Abstract
In kidney transplantation, a biopsy is currently the gold standard for monitoring the transplanted organ. However, this is far from an ideal screening method given its invasive nature and the discomfort it can cause the patient. Large-scale studies in renal transplantation show that approximately 1% of biopsies generate major complications, with a risk of macroscopic hematuria greater than 3.5%. It would not be until 2011 that a method to detect donor-derived cell-free DNA (dd-cfDNA) employing digital PCR was devised based on analyzing the differences in SNPs between the donor and recipient. In addition, since the initial validation studies were carried out at the specific moments in which rejection was suspected, there is still not a good understanding of how dd-cfDNA levels naturally evolve post-transplant. In addition, various factors, both in the recipient and the donor, can influence dd-cfDNA levels and cause increases in the levels of dd-cfDNA themselves without suspicion of rejection. All that glitters in this technology is not gold; therefore, in this article, we discuss the current state of clinical studies, the benefits, and disadvantages.
1. Introduction
In kidney transplantation, a biopsy is currently the gold standard for monitoring the transplanted organ. A biopsy involves taking a small tissue sample from the transplanted kidney, which is then examined under a microscope to assess organ health. This is done to assess for any signs of rejection or other problems that may be occurring in the transplanted kidney. However, this is far from an ideal screening method given its invasive nature and the discomfort it can cause the patient. Large-scale studies in renal transplantation show that approximately 1% of biopsies generate major complications, with a risk of macroscopic hematuria greater than 3.5% [1]. Most biopsy-related complications, such as pain and bleeding, are minor and localized, and can be managed conservatively [2]. The most severe complication, however, is the risk of perforation of the collecting system or the kidney itself, which can result in severe hemorrhage, sepsis, and even death. To minimize the risk of complications, careful patient selection, proper imaging guidance, and specialized instruments and techniques are essential [3,4]. Imaging techniques, such as ultrasound and computed tomography (CT), are essential for accurate needle placement. Ultrasound imaging is the most commonly used modality for needle guidance due to its versatility, cost-effectiveness, and relative safety. It allows for real-time visualization of the renal transplant and the surrounding anatomy, making it ideal for guiding percutaneous needle placement. CT imaging can also be used but is typically reserved for more complex cases with insufficient ultrasound imaging.
In addition, with current immunosuppressive therapies, the detection of subclinical rejection is too infrequent to justify this risk, which has meant that many units no longer perform these routine biopsies [5], thus raising the urgent need to find a new non-invasive biomarker that allows the detection of said rejection in order to intervene in time or modify immunosuppression. In response to this need, recent studies have shown that non-invasive biomarkers could be a viable option for detecting subclinical rejection [6,7,8,9]. These non-invasive biomarkers include urinary and serum markers such as urinary albumin-to-creatinine ratio (UACR) and donor-specific antibodies (DSA). Additionally, imaging techniques, such as magnetic resonance imaging (MRI) and ultrasound (US), have been used to detect graft changes that may signal rejection [10,11,12]. Finally, genetic and epigenetic biomarkers, such as microRNAs, have also been used to detect subclinical rejection [13,14,15]. Ultimately, using these non-invasive biomarkers could help identify and intervene in cases of subclinical rejection earlier, thus avoiding more serious complications.
Currently, serum creatinine and urinary indicators are the ones that fulfill this role because they are cheap, easily interpretable, and relatively reliable [16]. The creatinine level in the blood measures kidney function, and an increased creatinine level indicates a decrease in kidney function [17]. Urinary indicators, such as the urine protein/creatinine ratio, are used to assess kidney function [18]. These tests can indicate the health of the kidneys. Other tests, such as blood urea nitrogen and glomerular filtration rate, are also used to assess kidney function [19]. However, these tests are more expensive and require specialized equipment. They are also more challenging to interpret and can be affected by factors such as hydration status.
In conclusion, serum creatinine and urinary indicators are the most commonly used tests for assessing kidney function due to their affordability, ease of interpretation, and reliability. However, its sensitivity and specificity in detecting allograft damage are poor. The recent approval of tests to detect donor-derived cell-free DNA (dd-cfDNA) has given great hope in the non-invasive recognition of allogeneic damage.
2. Types de Cell-Free DNA
Degraded DNA fragments released into the blood or other fluids are known as cfDNA. Its first detection dates back to 1948 in patients with systemic lupus erythematosus [20]. It would not be until 1970 that this new biomarker would begin to be considered helpful for the clinic, as researchers observed differences in its concentration depending on the health status of the individual studied and began to see its application in cancer patients by allowing the detection of fragments of tumor DNA in the blood [21]. Its interest increased when it was discovered that tumor cells not only released cfDNA into the bloodstream but that these fragments also had the genetic and epigenetic changes of the tumor cells from which they had originated [22]. Shortly after, analysis of fetal cfDNA in maternal plasma began to be used to detect Rh mismatches and chromosomal aneuploidies [23].
