Evaluating Scoliosis Severity Based on Posturographic X-ray Images Using a Contrastive Language–Image Pretraining Model
Abstract
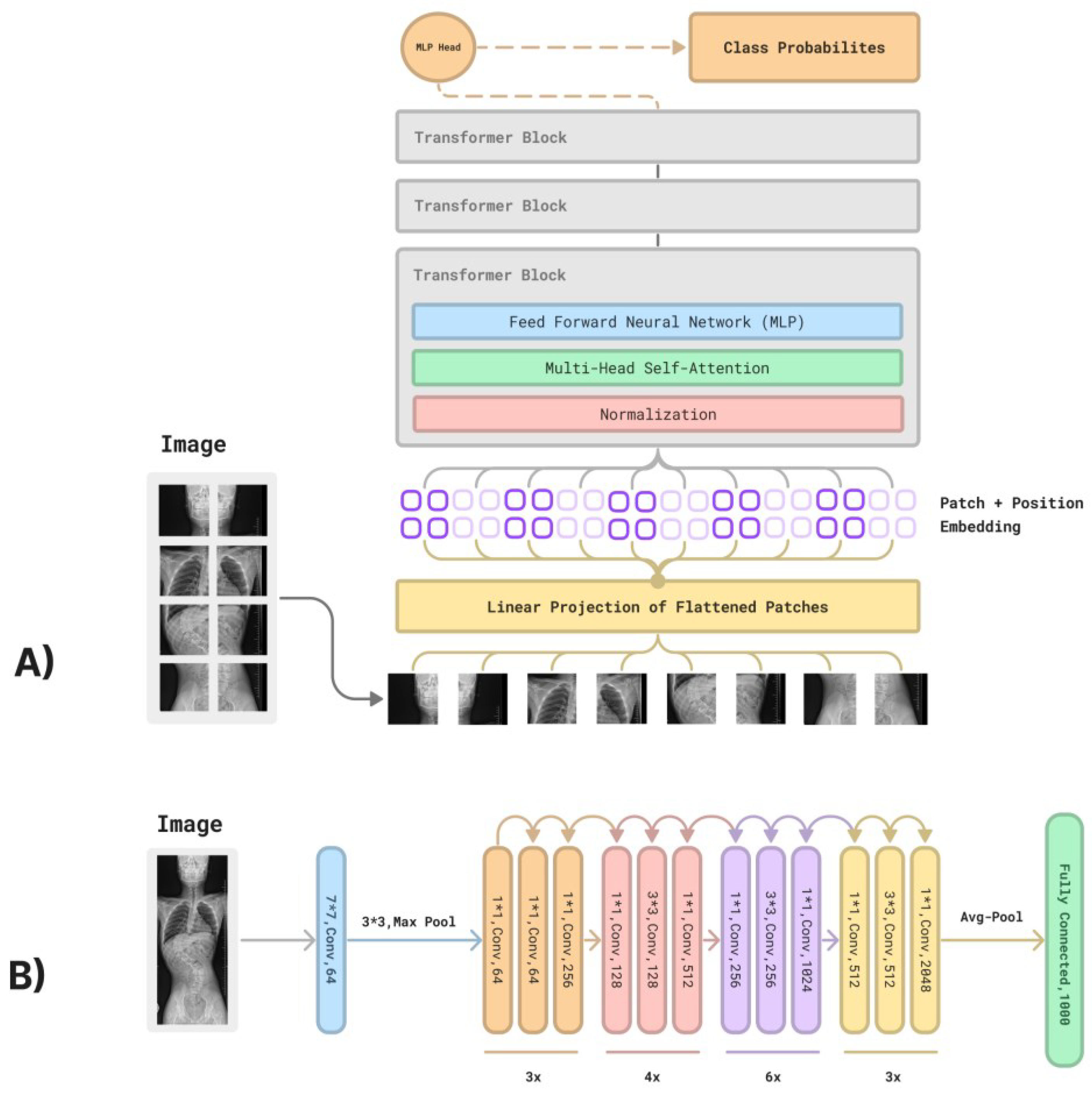
1. Introduction
- RN50;
- RN101;
- RN50×4;
- RN50×16;
- RN50×64.
