An Invasive Ductal Carcinomas Breast Cancer Grade Classification Using an Ensemble of Convolutional Neural Networks
Abstract
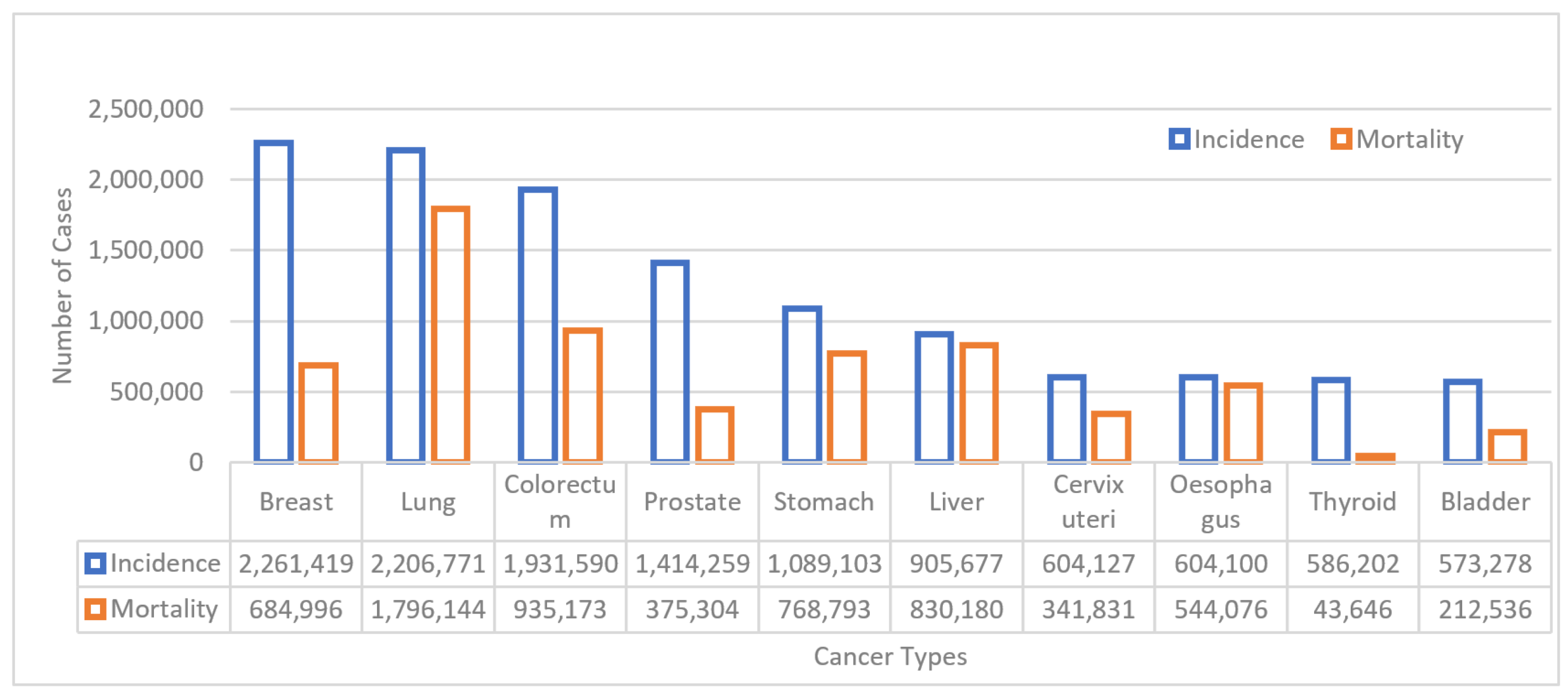
1. Introduction
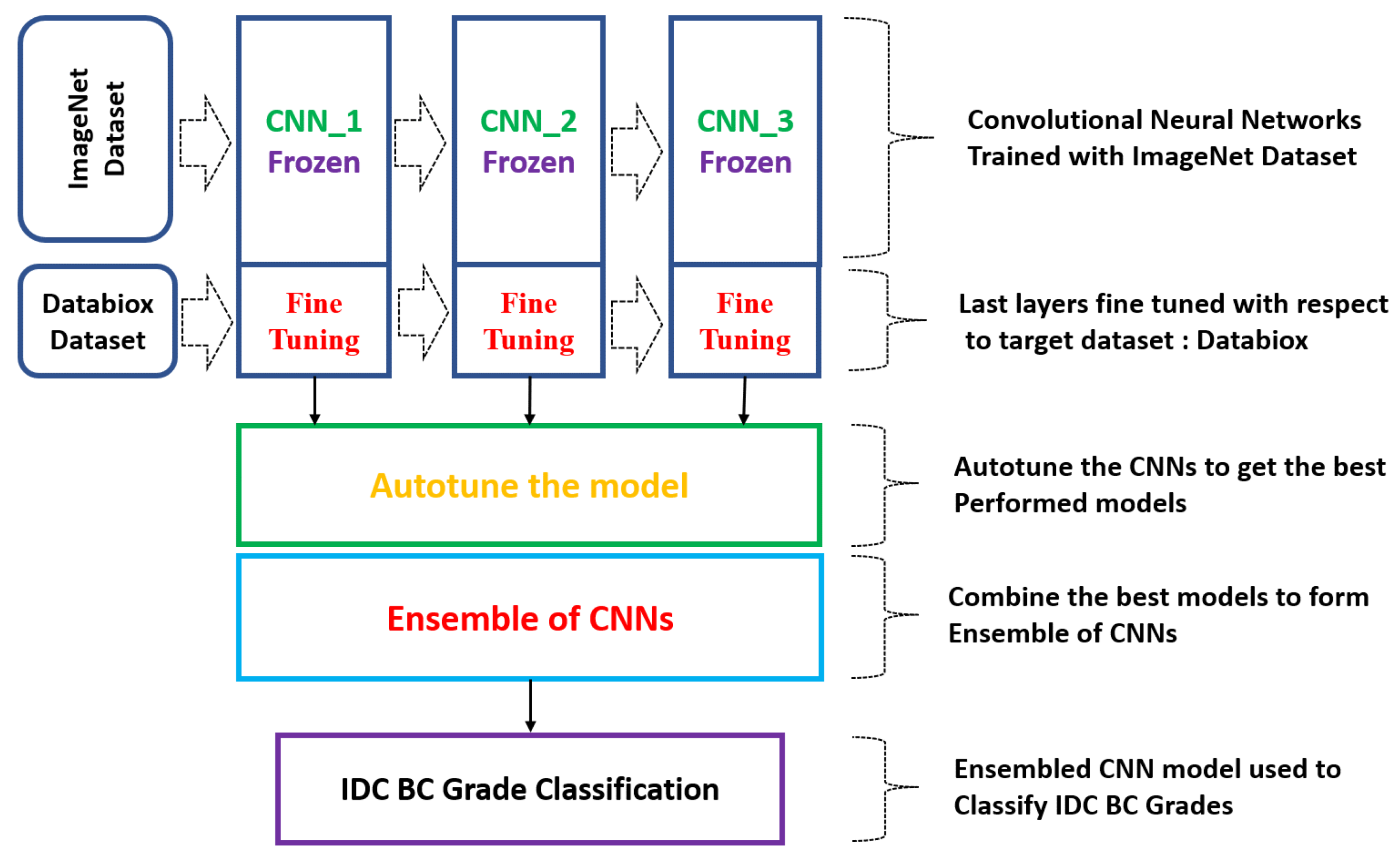
- In the present study, an ensemble model comprising three pre-trained convolutional neural networks (CNNs) was designed to make grading predictions for the Databiox dataset, which consists of histopathological images of IDC-diagnosed patients for this grade classification. Different pre-trained base models were analysed for their performances individually and in combination to determine the most optimal and coherent solution for breast cancer grade classification.
- In Databiox, the dataset is imbalanced and the distribution of images among the different grades and total number of images in each grade are insufficient for training a CNN. This may lead the problem of bias towards one particular class with more images. To overcome this issue, data augmentation techniques were employed to balance the dataset. The performance of the best model was compared for three different balanced datasets of Databiox (i.e., 1200, 1400, and 1600 images) to ensure the limit of the data augmentation.
- Additionally, the implications of the number of epochs were also demonstrated throughout this experimental work. The performances of the models were observed for four different numbers of epochs, which further determined the robustness and coherency of the proposed ensemble model. The performance of the proposed models was analysed by utilizing the evaluation parameters, namely, precision, recall, f1 score, accuracy, ROC curve, and the area under the ROC curve (AUC).