Recent literature shows that different cfDNA types can be used as biomarkers of various disease states [16,24]. The following stand out for their relevance: ccf mtDNA (circulating cell-free mitochondrial DNA), ctDNA (circulating tumor DNA), cffDNA (cell-free fetal DNA), and dd-cfDNA (donor-derived cell-free DNA). These types and their applications are listed in Table 1.

Table 1.
Different types of cfDNA and their main clinical applications.
3. History of Donor-Derived Cell-Free DNA
Non-patient cfDNA from a transplanted organ is known as circulating dd-cfDNA and can be detected in both blood and urine. Initially, the dd-cfDNA concentration increases to values greater than 5% after transplantation; however, these decrease rapidly and are practically undetectable after a week. Therefore, the presence of dd-cfDNA in the blood of the transplanted patient after a short period may be related to possible complications [1,26]. If the transplanted individual rejects the graft, the concentration of dd-cfDNA increases up to five times more than in healthy controls. In addition, these increased levels can be identified before any other clinical or biochemical symptom or complication, which is why it is of great interest for the early approach to subclinical rejection [27].
In 1998, this interesting biomarker was detected for the first time in the plasma and urine of solid organ recipients, identifying DNA from the Y chromosome in women transplanted with organs from male donors [27,28]. After this discovery, the technology began to evolve, emerging quantitative PCR techniques oriented to typing HLA genes. However, they had reproducibility problems and could not distinguish donor and recipient if they shared typings [5,29].
It would not be until 2011 that a method to detect dd-cfDNA employing digital PCR was devised based on analyzing the differences in SNPs between donor and recipient [30]. This strategy was optimized in 2016 with other authors [31] who, based on the principles of allelic imbalance, were able to measure dd-cfDNA levels by genotyping only the recipientm and between 150,000 and 600,000 SNPs from the donor, therefore, not being necessary to genotype the latter [15] altogether. Later that same year, other authors demonstrated that it was possible to quantify cfDNA levels by analyzing only 266 carefully chosen SNPs to minimize the probability that two unrelated individuals share them [32]. However, although these discoveries have laid the foundations for commercial dd-cfDNA detection kits, they have a series of limitations that have not yet been resolved, such as the impossibility of detecting dd-cfDNA levels in the transplantation of identical twins or differentiating the presence of more than two different genomes, as occurs in the case of retransplanted patients [16,33].
4. Commercial Tests for dd-cfDNA Detection
Three commercial dd-cfDNA detection kits are available for clinical use in kidney transplantation; Allosure from CareDx, Prospera from Natera, and Trac from Viracor Eurofins [25,29,34]. The most commonly used and described in various publications is CareDX, where a panel of 266 SNPs in 22 somatic chromosomes was used to study 102 kidney transplant recipients, 27 with rejection confirmed by biopsy [33]. In the said test, a 1% dd-cfDNA was established as a cut-off to discriminate the presence or absence of active rejection, and it had a specificity of 85%, a sensitivity of 59%, a positive predictive value (PPV) of 61% and an 84% of accuracy negative predictive value (NPV). In this study, TCMR was insufficiently detected, as confirmed by Huang in an independent study with the same test [35]. One reason may be using relatively long amplicons (100–130 bp) in the employed test. According to Clausen et al. [36], the recommended amplicon length is 85.4 bp (66–103 bp).
Prospera de Natera’s dd-cfDNA levels were determined in the following study by detecting 13,392 SNPs on four chromosomes [30,37], and a retrospective study was performed on 178 kidney transplant patients diagnosed with rejection. Acute T-lymphocyte-mediated (TCMR) or antibody-mediated (ABMR). Based on the results obtained, a cut-off for the presence of rejection more significant than 1% of dd-cfDNA was established, as in the Allosure method, with a specificity of 73%, a sensitivity of 89%, a PPV of 52%, and an NPV of 95% [38]. TCMR was well detected, presumably due to shorter amplicon size. Similar results were obtained in the Trifecta study [39].
These two studies would be the most relevant to date at the level of bibliographic background; however, a large number of companies are trying to develop dd-cfDNA kits, and that is where the Eurofins TRAC (Transplant Rejection Allograft Check) study would come in, which uses NGS techniques and recipient genotype data to determine the percentage of dd-cfDNA from the donated organ. In said study, biomarkers were determined in 77 kidney transplant patients with a cut-off of 0.70% to discriminate active rejection and sensitivity values of 58%, specificity of 85%, PPV of 55%, and NPV of 86% (Eurofins study) [38].
To clarify and inform the particular process, a schematic illustration of cfDNA isolation and analysis is shown in Figure 1.
