Cell-Free Methylated PTGER4 and SHOX2 Plasma DNA as a Biomarker for Therapy Monitoring and Prognosis in Advanced Stage NSCLC Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Plasma Preparation
2.3. DNA Isolation, Purification, and Bisulfite Conversion
2.4. Statistical Analysis
3. Results
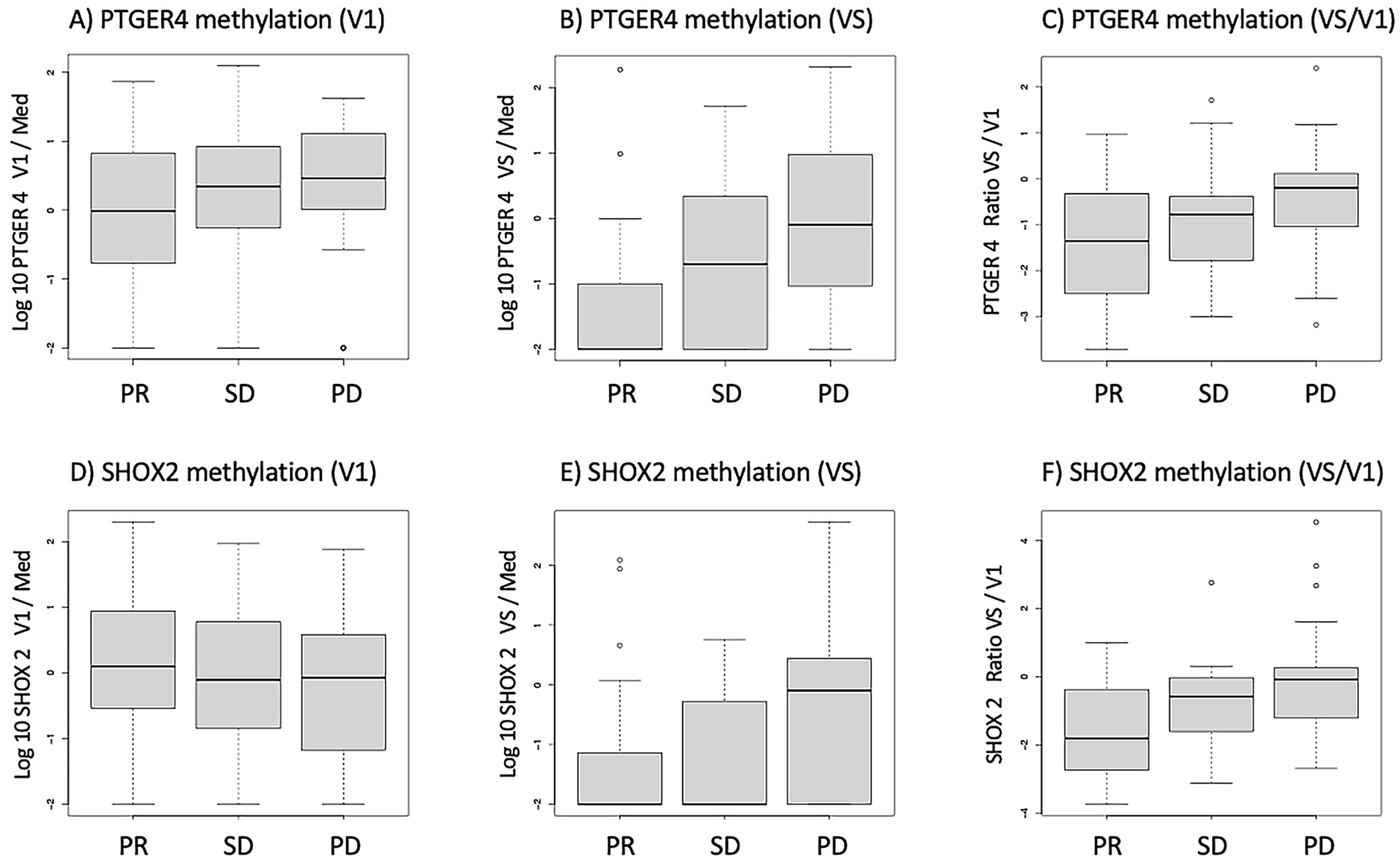
3.1. Distribution of mPTGER4 and mSHOX2 Methylation Values in Response Groups
