10−4 | ** 1.99 × 10−4 | 5.49 × 10−1 | 4.70 × 10−1 |
References
- Carneiro, C.; Brito, J.; Bilreiro, C.; Barros, M.; Bahia, C.; Santiago, I.; Caseiro-Alves, F. All about portal vein: A pictorial display to anatomy, variants and physiopathology. Insights Into Imaging 2019, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Gallego, C.; Velasco, M.; Marcuello, P.; Tejedor, D.; De Campo, L.; Friera, A. Congenital and acquired anomalies of the portal venous system. Radiographics 2002, 22, 141–159. [Google Scholar] [CrossRef]
- Neil Granger, D.; Holm, L.; Kvietys, P. The gastrointestinal circulation: Physiology and pathophysiology. Compr. Physiol. 2011, 5, 1541–1583. [Google Scholar]
- Lebre, M.A.; Vacavant, A.; Grand-Brochier, M.; Rositi, H.; Abergel, A.; Chabrot, P.; Magnin, B. Automatic segmentation methods for liver and hepatic vessels from CT and MRI volumes, applied to the Couinaud scheme. Comput. Biol. Med. 2019, 110, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wu, X.; Wang, S.; Xu, L.; Xu, H.; Zheng, D.; Yu, N.; Hong, Y.; Yu, Z.; Yang, D.; et al. Automated segmentation of liver segment on portal venous phase MR images using a 3D convolutional neural network. Insights Imaging 2022, 13, 26. [Google Scholar] [CrossRef]
- Guo, Q.; Song, H.; Fan, J.; Ai, D.; Gao, Y.; Yu, X.; Yang, J. Portal Vein and Hepatic Vein Segmentation in Multi-Phase MR Images Using Flow-Guided Change Detection. IEEE Trans. Image Process. 2022, 31, 2503–2517. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Cheng, W.H.; Lee, C.W.; Yang, C.Y.; Liu, H.M. Multiphase computed tomographic angiography with bone subtraction using 3D multichannel convolution neural networks. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1274–1277. [Google Scholar]
- Bharati, S.; Mondal, M.; Podder, P.; Prasath, V. Deep learning for medical image registration: A comprehensive review. arXiv 2022, arXiv:2204.11341. [Google Scholar]
- Ibragimov, B.; Toesca, D.; Chang, D.; Koong, A.; Xing, L. Combining deep learning with anatomical analysis for segmentation of the portal vein for liver SBRT planning. Phys. Med. Biol. 2017, 62, 8943. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; Part II 19. Springer: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar]
- Kitrungrotsakul, T.; Han, X.H.; Iwamoto, Y.; Lin, L.; Foruzan, A.H.; Xiong, W.; Chen, Y.W. VesselNet: A deep convolutional neural network with multi pathways for robust hepatic vessel segmentation. Comput. Med. Imaging Graph. 2019, 75, 74–83. [Google Scholar] [CrossRef]
- Yu, W.; Fang, B.; Liu, Y.; Gao, M.; Zheng, S.; Wang, Y. Liver vessels segmentation based on 3d residual U-NET. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 250–254. [Google Scholar]
- Xu, Z.; Lu, D.; Wang, Y.; Luo, J.; Jayender, J.; Ma, K.; Zheng, Y.; Li, X. Noisy labels are treasure: Mean-teacher-assisted confident learning for hepatic vessel segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention– MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer: Cham, Switzerland, 2021; pp. 3–13. [Google Scholar]
- Tao, G.; Singh, A.; Bidaut, L. Liver segmentation from registered multiphase CT data sets with EM clustering and GVF level set. In Proceedings of the Medical Imaging 2010: Image Processing, San Diego, CA, USA, 14–16 February 2010; Volume 7623, pp. 300–308. [Google Scholar]
- Zhang, X.; Tan, X.; Gao, X.; Wu, D.; Zhou, X.; Fujita, H. Non-rigid registration of multi-phase liver CT data using fully automated landmark detection and TPS deformation. Clust. Comput. 2019, 22, 15305–15319. [Google Scholar] [CrossRef]
- Rameshbabu, C.; Wani, Z.A.; Rai, P.; Abdulqader, A.; Garg, S.; Sharma, M. Standard imaging techniques for assessment of portal venous system and its tributaries by linear endoscopic ultrasound: A pictorial essay. Endosc. Ultrasound 2013, 2, 16. [Google Scholar] [PubMed]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Patel, V.M.; Gopalan, R.; Li, R.; Chellappa, R. Visual domain adaptation: A survey of recent advances. IEEE Signal Process. Mag. 2015, 32, 53–69. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Jiang, J.; Rimner, A.; Deasy, J.O.; Veeraraghavan, H. Unpaired cross-modality educed distillation (CMEDL) for medical image segmentation. IEEE Trans. Med. Imaging 2021, 41, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Xu, K.; Li, S.; Zeng, Z.; Guan, C. Mt-uda: Towards unsupervised cross-modality medical image segmentation with limited source labels. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer: Cham, Switzerland, 2021; pp. 293–303. [Google Scholar]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Unsupervised bidirectional cross-modality adaptation via deeply synergistic image and feature alignment for medical image segmentation. IEEE Trans. Med. Imaging 2020, 39, 2494–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weston, J.; Ratle, F.; Collobert, R. Deep learning via semi-supervised embedding. In Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 5–9 July 2008; pp. 1168–1175. [Google Scholar]