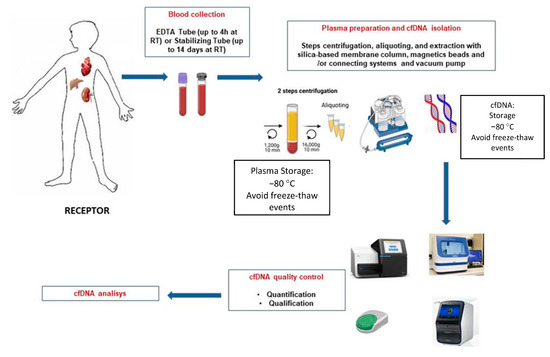
Figure 1.
Schematic overview of the different steps for blood sample processing of graft recipients and cfDNA extraction.
These commercial kits for determining dd-cfDNA represent a significant advance in the early detection of subclinical rejection; however, their main drawback is that their prohibitive price means that they are still not valid as a detection technique screening [33,40]. Another group carried out an economic analysis in which commercial dd-cfDNA tests were used as screening for subclinical rejection in kidney transplant patients and concluded that this system was not cost-efficient and that, in order to reach so, the price of each test should be less than US$700, instead of the US$2200–2800 it currently ranges from [38]. That is why new technologies aimed at minimizing this cost are beginning to emerge, such as those based on dd-PCR [41], which reduce the price to less than US$400. However, this technology is relatively recent and has few studies that endorse it [42].
5. Current Status of dd-cfDNA
Leaving economic aspects aside, dd-cfDNA has the problem of having a relatively low PPV (although it should be mentioned that predictive values also depend on the variable prevalence of rejection in the study populations), which makes it difficult to interpret a high level of dd-cfDNA in the absence of additional clinical information, which is why it is often used, together with the analysis of leukocyte expression markers. However, its NPV is much higher, which allows it to play an essential role in avoiding unnecessary biopsies [43]. In addition, it is important to remember that other types of pathologies, independent of rejection, can raise dd-cfDNA levels, so it is essential to evaluate the results within the patient’s clinical context [33,44,45,46,47,48]. The absolute quantification of dd-cfDNA (cp/mL) has the advantage of not being affected by changes in the cfDNA receptor [49,50]. Leukopenia and leukocytosis can alter the dd-cfDNA fraction and produce false positive or negative results [41,51,52]. An example of these facts was the studies carried out by another group in 2020, which retrospectively studied elevations in dd-cfDNA levels in patients with BK viremia; however, these studies did not reach statistical significance [53]. Another group worked along the same lines, observing a positive correlation between BK viremia and increased dd-cfDNA levels in a small cohort of 10 patients [54].
In addition, since the initial validation studies were carried out at the specific moments in which rejection was suspected, there is still not a good understanding of how dd-cfDNA levels naturally evolve post-transplant. In addition, various factors both in the recipient (such as the panel reactive antibody [PRA]) and in the donor (living transplant or after circulatory death [DCD]) can influence dd-cfDNA levels and cause increases in the levels of dd-cfDNA themselves in the absence of suspicion of rejection [33]. Another physiological factor influencing dd-cfDNA that is being studied is obesity, with a retrospective study (in 2020) showing an inverse relationship between morbid obesity and dd-cfDNA levels [55].
The predictive value of dd-cfDNA has been extensively analyzed in acute cellular rejection (ACR) cases and AMR, the two main mechanisms of allograft damage. Among the multiple studies carried out, the most relevant was the so-called DART (Diagnosing Acute Rejection in Kidney Transplant), carried out in the United States with a representative sample of the kidney transplant population (n = 384) [37], which reports a better predictor of AMR than of ACR, since the levels of the biomarker in the first case are much higher, ranging between 1.4% and 2.9%. In comparison, for ACR, they are around 1.2%.
6. Active Studies
Given the great potential of dd-cfDNA, many active clinical trials are delving into it, both by commercial houses to improve their products and by hospitals and university entities seeking to expand knowledge and technology, and the clinical utility of this new biomarker is shown in Table 2.

Table 2.
Compilation of currently active clinical trials.
The different methods of cfDNA analysis have also advantages and limitations. For example, in the case of PCR-based methods, these analyses could be performed by qPCR and digital PCR. In the first case of qPCR, high specificity and sensitivity, cost-effectiveness, rapid, and ease of use are essential characteristics, and its limitations are no multiplexing and limited to detecting known mutations. In the second case of digital PCR, the particular advantages would be up to five targets, high sensitivity and specificity, absolute quantification, single molecule analysis, cost-effective, rapid and ease of use, and its limitations are limited multiplexing (number of fluorescent colors) and limited to detection of known mutations.
In the case of PCR coupled to spectrometry, the advantage is multiplexing capacity, and its limitation would be that this method is also limited to detecting known mutations as previous methods.