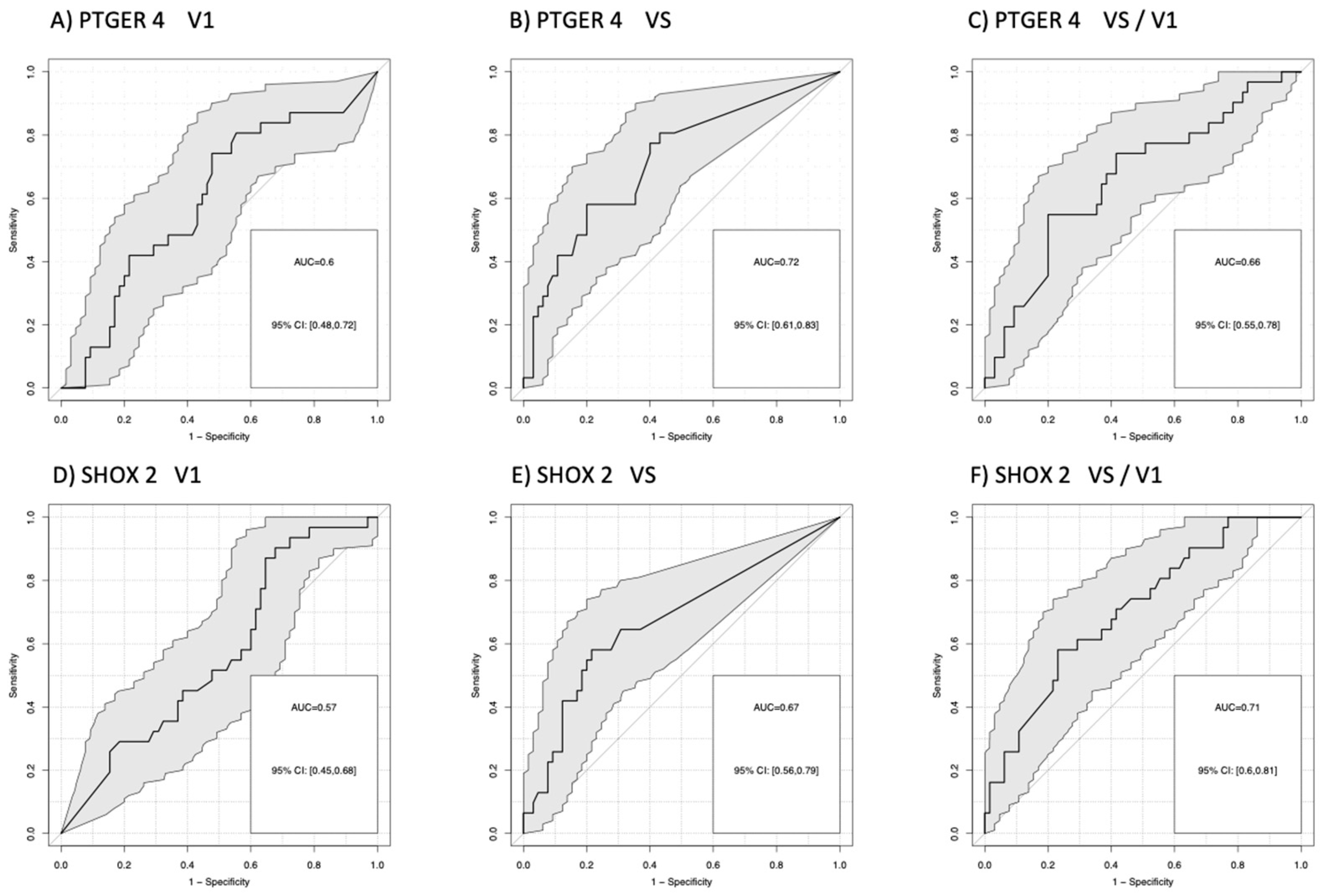
3.2. Differentiation of Patients According to Their Therapy Response
3.3. Combination of Markers in Decision Trees
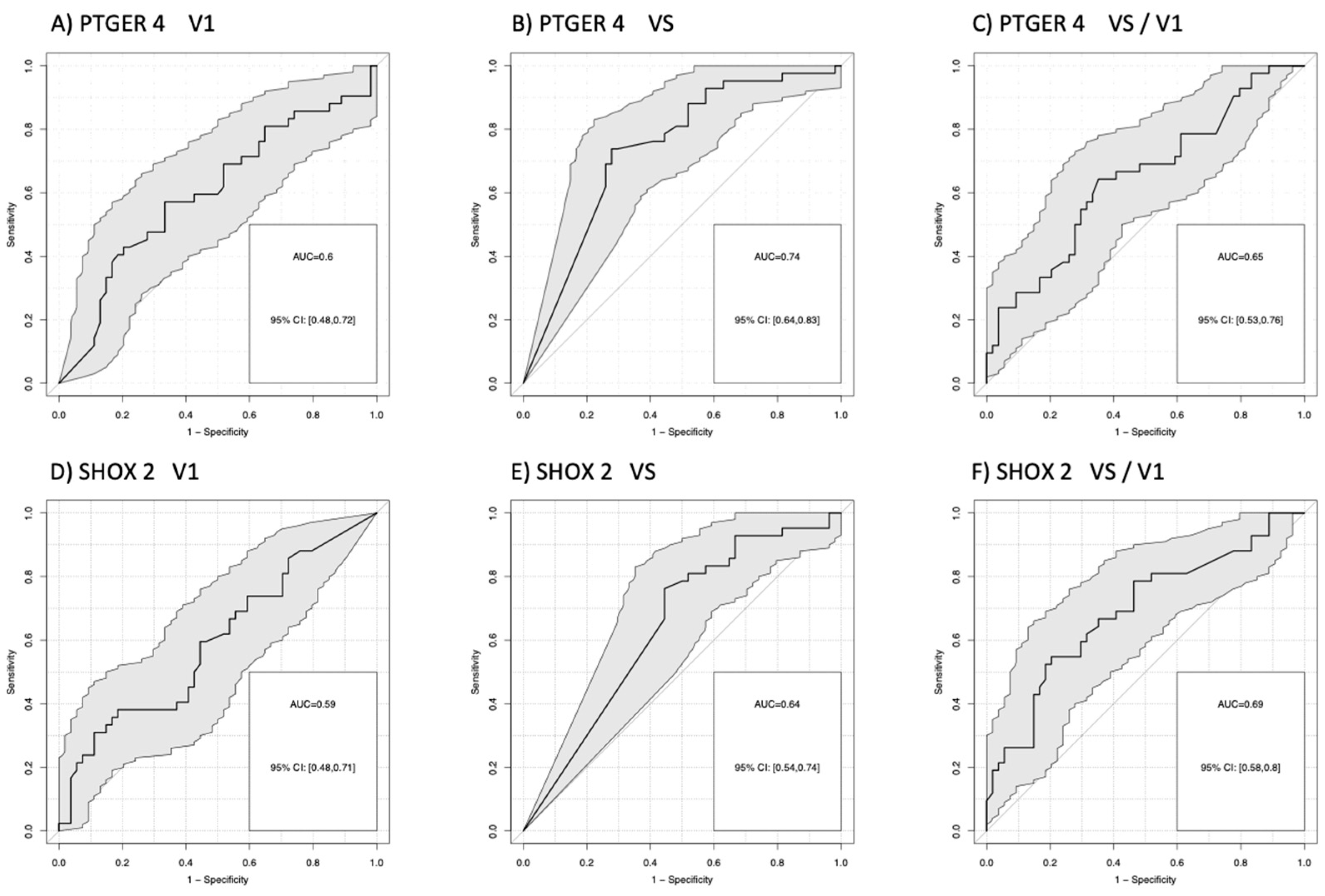
3.4. Relevance of PTGER4 and SHOX2 for Prognosis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fois, S.S.; Paliogiannis, P.; Zinellu, A.; Fois, A.G.; Cossu, A.; Palmieri, G. Molecular Epidemiology of the Main Druggable Genetic Alterations in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 612. [Google Scholar] [CrossRef] [PubMed]
- S3-Leitlinie Prävention, Diagnostik, Therapie und Nachsorge des Lungenkarzinoms. Available online: https://www.leitlinienprogramm-onkologie.de/leitlinien/lungenkarzinom/ (accessed on 28 February 2023).