- Qu, L.; Liu, S.; Liu, X.; Wang, M.; Song, Z. Towards label-efficient automatic diagnosis and analysis: A comprehensive survey of advanced deep learning-based weakly-supervised, semi-supervised and self-supervised techniques in histopathological image analysis. Phys. Med. Biol. 2022, 67, 20. [Google Scholar] [CrossRef] [PubMed]
- Raju, A.; Cheng, C.T.; Huo, Y.; Cai, J.; Huang, J.; Xiao, J.; Lu, L.; Liao, C.; Harrison, A.P. Co-heterogeneous and adaptive segmentation from multi-source and multi-phase CT imaging data: A study on pathological liver and lesion segmentation. In Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020; Proceedings, Part XXIII. Springer: Cham, Switzerland, 2020; pp. 448–465. [Google Scholar]
- Qu, T.; Wang, X.; Fang, C.; Mao, L.; Li, J.; Li, P.; Qu, J.; Li, X.; Xue, H.; Yu, Y.; et al. M3Net: A multi-scale multi-view framework for multi-phase pancreas segmentation based on cross-phase non-local attention. Med. Image Anal. 2022, 75, 102232. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells, W.M., III; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index1: Scientific reports. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Methods | IA | IV | TrueA | TrueV | H-Phase | E-Phase | ||
|---|---|---|---|---|---|---|---|---|
| DSC | Jaccard | DSC | Jaccard | |||||
| nnUNet (Supervised) | ✔ | ✔ | 0.832 ± 0.001 | 0.724 ± 0.001 | 0.894 ± 0.000 | 0.816 ± 0.001 | ||
| 3D U-Net (Supervised) | ✔ | ✔ | 0.724 ± 0.000 | 0.581 ± 0.000 | 0.832 ± 0.000 | 0.723 ± 0.001 | ||
| 3D U-Net | ✔ | ✔ | 0.590 ± 0.001 | 0.437 ± 0.001 | 0.650 ± 0.001 | 0.500 ± 0.001 | ||
| 3D U-Net | ✔ | ✔ | 0.117 ± 0.000 | 0.079 ± 0.000 | 0.801 ± 0.001 | 0.680 ± 0.001 | ||
| CycleGan | ✔ | ✔ | ✔ | 0.633 ± 0.001 | 0.482 ± 0.001 | 0.643 ± 0.001 | 0.493 ± 0.001 | |
| PVSegNet | ✔ | ✔ | ✔ | 0.689 ± 0.001 | 0.546 ± 0.001 | 0.826 ± 0.001 | 0.712 ± 0.001 | |
| # | DB | G&DA | PL | H-Phase | E-Phase | |||
|---|---|---|---|---|---|---|---|---|
| DSC | Jaccard | DSC | Jaccard | |||||
| PVSegNet based | 1 | 0.643 ± 0.000 | 0.486 ± 0.000 | 0.758 ± 0.003 | 0.672 ± 0.007 | |||
| 2 | ✔ | ✔ | 0.666 ± 0.000 | 0.485 ± 0.002 | 0.819 ± 0.000 | 0.702 ± 0.001 | ||
| 3 | ✔ | ✔ | 0.648 ± 0.000 | 0.493 ± 0.000 | 0.759 ± 0.003 | 0.624 ± 0.004 | ||
| 4 | ✔ | ✔ | 0.676 ± 0.002 | 0.534 ± 0.002 | 0.823 ± 0.001 | 0.709 ± 0.002 | ||
| PVSegNet | 5 | ✔ | ✔ | ✔ | 0.689 ± 0.001 | 0.546 ± 0.001 | 0.826 ± 0.001 | 0.712 ± 0.001 |
| H-Phase | E-Phase | |||
|---|---|---|---|---|
| DSC | Jaccard | DSC | Jaccard | |
| 0.1 | 0.689 ± 0.001 | 0.546 ± 0.001 | 0.818 ± 0.000 | 0.708 ± 0.000 |
| 0.3 | 0.665 ± 0.001 | 0.529 ± 0.001 | 0.815 ± 0.001 | 0.698 ± 0.002 |
| 0.5 | 0.681 ± 0.002 | 0.520 ± 0.002 | 0.819 ± 0.001 | 0.703 ± 0.001 |
| 0.7 | 0.621 ± 0.002 | 0.474 ± 0.002 | 0.826 ± 0.001 | 0.712 ± 0.001 |
| 0.9 | 0.656 ± 0.000 | 0.498 ± 0.000 | 0.824 ± 0.001 | 0.708 ± 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, G.; Xie, Z.; Wang, H.; Li, S.; Yao, D.; Chen, S.; Shi, Y. Segmentation of Portal Vein in Multiphase CTA Image Based on Unsupervised Domain Transfer and Pseudo Label. Diagnostics 2023, 13, 2250. https://doi.org/10.3390/diagnostics13132250
Song G, Xie Z, Wang H, Li S, Yao D, Chen S, Shi Y. Segmentation of Portal Vein in Multiphase CTA Image Based on Unsupervised Domain Transfer and Pseudo Label. Diagnostics. 2023; 13(13):2250. https://doi.org/10.3390/diagnostics13132250
Chicago/Turabian StyleSong, Genshen, Ziyue Xie, Haoran Wang, Shiman Li, Demin Yao, Shiyao Chen, and Yonghong Shi. 2023. "Segmentation of Portal Vein in Multiphase CTA Image Based on Unsupervised Domain Transfer and Pseudo Label" Diagnostics 13, no. 13: 2250. https://doi.org/10.3390/diagnostics13132250
APA StyleSong, G., Xie, Z., Wang, H., Li, S., Yao, D., Chen, S., & Shi, Y. (2023). Segmentation of Portal Vein in Multiphase CTA Image Based on Unsupervised Domain Transfer and Pseudo Label. Diagnostics, 13(13), 2250. https://doi.org/10.3390/diagnostics13132250