Finally, the NGS-based methods can be targeted or untargeted. In the first case of targeted, the methods or techniques [Tam-Seq, eTam-Seq, Safe-SeqS, duplex sequencing, TEC-Seq, Single primer extension (SPE), SPE-duplex UMI, CAPP-Seq, iDES eCAPP-Seq, or Ig-HTS] have advantages as high and very high specificity, error correction, error correction by SSCS (singe stranded consensus sequence), error correction by DSCS (double-stranded consensus sequence), hybrid capture method (not dependent on fragment size), amplicon methods by SPE (not dependent on fragment size), error correction by SSCS and/or error correction by DSCS and correction of stereotypical error and their limitation are amplicon methods by multiplex PCR (depend on fragment size), less comprehensive than WGS (whole genome sequencing) or WES (whole exome sequencing), need extensive input, allelic bias (capture), stereotypical errors (hybridization steps) or tissue biopsy needed.
In the case of untargeted, the advantages are mutation discovery and signatures, CNV (copy number variation), fusion genes, rearrangements, predicted neoantigens, tumor mutational burden, shallow sequencing, genome-wide, profiling, and identification of cancer signatures. The limitations would be low sensitivity (increasing depth leads to high cost), need for bioinformatics expertise, expensive, variable sensitivity, and lots of data generated.
7. Future Directions
In the field of kidney transplantation, a biopsy is currently the only reliable method to detect graft damage, so there is an urgent need to find a non-invasive technique or a biomarker that allows anticipating the deterioration of kidney function in order to intervene early or modify immunosuppression [56,57,58]. Currently, serum creatinine and urinary indicators are the ones that fulfill this role because they are cheap, easily interpretable, and relatively reliable. However, their sensitivity and specificity in detecting allograft damage are poor. For this reason, the recent approval of tests to detect dd-cfDNA has given great hope in the non-invasive recognition of allogeneic damage. The use of dd-cfDNA tests is growing in importance due to their sensitivity and specificity, which allow early and reliable detection of allograft damage. Additionally, dd-cfDNA tests are easier to use and interpret than conventional tests, as they do not require sample manipulation and can be performed with a single sample. Furthermore, they provide a more comprehensive range of information, such as the presence of donor-specific antibodies (DSA), which can be used to tailor treatment in the case of allograft injury.
After a kidney transplant, various causes can cause graft failure. On the one hand, a rejection of the transplanted kidney can occur. Rejection episodes can be mild, moderate, or severe. In addition, the risk of infection is increased after a transplant due to the use of immunosuppressive drugs. Infections can lead to graft failure if not treated on time.
On the other hand, if the transplanted kidney does not receive enough blood, it may not work correctly, and the graft may fail. Damage to transplanted kidney tissue can also occur from physical trauma, surgery, or certain medications. Blood clots can form in the transplanted kidney, leading to graft failure. Furthermore, using immunosuppressive drugs can cause long-term toxicity, leading to graft failure. In these cases, dd-cfDNA levels increase rapidly, which makes it possible to act in the face of subclinical rejection more quickly and efficiently than just measuring creatinine. This can allow for earlier intervention when subclinical rejection occurs, preventing further damage to the organ. In addition, because dd-cfDNA levels are a better indicator of subclinical rejection than creatinine, clinicians can be more confident in their diagnosis and treatment. In this way, identifying an early increase in dd-cfDNA levels in the receptor would not only reduce the number of unnecessary biopsies. However, it would also provide the clinician with essential information to modify the immunosuppressive therapy at the appropriate time and prevent the progression of the damage [59,60]. Identifying an early increase in dd-cfDNA levels in the receptor could reduce unnecessary biopsies and provide the clinician with essential information to modify the immunosuppressive therapy at the appropriate time and prevent the progression of the damage. This would benefit both the patient and the healthcare system, reducing costs and improving patient outcomes.
However, despite the eventual promise of this biomarker, many aspects of it and its implementation are still not fully controlled, so further investigations are necessary. As previously highlighted, it is still unclear how different donor and recipient parameters can influence this measurement and alter its results. In addition, there is still no consensus on which clinical data should prevail and whether the absolute value of dd-cfDNA or the percentage should be standardized to optimize clinical practice [8,11,16,29].
Another problem currently facing the use of dd-cfDNA as a biomarker in clinical practice is that most commercial kits require sending blood samples to an external laboratory for analysis, leading to inconsistent results. cf-DNA is quite unstable, and the transfer and transport processes and the delay in sample processing can alter the results, so it would be necessary to establish the possibility of performing these tests in the transplant center itself [33].
Lastly, another critical factor for health management is the current high economic cost of cfDNA determination, especially with NGS technology, which makes eventual implementation in a clinical service more expensive.
Therefore, despite the strong demand for a new biomarker to predict rejection and the great hope placed on dd-cfDNA, today, the number of variables that surround it means that it cannot yet reach the clinical relevance that is expected, as highlighted in two recent meta-analyses that conclude that, even though dd-cfDNA may become a very valuable marker for detecting AMR in kidney transplant patients, it is not yet fully demonstrated or established when it comes to coping with cell rejection [60,61]. This does not mean that research should be abandoned, but rather that more studies should be conducted to understand better the role that dd-cfDNA can play in predicting rejection and to develop better methods for interpreting the results obtained from the tests. The potential of longitudinal monitoring for personalized immunosuppression should be addressed [62]. Medicare provides coverage for dd-cfDNA routine testing in the US based on current evidence [35].