2. Material and Method Used
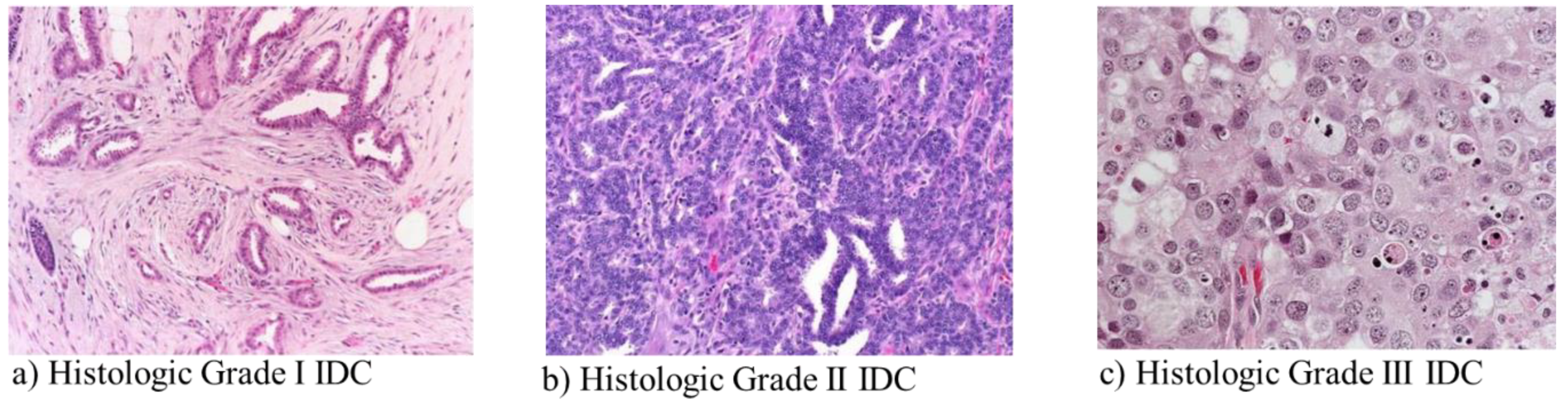
2.1. Dataset
2.2. Data Pre-Processing and Augmentation
2.3. Convolutional Neural Networks (CNNs)
2.4. Ensemble of CNNs
3. Results and Discussion
3.1. Confusion Matrix
Time Complexity
3.2. Comparison of the State-of-the-Art Techniques
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Cancer Observatory. Available online: https://gco.iarc.fr/today/online-analysis-multi-bars (accessed on 25 March 2023).
- American Cancer Society. Available online: https://www.cancer.org/cancer/breast-cancer/screening-tests-and-early-detection/breast-cancer-signs-and-symptoms.html (accessed on 25 March 2023).
- Sharma, A.; Ives, C. Management of breast cancer: Basic principles. Surgery 2022, 40, 113–120. [Google Scholar] [CrossRef]
- Breast Cancer India. Available online: https://www.breastcancerindia.net/statistics/trends.html (accessed on 25 March 2023).
- Miles, R.C.; Chou, S.-H.; Vijapura, C.; Patel, A. Breast Cancer Screening in Women With Dense Breasts: Current Status and Future Directions for Appropriate Risk Stratification and Imaging Utilization. J. Breast Imaging 2022, 4, 559–567. [Google Scholar] [CrossRef]
- Allweis, T.M.; Hermann, N.; Berenstein-Molho, R.; Guindy, M. Personalized screening for breast cancer: Rationale, present practices, and future directions. Ann. Surg. Oncol. 2021, 28, 4306–4317. [Google Scholar] [CrossRef] [PubMed]
- Mehra, R. Breast cancer histology images classification: Training from scratch or transfer learning? ICT Express 2018, 4, 247–254. [Google Scholar]
- Tan, M.; Le, Q. Efficientnetv2: Smaller Models and Faster Training. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10096–10106. [Google Scholar]
- Sarwinda, D.; Paradisa, R.H.; Bustamam, A.; Anggia, P. Deep learning in image classification using residual network (ResNet) variants for detection of colorectal cancer. Procedia Comput. Sci. 2021, 179, 423–431. [Google Scholar] [CrossRef]
- Jaiswal, A.; Gianchandani, N.; Singh, D.; Kumar, V.; Kaur, M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2021, 39, 5682–5689. [Google Scholar] [CrossRef] [PubMed]
- Bolhasani, H.; Amjadi, E.; Tabatabaeian, M.; Jassbi, S.J. A histopathological image dataset for grading breast invasive ductal carcinomas. Inform. Med. Unlocked 2020, 19, 100341. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, S. Sub-classification of invasive and non-invasive cancer from magnification independent histopathological images using hybrid neural networks. Evol. Intell. 2022, 15, 1531–1543. [Google Scholar] [CrossRef]
- Rakha, E.A.; Reis-Filho, J.S.; Baehner, F.; Dabbs, D.J.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R. Breast cancer prognostic classification in the molecular era: The role of histological grade. Breast Cancer Res. 2010, 12, 207. [Google Scholar] [CrossRef] [PubMed]
- Canadian Cancer Society. Available online: https://cancer.ca/en/cancer-information/cancer-types/breast/grading (accessed on 25 March 2023).