- ViT-B/32;
- ViT-B/16;
- ViT-L/14;
- ViT-L/14@336px.
2. Materials and Methods
2.1. Manual Measurement
2.2. CLIP Methodology
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daeschler, S.C.; Bourget, M.H.; Derakhshan, D.; Sharma, V.; Asenov, S.I.; Gordon, T.; Cohen-Adad, J.; Borschel, G.H. Rapid, automated nerve histomorphometry through open-source artificial intelligence. Sci. Rep. 2022, 12, 5975. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, S.; Kobs, K.; Hotho, A. CLIP knows image aesthetics. Front. Artif. Intell. 2022, 5, 976235. [Google Scholar] [CrossRef] [PubMed]
- OpenAI. CLIP: Connecting Text and Images. Available online: https://openai.com/research/clip (accessed on 29 April 2023).
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning transferable visual models from natural language supervision. arXiv 2021, arXiv:2103.00020. [Google Scholar] [CrossRef]
- Yang, F.; He, Y.; Deng, Z.S.; Yan, A. Improvement of automated image stitching system for DR X-ray images. Comput. Biol. Med. 2016, 71, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.S.; Lai, P.L.; Tsai, H.Y.; Kung, Y.C.; Lin, Y.Y.; He, R.J.; Wu, C.T. Radiation dose for pediatric scoliosis patients undergoing whole spine radiography: Effect of the radiographic length in an auto-stitching digital radiography system. Eur. J. Radiol. 2018, 108, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Maharathi, S.; Iyengar, R.; Chandrasekhar, P. Biomechanically designed Curve Specific Corrective Exercise for Adolescent Idiopathic Scoliosis gives significant outcomes in an Adult: A case report. Front. Rehabil. Sci. 2023, 4, 1127222. [Google Scholar] [CrossRef] [PubMed]
- Hey, H.W.D.; Ramos, M.R.D.; Lau, E.T.; Tan, J.H.J.; Tay, H.W.; Liu, G.; Wong, H.K. Risk Factors Predicting C- Versus S-shaped Sagittal Spine Profiles in Natural, Relaxed Sitting: An Important Aspect in Spinal Realignment Surgery. Spine 2020, 45, 1704–1712. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, H.; Zateri, C.; Kusvuran Ozkan, A.; Kayalar, G.; Berk, H. Prevalence of adolescent idiopathic scoliosis in Turkey: An epidemiological study. Spine J. 2020, 20, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Kuznia, A.L.; Hernandez, A.K.; Lee, L.U. Adolescent Idiopathic Scoliosis: Common Questions and Answers. Am. Fam. Physician 2020, 101, 19–23. [Google Scholar] [PubMed]
- Meng, N.; Cheung, J.P.Y.; Wong, K.K.; Dokos, S.; Li, S.; Choy, R.W.; To, S.; Li, R.J.; Zhang, T. An artificial intelligence powered platform for auto-analyses of spine alignment irrespective of image quality with prospective validation. EClinicalMedicine 2022, 43, 101252. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Zhou, M.; Wang, H.; Gevaert, O.; Metaxas, D.; Zhang, S. A Large-scale Synthetic Pathological Dataset for Deep Learning-enabled Segmentation of Breast Cancer. Sci. Data 2023, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Moazemi, S.; Vahdati, S.; Li, J.; Kalkhoff, S.; Castano, L.J.V.; Dewitz, B.; Bibo, R.; Sabouniaghdam, P.; Tootooni, M.S.; Bundschuh, R.A.; et al. Artificial intelligence for clinical decision support for monitoring patients in cardiovascular ICUs: A systematic review. Front. Med. 2023, 10, 1109411. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.; Kay, K.; Naselaris, T.; Tarr, M.J.; Wehbe, L. Incorporating natural language into vision models improves prediction and understanding of higher visual cortex. bioRxiv 2022. [Google Scholar] [CrossRef]