With further research, it is possible that dd-cfDNA could become a reliable and clinically relevant tool for predicting rejection and improving patient outcomes in kidney transplantation.
In conclusion, dd-cfDNA is a promising biomarker capable of predicting acute rejection organ transplants, but more and more studies should be performed to complete its implementation in routine clinical practice.
Funding
Our work was possible thanks to support from Instituto de Salud Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness. Grant Number P19/01194 and co-funding of the European Union with European Fund of Regional Development (FEDER) with the principle of “A manner to build Europe”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Schwarz, A.; Gwinner, W.; Hiss, M.; Radermacher, J.; Mengel, M.; Haller, H. Safety and Adequacy of Renal Transplant Protocol Biopsies. Am. J. Transplant. 2005, 5, 1992–1996. [Google Scholar] [CrossRef] [PubMed]
- Volpe, A.; Kachura, J.R.; Geddie, W.R.; Evans, A.J.; Gharajeh, A.; Saravanan, A.; Jewett, M.A.S. Techniques, Safety and Accuracy of Sampling of Renal Tumors by Fine Needle Aspiration and Core Biopsy. J. Urol. 2007, 178, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.R.; Thomas, S. Complications of image-guided thermal ablation of liver and kidney neoplasms. Semin. Intervent. Radiol. 2014, 31, 138–148. [Google Scholar] [PubMed]
- Michel, M.S.; Trojan, L.; Rassweiler, J.J. Complications in Percutaneous Nephrolithotomy. Eur. Urol. 2007, 51, 899–906. [Google Scholar] [CrossRef]
- Rush, D.N.; Gibson, I.W. Subclinical inflammation in renal transplantation. Transplantation 2019, 103, E139–E145. [Google Scholar] [CrossRef]
- Mac, Q.D.; Mathews, D.V.; Kahla, J.A.; Stoffers, C.M.; Delmas, O.M.; Holt, B.A.; Adams, A.B.; Kwong, G.A. Non-invasive early detection of acute transplant rejection via nanosensors of granzyme B activity. Nat. Biomed. Eng. 2019, 3, 281–291. [Google Scholar] [CrossRef]
- Hirt-Minkowski, P.; De Serres, S.A.; Ho, J. Developing renal allograft surveillance strategies-urinary biomarkers of cellular rejection. Can. J. Kidney Health Dis. 2015, 2, 28. [Google Scholar] [CrossRef]
- Jamshaid, F.; Froghi, S.; Di Cocco, P.; Dor, F.J.M.F. Novel non-invasive biomarkers diagnostic of acute rejection in renal transplant recipients: A systematic review. Int. J. Clin. Pract. 2018, 72, e13220. [Google Scholar] [CrossRef]
- Baumann, A.K.; Beck, J.; Kirchner, T.; Hartleben, B.; Schütz, E.; Oellerich, M.; Wedemeyer, H.; Jaeckel, E.; Taubert, R. Elevated fractional donor-derived cell-free DNA during subclinical graft injury after liver transplantation. Liver Transplant. 2022, 28, 1911–1919. [Google Scholar] [CrossRef]
- Neimatallah, M.A.; Dong, Q.; Schoenberg, S.O.; Cho, K.J.; Prince, M.R. Magnetic Resonance Imaging in Renal Transplantation. J. Magn. Reson. Imaging Off. J. Int. Soc. Magn. Reson. Med. 1999, 10, 357–368. [Google Scholar] [CrossRef]
- Hollis, E.; Shehata, M.; Khalifa, F.; Abou El-Ghar, M.; El-Diasty, T.; El-Baz, A. Towards non-invasive diagnostic techniques for early detection of acute renal transplant rejection: A review. Egypt. J. Radiol. Nucl. Med. 2017, 48, 257–269. [Google Scholar] [CrossRef]
- Farag, A.; El-Baz, A.; Yuksel, S.E.; El-Ghar, M.A.; Eldiasty, T. A framework for the detection of acute renal rejection with dynamic contrast enhanced magnetic resonance imaging. In Proceedings of the 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro, Arlington, VA, USA, 6–9 April 2006; 2006, pp. 418–421. [Google Scholar]
- Bontha, S.V.; Maluf, D.G.; Mueller, T.F.; Mas, V.R. Systems Biology in Kidney Transplantation: The Application of Multi-Omics to a Complex Model. Am. J. Transplant. 2017, 17, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Asvapromtada, S.; Sonoda, H.; Kinouchi, M.; Oshikawa, S.; Takahashi, S.; Hoshino, Y.; Sinlapadeelerdkul, T.; Yokota-Ikeda, N.; Matsuzaki, T.; Ikeda, M. Recent Advances on Biomarkers of Early and Late Kidney Graft Dysfunction. Int. J. Mol. Sci. 2020, 21, 5404. [Google Scholar]
- Salvadori, M.; Tsalouchos, A. Biomarkers in renal transplantation: An updated review. World J. Transplant. 2017, 7, 161. [Google Scholar] [CrossRef]
- Jimenez-Coll, V.; Llorente, S.; Boix, F.; Alfaro, R.; Galián, J.A.; Martinez-Banaclocha, H.; Botella, C.; Moya-Quiles, M.R.; Muro-Pérez, M.; Minguela, A.; et al. Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome. Int. J. Mol. Sci. 2023, 24, 3908. [Google Scholar] [CrossRef]