- Mandel, P.; Metais, P. Les Acides nucleiques du plasma sangiun chez l’Homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Stroun, M.; Anker, P.; Maurice, P.; Gahan, P.B. Circulating nucleic acids in higher organisms. Int. Rev. Cytol. 1977, 51, 1–48. [Google Scholar] [PubMed]
- Stroun, M.; Anker, P.; Lyautey, J.; Lederrey, C.; Maurice, P.A. Isolation and characterization of DNA from the plasma of cancer patients. Eur. J. Cancer Clin. Oncol. 1987, 23, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Q.; Stroun, M.; Magnenat, J.-L.; Nicod, L.P.; Kurt, A.-M.; Lyautey, J.; Lederrey, C.; Anker, P. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat. Med. 1996, 2, 1033–1035. [Google Scholar] [CrossRef]
- Huang, J.; Wang, L. Cell-Free DNA Methylation Profiling Analysis—Technologies and Bioinformatics. Cancers 2019, 11, 1741. [Google Scholar] [CrossRef] [Green Version]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Barault, L.; Amatu, A.; Bleeker, F.E.; Moutinho, C.; Falcomatà, C.; Fiano, V.; Cassingena, A.; Siravegna, G.; Milione, M.; Cassoni, P.; et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1994–1999. [Google Scholar] [CrossRef]
- Huang, G.; Krocker, J.D.; Kirk, J.L.; Merwat, S.N.; Ju, H.; Soloway, R.D.; Wieck, L.R.; Li, A.; Okorodudu, A.O.; Petersen, J.R.; et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin. Chem. Lab. Med. 2014, 52, 899–909. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zheng, Y.; Wu, L.; Li, J.; Ji, J.; Yu, Q.; Dai, W.; Feng, J.; Wu, J.; Guo, C. Current status of ctDNA in precision oncology for hepatocellular carcinoma. J. Exp. Clin. Canc. Res. 2021, 40, 140. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Biziota, E.; Koukaki, T.; Karamitrousis, E.; Nena, E.; Tsamardinos, I.; Kolios, G.; Lianidou, E.; et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2018, 38, 3387–3401. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, B.; Chen, D.; Xia, W.; Zhang, J.; Wang, F.; Xu, J.; Zhang, Y.; Zhang, M.; Zhang, L.; et al. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin. Epigenetics 2015, 7, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, B.; Beyer, J.; Dietrich, D.; Bork, I.; Liebenberg, V.; Fleischhacker, M. Quantification of Cell-Free mSHOX2 Plasma DNA for Therapy Monitoring in Advanced Stage Non-Small Cell (NSCLC) and Small-Cell Lung Cancer (SCLC) Patients. PLoS ONE 2015, 10, e0118195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, G.; Schlegel, A.; Kottwitz, D.; König, T.; Tetzner, R. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J. Thorac. Oncol. 2017, 12, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Kneip, C.; Schmidt, B.; Seegebarth, A.; Weickmann, S.; Fleischhacker, M.; Liebenberg, V.; Field, J.K.; Dietrich, D. SHOX2 DNA Methylation Is a Biomarker for the Diagnosis of Lung Cancer in Plasma. J. Thorac. Oncol. 2011, 6, 1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Globus, O.; Sagie, S.; Lavine, N.; Barchana, D.I.; Urban, D. Early death after a diagnosis of metastatic solid cancer—Raising awareness and identifying risk factors from the SEER database. medRxiv 2023. [Google Scholar] [CrossRef]
- Ponomaryova, A.A.; Rykova, E.Y.; Cherdyntseva, N.V.; Skvortsova, T.E.; Dobrodeev, A.Y.; Zav’yalov, A.A.; Bryzgalov, L.O.; Tuzikov, S.A.; Vlassov, V.V.; Laktionov, P.P. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013, 81, 397–403. [Google Scholar] [CrossRef]
- Janke, F.; Angeles, A.K.; Riediger, A.L.; Bauer, S.; Reck, M.; Stenzinger, A.; Schneider, M.A.; Muley, T.; Thomas, M.; Christopoulos, P.; et al. Longitudinal monitoring of cell-free DNA methylation in ALK-positive non-small cell lung cancer patients. Clin. Epigenetics 2022, 14, 163. [Google Scholar] [CrossRef]