- Palepu, K.; Ponnapati, M.; Bhat, S.; Tysinger, E.; Stan, T.; Brixi, G.; Koseki, S.R.T.; Chatterjee, P. Design of Peptide-Based Protein Degraders via Contrastive Deep Learning. bioRxiv 2022. [Google Scholar] [CrossRef]
- Tan, H.; Bansal, M. LXMERT: Learning Cross-Modality Encoder Representations from Transformers. arXiv 2019, arXiv:1908.07490. [Google Scholar]
- Lu, J.; Batra, D.; Parikh, D.; Lee, S. ViLBERT: Pretraining Task-Agnostic Visiolinguistic Representations for Vision-and-Language Tasks. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 13–23. [Google Scholar]
- Jia, X.; Goyal, N.; Wehrmann, J.; Gupta, A.; Paluri, M.; Shih, K. Scaling Up Visual and Vision-Language Representation Learning With Noisy Text Supervision. arXiv 2021, arXiv:2102.05918. [Google Scholar]

| CLIP-ResNet Hyperparameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Learning Rate | Embedding Dimension | Input Resolution | ResNet Blocks | ResNet Width | Text Transformer Layers | Text Transformer Width | Text Transformer Heads | |
| RN50 | 5 × 10−4 | 1024 | 224 | (3, 4, 6, 3) | 2048 | 12 | 512 | 8 | |
| RN101 | 5 × 10−4 | 512 | 224 | (3, 4, 23, 3) | 2048 | 12 | 512 | 8 | |
| RN50×4 | 5 × 10−4 | 640 | 288 | (4, 6, 10, 6) | 2560 | 12 | 640 | 10 | |
| RN50×16 | 4 × 10−4 | 768 | 384 | (6, 8, 18, 8) | 3072 | 12 | 768 | 12 | |
| RN50×64 | 3.6 × 10−4 | 1024 | 448 | (3, 15, 36, 10) | 4096 | 12 | 1024 | 16 | |
| CLIP-ViT Hyperparameters | |||||||||
| Model | Learning Rate | Embedding Dimension | Input Resolution | Vision Transformer Layers | Vision Transformer Width | Vision Transformer Heads | Text Transformer Layers | Text Transformer Width | Text Transformer Heads |
| VitB16 | 5 × 10−4 | 512 | 224 | 12 | 768 | 12 | 12 | 512 | 8 |
| VitB32 | 5 × 10−4 | 512 | 224 | 12 | 768 | 12 | 12 | 512 | 8 |
| VitL14 | 4 × 10−4 | 768 | 224 | 24 | 1024 | 16 | 12 | 768 | 12 |
| VitL14@336px | 2 × 10−5 | 768 | 336 | 24 | 1024 | 16 | 12 | 768 | 12 |
| Common CLIP Hyperparameters | |||||||||
| Hyperparameter | Value | ||||||||
| Batch size | 32,768 | ||||||||
| Vocabulary size | 49,408 | ||||||||
| Training epochs | 32 | ||||||||
| Maximum temperature | 100.0 | ||||||||
| Weight decay | 0.2 | ||||||||
| Warm-up iterations | 2000 | ||||||||
| Adam β1 | 0.9 | ||||||||
| Adam β2 | 0.999 (ResNet), 0.98 (ViT) | ||||||||
| Adam epsilon | 10−8 (ResNet), 10−6 (ViT) | ||||||||
| Area of Interests | Questions |
|---|---|
| Scoliosis/no scoliosis (further—scoliosis) | “Is there any scoliosis on the radiograph?” “Is there no scoliosis on the radiograph?” |
| C-shape scoliosis/noC-shape scoliosis (further—C-shape) | “Is there a C-shape scoliosis on the radiograph?” “Is there no scoliosis C-shape on the radiograph?” |
| Single curve/non-single curve (further—single-curve) | “Is there a single curve scoliosis on the radiograph?” “Is there no single curve scoliosis on the radiograph?” |