- Bostom, A.G.; Kronenberg, F.; Ritz, E. Predictive performance of renal function equations for patients with chronic kidney disease and normal serum creatinine levels. J. Am. Soc. Nephrol. 2002, 13, 2140–2144. [Google Scholar] [CrossRef]
- Yang, C.Y.; Chen, F.A.; Chen, C.F.; Liu, W.S.; Shih, C.J.; Ou, S.M.; Yang, W.C.; Lin, C.C.; Yang, A.H. Diagnostic Accuracy of Urine Protein/Creatinine Ratio Is Influenced by Urine Concentration. PLoS ONE 2015, 10, e0137460. [Google Scholar] [CrossRef]
- Gowda, S.; Desai, P.B.; Kulkarni, S.S.; Hull, V.V.; Math, A.A.K.; Vernekar, S.N. Markers of renal function tests. N. Am. J. Med. Sci. 2010, 2, 170. [Google Scholar]
- Tug, S.; Helmig, S.; Menke, J.; Zahn, D.; Kubiak, T.; Schwarting, A.; Simon, P. Correlation between cell free DNA levels and medical evaluation of disease progression in systemic lupus erythematosus patients. Cell Immunol. 2014, 292, 32–39. [Google Scholar] [CrossRef]
- Ulrich, B.C.; Guibert, N. Towards a comprehensive framework for cell-free DNA analysis: Lessons from TRACERx. Ann. Transl. Med. 2017, 5, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B. Update on the types and usage of liquid biopsies in the clinical setting: A systematic review. BMC Cancer 2018, 18, 527. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.H.; Lee, H.J.; Bae, K.; Yoon, K.A.; Lee, E.S.; Cho, Y. Efficient Capture and Isolation of Tumor-Related Circulating Cell-Free DNA from Cancer Patients Using Electroactive Conducting Polymer Nanowire Platforms. Theranostics 2016, 6, 828. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Bloom, R.D.; Bromberg, J.S.; Poggio, E.D.; Bunnapradist, S.; Langone, A.J.; Sood, P.; Matas, A.J.; Mehta, S.; Mannon, R.B.; Sharfuddin, A.; et al. Cell-Free DNA and active rejection in kidney allografts. J. Am. Soc. Nephrol. 2017, 28, 2221–2232. [Google Scholar] [CrossRef]
- Starzl, T.E.; Murase, N.; Ildstad, S.; Ricordi, C.; Demetris, A.J.; Trucco, M. Cell migration, chimerism, and graft acceptance. Lancet 1992, 339, 1579. [Google Scholar] [CrossRef]
- Beck, J.; Bierau, S.; Balzer, S.; Andag, R.; Kanzow, P.; Schmitz, J.; Gaedcke, J.; Moerer, O.; Slotta, J.E.; Walson, P.; et al. Digital Droplet PCR for Rapid Quantification of Donor DNA in the Circulation of Transplant Recipients as a Potential Universal Biomarker of Graft Injury. Clin. Chem. 2013, 59, 1732–1741. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Tein, M.S.C.; Pang, C.C.P.; Yeung, C.K.; Tong, K.L.; Magnus Hjelm, N. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet 1998, 351, 1329–1330. [Google Scholar] [CrossRef]
- Kueht, M.L.; Dongur, L.P.; Cusick, M.; Stevenson, H.L.; Mujtaba, M. The Current State of Donor-Derived Cell-Free DNA Use in Allograft Monitoring in Kidney Transplantation. J. Pers. Med. 2022, 12, 1700. [Google Scholar] [CrossRef]
- Gadi, V.K.; Nelson, J.L.; Boespflug, N.D.; Guthrie, K.A.; Kuhr, C.S. Soluble Donor DNA Concentrations in Recipient Serum Correlate with Pancreas-Kidney Rejection. Clin. Chem. 2006, 52, 379–382. [Google Scholar] [CrossRef]
- Snyder, T.M.; Khush, K.K.; Valantine, H.A.; Quake, S.R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. USA 2011, 108, 6229–6234. [Google Scholar] [CrossRef] [PubMed]
- Sharon, E.; Shi, H.; Kharbanda, S.; Koh, W.; Martin, L.R.; Khush, K.K.; Valantine, H.; Pritchard, J.K.; De Vlaminck, I. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput. Biol. 2017, 13, e1005629. [Google Scholar] [CrossRef] [PubMed]
- Grskovic, M.; Hiller, D.J.; Eubank, L.A.; Sninsky, J.J.; Christopherson, C.; Collins, J.P.; Thompson, K.; Song, M.; Wang, Y.S.; Ross, D.; et al. Validation of a Clinical-Grade Assay to Measure Donor-Derived Cell-Free DNA in Solid Organ Transplant Recipients. J. Mol. Diagn. 2016, 18, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Jordan, S.C. Donor-derived cell-free DNA in kidney transplantation: Evolving concepts and potential limitations. Kidney Int. 2022, 101, 676–677. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Sethi, S.; Peng, A.; Najjar, R.; Mirocha, J.; Haas, M.; Vo, A.; Jordan, S.C. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am. J. Transplant. 2019, 19, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Clausen, F.B.; Jørgensen, K.M.C.L.; Wardil, L.W.; Nielsen, L.K.; Krog, G.R. Droplet digital PCR-based testing for donor-derived cell-free DNA in transplanted patients as noninvasive marker of allograft health: Methodological aspects. PLoS ONE 2023, 18, e0282332. [Google Scholar] [CrossRef] [PubMed]
- 30876—Viracor TRAC® Kidney dd-cfDNA | Clinical | Eurofins-Viracor. Available online: https://www.eurofins-viracor.com/clinical/test-menu/30876-viracor-trac-kidney-dd-cfdna/ (accessed on 17 February 2023).