- Metzenmacher, M.; Hegedues, B.; Forster, J.; Schramm, A.; Horn, P.A.; Klein, C.A.; Bielefeld, N.; Ploenes, T.; Aigner, C.; Theegarten, D.; et al. Combined multimodal ctDNA analysis and radiological imaging for tumor surveillance in Non-small cell lung cancer. Transl. Oncol. 2022, 15, 101279. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Malapelle, U.; Troncone, G. Liquid Biopsy and Lung Cancer. Acta Cytol. 2019, 63, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.; Thompson, J.C.; Carpenter, E.L.; Elkhouly, E.; Aggarwal, C. Plasma Cell-Free DNA Genotyping: From an Emerging Concept to a Standard-of-Care Tool in Metastatic Non-Small Cell Lung Cancer. Oncologist 2021, 26, e1812–e1821. [Google Scholar] [CrossRef] [PubMed]
- Shields, M.D.; Chen, K.; Dutcher, G.; Patel, I.; Pellini, B. Making the Rounds: Exploring the Role of Circulating Tumor DNA (ctDNA) in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 9006. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Klotzek, S.; Lewandowski, M.; Fleischhacker, M.; Jung, K. Changes in Concentration of DNA in Serum and Plasma during Storage of Blood Samples. Clin. Chem. 2003, 49, 1028–1029. [Google Scholar] [CrossRef] [Green Version]
- Warton, K.; Yuwono, N.L.; Cowley, M.J.; McCabe, M.J.; So, A.; Ford, C.E. Evaluation of Streck BCT and PAXgene Stabilised Blood Collection Tubes for Cell- Free Circulating DNA Studies in Plasma. Mol. Diagn. Ther. 2017, 21, 563–570. [Google Scholar] [CrossRef]
- Schmidt, B.; Reinicke, D.; Reindl, I.; Bork, I.; Wollschläger, B.; Lambrecht, N.; Fleischhacker, M. Liquid biopsy—Performance of the PAXgene® Blood ccfDNA Tubes for the isolation and characterization of cell-free plasma DNA from tumor patients. Clin. Chim. Acta 2017, 469, 94–98. [Google Scholar] [CrossRef]


| Patient | Number |
|---|---|
| Sex | |
| female | 62 |
| male | 34 |
| Age | |
| range | 48–81 |
| median | 63 |
| Smoker | |
| yes | 41 |
| no | 4 |
| ex-smoker | 30 |
| unknown | 21 |
| Histology | |
| NSCLC (unclassified) | 25 |
| Adenocarcinoma | 54 |
| Squamous cell carcinoma | 17 |
| Treatment | |
| Chemo +/− Radiotherapy | 83 |
| Chemo + Immunotherapy | 9 |
| Immunotherapy | 4 |
| Therapy response | |
| partial remission | 42 |
| stable disease | 23 |
| progressive disease | 31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fleischhacker, M.; Arslan, E.; Reinicke, D.; Eisenmann, S.; Theil, G.; Kollmeier, J.; Schäper, C.; Grah, C.; Klawonn, F.; Holdenrieder, S.; et al. Cell-Free Methylated PTGER4 and SHOX2 Plasma DNA as a Biomarker for Therapy Monitoring and Prognosis in Advanced Stage NSCLC Patients. Diagnostics 2023, 13, 2131. https://doi.org/10.3390/diagnostics13132131
Fleischhacker M, Arslan E, Reinicke D, Eisenmann S, Theil G, Kollmeier J, Schäper C, Grah C, Klawonn F, Holdenrieder S, et al. Cell-Free Methylated PTGER4 and SHOX2 Plasma DNA as a Biomarker for Therapy Monitoring and Prognosis in Advanced Stage NSCLC Patients. Diagnostics. 2023; 13(13):2131. https://doi.org/10.3390/diagnostics13132131
Chicago/Turabian StyleFleischhacker, Michael, Erkan Arslan, Dana Reinicke, Stefan Eisenmann, Gerit Theil, Jens Kollmeier, Christoph Schäper, Christian Grah, Frank Klawonn, Stefan Holdenrieder, and et al. 2023. "Cell-Free Methylated PTGER4 and SHOX2 Plasma DNA as a Biomarker for Therapy Monitoring and Prognosis in Advanced Stage NSCLC Patients" Diagnostics 13, no. 13: 2131. https://doi.org/10.3390/diagnostics13132131
APA StyleFleischhacker, M., Arslan, E., Reinicke, D., Eisenmann, S., Theil, G., Kollmeier, J., Schäper, C., Grah, C., Klawonn, F., Holdenrieder, S., & Schmidt, B. (2023). Cell-Free Methylated PTGER4 and SHOX2 Plasma DNA as a Biomarker for Therapy Monitoring and Prognosis in Advanced Stage NSCLC Patients. Diagnostics, 13(13), 2131. https://doi.org/10.3390/diagnostics13132131