| Cobb angle | “Cobb angle is 0–10 Degrees?” “Cobb angle is 11–20 Degrees?” “Cobb angle is 21–30 Degrees?” “Cobb angle is 31–40 Degrees?” “Cobb angle is 41–50 Degrees?” “Cobb angle is 51–60 Degrees?” “Cobb angle is 61–70 Degrees?” “Cobb angle is 71–80 Degrees?” “Cobb angle is above 81 Degrees?” |
| Neural Network | Set of Questions | n | TPs | FNs | Sensitivity |
|---|---|---|---|---|---|
| RN50 | Scoliosis | 23 | 23 | 0 | 100% |
| C-shape | 23 | 0 | 23 | 0% | |
| Single-curve | 23 | 23 | 0 | 100% | |
| Cobb angle | 23 | 0 | 23 | 0 | |
| Overall | 92 | 46 | 46 | 50.0% | |
| RN101 | Scoliosis | 23 | 23 | 0 | 100% |
| C-shape | 23 | 0 | 23 | 0% | |
| Single-curve | 23 | 0 | 23 | 0% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 23 | 69 | 25.0% | |
| RN50×4 | Scoliosis | 23 | 23 | 0 | 100% |
| C-shape | 23 | 0 | 23 | 0% | |
| Single-curve | 23 | 2 | 21 | 8.7% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 25 | 67 | 37.3% | |
| RN50×16 | Scoliosis | 23 | 23 | 0 | 100% |
| S-shape | 23 | 0 | 23 | 0% | |
| Single-curve | 23 | 1 | 22 | 4.3% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 24 | 68 | 26.1% | |
| RN50×64 | Scoliosis | 23 | 23 | 0 | 100% |
| C-shape | 23 | 14 | 9 | 60.9% | |
| Single-curve | 23 | 3 | 20 | 13.0% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 40 | 52 | 43.5% | |
| VitB16 | Scoliosis | 23 | 23 | 0 | 100% |
| C-shape | 23 | 0 | 23 | 0% | |
| Single-curve | 23 | 5 | 18 | 21.7% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 28 | 64 | 28.9% | |
| VitB32 | Scoliosis | 23 | 23 | 0 | 100% |
| C-shape | 23 | 15 | 8 | 65.2% | |
| Single-curve | 23 | 0 | 23 | 0% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 38 | 54 | 41.3% | |
| VitL14 | Scoliosis | 23 | 2 | 21 | 9.5% |
| C-shape | 23 | 2 | 21 | 9.5% | |
| Single-curve | 23 | 23 | 0 | 100% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 27 | 65 | 29.3% | |
| VitL14@336px | Scoliosis | 23 | 1 | 22 | 4.3% |
| C-shape | 23 | 6 | 17 | 26.1% | |
| Single-curve | 23 | 23 | 0 | 100% | |
| Cobb angle | 23 | 0 | 23 | 0% | |
| Overall | 92 | 30 | 62 | 32.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabijan, A.; Fabijan, R.; Zawadzka-Fabijan, A.; Nowosławska, E.; Zakrzewski, K.; Polis, B. Evaluating Scoliosis Severity Based on Posturographic X-ray Images Using a Contrastive Language–Image Pretraining Model. Diagnostics 2023, 13, 2142. https://doi.org/10.3390/diagnostics13132142
Fabijan A, Fabijan R, Zawadzka-Fabijan A, Nowosławska E, Zakrzewski K, Polis B. Evaluating Scoliosis Severity Based on Posturographic X-ray Images Using a Contrastive Language–Image Pretraining Model. Diagnostics. 2023; 13(13):2142. https://doi.org/10.3390/diagnostics13132142
Chicago/Turabian StyleFabijan, Artur, Robert Fabijan, Agnieszka Zawadzka-Fabijan, Emilia Nowosławska, Krzysztof Zakrzewski, and Bartosz Polis. 2023. "Evaluating Scoliosis Severity Based on Posturographic X-ray Images Using a Contrastive Language–Image Pretraining Model" Diagnostics 13, no. 13: 2142. https://doi.org/10.3390/diagnostics13132142
APA StyleFabijan, A., Fabijan, R., Zawadzka-Fabijan, A., Nowosławska, E., Zakrzewski, K., & Polis, B. (2023). Evaluating Scoliosis Severity Based on Posturographic X-ray Images Using a Contrastive Language–Image Pretraining Model. Diagnostics, 13(13), 2142. https://doi.org/10.3390/diagnostics13132142