- Sigdel, T.K.; Archila, F.A.; Constantin, T.; Prins, S.A.; Liberto, J.; Damm, I.; Towfighi, P.; Navarro, S.; Kirkizlar, E.; Demko, Z.P.; et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. J. Clin. Med. 2018, 8, 19. [Google Scholar] [CrossRef]
- Halloran, P.F.; Reeve, J.; Madill-Thomsen, K.S.; Demko, Z.; Prewett, A.; Billings, P. The Trifecta Study: Comparing Plasma Levels of Donor-derived Cell-Free DNA with the Molecular Phenotype of Kidney Transplant Biopsies. J. Am. Soc. Nephrol. 2022, 33, 387–400. [Google Scholar]
- Muro, M. ¿Es el ADN libre circulante (cfDNA) derivado del donante (dd-cfDNA) un nuevo biomarcador para el rechazo de aloinjertos en trasplante? Inmunología 2020, 39, 23–26. [Google Scholar]
- Oellerich, M.; Sherwood, K.; Keown, P.; Schütz, E.; Beck, J.; Stegbauer, J.; Rump, L.C.; Walson, P.D. Liquid biopsies: Donor-derived cell-free DNA for the detection of kidney allograft injury. Nat. Rev. Nephrol. 2021, 17, 591–603. [Google Scholar] [CrossRef]
- Puttarajappa, C.M.; Mehta, R.B.; Roberts, M.S.; Smith, K.J.; Hariharan, S. Economic analysis of screening for subclinical rejection in kidney transplantation using protocol biopsies and noninvasive biomarkers. Am. J. Transplant. 2021, 21, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Porrini, E.; Ruggenenti, P.; Luis-Lima, S.; Carrara, F.; Jiménez, A.; de Vries, A.P.J.; Torres, A.; Gaspari, F.; Remuzzi, G. Estimated GFR: Time for a critical appraisal. Nat. Rev. Nephrol. 2018, 15, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Filippone, E.J.; Farber, J.L. The Monitoring of Donor-derived Cell-free DNA in Kidney Transplantation. Transplantation 2021, 105, 509–516. [Google Scholar] [CrossRef]
- Kataria, A.; Kumar, D.; Gupta, G. Donor-derived Cell-free DNA in Solid-organ Transplant Diagnostics: Indications, Limitations, and Future Directions. Transplantation 2021, 105, 1203–1211. [Google Scholar] [CrossRef]
- O’Callaghan, J.M.; Knight, S.R. Noninvasive biomarkers in monitoring kidney allograft health. Curr. Opin. Organ Transplant. 2019, 24, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Rosenheck, J.P.; Keller, B.C.; Fehringer, G.; Demko, Z.P.; Bohrade, S.M.; Ross, D.J. Why Cell-Free DNA Can Be a “Game Changer” for Lung Allograft Monitoring for Rejection and Infection. Curr. Pulmonol. Rep. 2022, 11, 75–85. [Google Scholar] [CrossRef]
- Oellerich, M.; Budde, K.; Osmanodja, B.; Bornemann-Kolatzki, K.; Beck, J.; Schütz, E.; Walson, P.D. Donor-derived cell-free DNA as a diagnostic tool in transplantation. Front. Genet. 2022, 13, 1031894. [Google Scholar] [CrossRef] [PubMed]
- Oellerich, M.; Shipkova, M.; Asendorf, T.; Walson, P.D.; Schauerte, V.; Mettenmeyer, N.; Kabakchiev, M.; Hasche, G.; Gröne, H.J.; Friede, T.; et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am. J. Transplant. 2019, 19, 3087–3099. [Google Scholar] [CrossRef]
- Whitlam, J.B.; Ling, L.; Skene, A.; Kanellis, J.; Ierino, F.L.; Slater, H.R.; Bruno, D.L.; Power, D.A. Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am. J. Transplant. 2019, 19, 1037–1049. [Google Scholar] [CrossRef]
- Oellerich, M.; Wu, A.; Halloran, P.F.; De Vlaminck, I.; Keller, M.; Agbor-Enoh, S. Molecular Approaches to Transplant Monitoring; Is the Horizon Here? Clin. Chem. 2021, 67, 1443–1449. [Google Scholar]
- Oellerich, M.; Budde, K.; Osmanodja, B.; Bornemann-Kolatzki, K.; Beck, J.; Schütz, E.; Walson, P.D. Donor-Derived Cell-free DNA for Personalized Immunosuppression in Renal Transplantation. Ther. Drug Monit. 2023, 45, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Lum, E.L.; Nieves-Borrero, K.; Homkrailas, P.; Lee, S.; Danovitch, G.; Bunnapradist, S. Single center experience comparing two clinically available donor derived cell free DNA tests and review of literature. Transplant. Rep. 2021, 6, 100079. [Google Scholar] [CrossRef]
- Goussous, N.; Xie, W.; Dawany, N.; Scalea, J.R.; Bartosic, A.; Haririan, A.; Kalil, R.; Drachenberg, C.; Costa, N.; Weir, M.R.; et al. Donor-derived Cell-free DNA in Infections in Kidney Transplant Recipients: Case Series. Transplant. Direct 2020, 6, e568. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Bromberg, J.; Haas, M.; Brennan, D. Donor-derived Cell-free DNA and the Prediction of BK Virus-associated Nephropathy. Transplant. Direct 2020, 6, e622. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, R.; Martínez-Banaclocha, H.; Llorente, S.; Jimenez-Coll, V.; Galián, J.A.; Botella, C.; Moya-Quiles, M.R.; Parrado, A.; Muro-Perez, M.; Minguela, A.; et al. Computational Prediction of Biomarkers, Pathways, and New Target Drugs in the Pathogenesis of Immune-Based Diseases Regarding Kidney Transplantation Rejection. Front. Immunol. 2021, 12, 5418. [Google Scholar] [CrossRef]
- Legaz, I.; Bernardo, M.V.; Alfaro, R.; Martínez-Banaclocha, H.; Galián, J.A.; Jimenez-Coll, V.; Boix, F.; Mrowiec, A.; Salmeron, D.; Botella, C.; et al. PCR Array Technology in Biopsy Samples Identifies Up-Regulated mTOR Pathway Genes as Potential Rejection Biomarkers After Kidney Transplantation. Front. Med. 2021, 8, 138. [Google Scholar] [CrossRef]
- Sureshkumar, K.K.; Aramada, H.R.; Chopra, B. Impact of body mass index and recipient age on baseline donor-derived cell free DNA (dd-cfDNA) in kidney transplant recipients. Clin. Transplant. 2020, 34, e14101. [Google Scholar] [CrossRef]
- Wijtvliet, V.P.W.M.; Plaeke, P.; Abrams, S.; Hens, N.; Gielis, E.M.; Hellemans, R.; Massart, A.; Hesselink, D.A.; De Winter, B.Y.; Abramowicz, D.; et al. Donor-derived cell-free DNA as a biomarker for rejection after kidney transplantation: A systematic review and meta-analysis. Transpl. Int. 2020, 33, 1626–1642. [Google Scholar] [CrossRef]
- Xiao, H.; Gao, F.; Pang, Q.; Xia, Q.; Zeng, X.; Peng, J.; Fan, L.; Liu, J.; Wang, Z.; Li, H. Diagnostic Accuracy of Donor-derived Cell-free DNA in Renal-allograft Rejection: A Meta-analysis. Transplantation 2021, 105, 1303–1310. [Google Scholar] [CrossRef]
- Alfaro, R.; Legaz, I.; Jimenez-Coll, V.; El Kaaoui El Band, J.; Martínez-Banaclocha, H.; Galián, J.A.; Parrado, A.; Mrowiec, A.; Botella, C.; Moya-Quiles, M.R.; et al. Microrna expression changes in kidney transplant: Diagnostic efficacy of mir-150-5p as potential rejection biomarker, pilot study. J. Clin. Med. 2021, 10, 2748. [Google Scholar] [CrossRef]
- Bu, L.; Gupta, G.; Pai, A.; Anand, S.; Stites, E.; Moinuddin, I.; Bowers, V.; Jain, P.; Axelrod, D.A.; Weir, M.R.; et al. Clinical outcomes from the Assessing Donor-derived cell-free DNA Monitoring Insights of kidney Allografts with Longitudinal surveillance (ADMIRAL) study. Kidney Int. 2022, 101, 793–803. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).