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, S. The Xception model: A potential feature extractor in breast cancer histology images classification. ICT Express 2022, 8, 101–108. [Google Scholar] [CrossRef]
- Ajit, A.; Acharya, K.; Samanta, A. A review of convolutional neural networks. In Proceedings of the 2020 International Conference on Emerging Trends in Information Technology and Engineering (ic-ETITE), Vellore, India, 24–25 February 2020; pp. 1–5. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Plis, S.M.; Calhoun, V.D.; Lee, J.-H. Task-specific feature extraction and classification of fMRI volumes using a deep neural network initialized with a deep belief network: Evaluation using sensorimotor tasks. NeuroImage 2017, 145, 314–328. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Zavareh, P.H.; Safayari, A.; Bolhasani, H. BCNet: A deep convolutional neural network for breast cancer grading. arXiv 2021, arXiv:2107.05037. [Google Scholar]
- Talpur, S.; Rashid, M.; Khan, S.J.; Syed, S.A. Automatic Detection System to Identify Invasive Ductal Carcinoma by Predicting Bloom Richardson Grading from Histopathological Images. J. Indep. Stud. Res. Comput. 2022, 20, 45–55. [Google Scholar] [CrossRef]
- Sujatha, R.; Chatterjee, J.M.; Angelopoulou, A.; Kapetanios, E.; Srinivasu, P.N.; Hemanth, D.J. A transfer learning-based system for grading breast invasive ductal carcinoma. IET Image Process. 2022, 17, 1979–1990. [Google Scholar] [CrossRef]
- Kumaraswamy, E.; Sharma, S.; Kumar, S. Invasive Ductal Carcinoma Grade Classification in Histopathological Images using Transfer Learning Approach. In Proceedings of the 2021 IEEE Bombay Section Signature Conference (IBSSC), Gwalior, India, 18–20 November 2021; pp. 1–6. [Google Scholar]
| Confusion Matrix | Predicted Class | |||
|---|---|---|---|---|
| Grade 0 | Grade 1 | Grade 2 | ||
| Actual Class | Grade 0 | GC00 | GC01 | GC02 |
| Grade 1 | GC10 | GC11 | GC12 | |
| Grade 2 | GC20 | GC21 | GC22 | |
| Number of Epochs = 5 | |||||
|---|---|---|---|---|---|
| CNN Model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.78 | 0.74 | 0.76 | 0.71 |
| Grade 1 | 0.72 | 0.62 | 0.67 | ||
| Grade 2 | 0.66 | 0.79 | 0.72 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.35 | 0.82 | 0.49 | 0.45 |
| Grade 1 | 0.66 | 0.24 | 0.36 | ||
| Grade 2 | 0.55 | 0.40 | 0.46 | ||
| Model 3: DenseNet201 | Grade 0 | 0.87 | 0.33 | 0.48 | 0.47 |
| Grade 1 | 0.73 | 0.12 | 0.20 | ||
| Grade 2 | 0.41 | 0.96 | 0.57 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.78 | 0.77 | 0.78 | 0.75 |
| Grade 1 | 0.67 | 0.82 | 0.74 | ||
| Grade 2 | 0.85 | 0.65 | 0.73 | ||
| Number of epochs = 10 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.70 | 0.87 | 0.77 | 0.76 |
| Grade 1 | 0.78 | 0.78 | 0.78 | ||
| Grade 2 | 0.80 | 0.67 | 0.73 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.66 | 0.61 | 0.63 | 0.61 |
| Grade 1 | 0.65 | 0.53 | 0.58 | ||
| Grade 2 | 0.55 | 0.69 | 0.61 | ||
| Model 3: DenseNet201 | Grade 0 | 0.56 | 0.75 | 0.64 | 0.69 |
| Grade 1 | 0.77 | 0.57 | 0.66 | ||
| Grade 2 | 0.75 | 0.78 | 0.76 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.78 | 0.80 | 0.79 | 0.78 |
| Grade 1 | 0.74 | 0.84 | 0.79 | ||
| Grade 2 | 0.84 | 0.69 | 0.76 | ||
| Number of epochs = 15 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.00 | 0.00 | 0.00 | 0.37 |
| Grade 1 | 0.38 | 0.83 | 0.52 | ||
| Grade 2 | 0.34 | 0.16 | 0.22 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.28 | 1.00 | 0.43 | 0.28 |
| Grade 1 | 0.00 | 0.00 | 0.00 | ||
| Grade 2 | 0.00 | 0.00 | 0.00 | ||
| Model 3: DenseNet201 | Grade 0 | 0.00 | 0.00 | 0.00 | 0.34 |
| Grade 1 | 0.00 | 0.00 | 0.00 | ||
| Grade 2 | 0.34 | 0.99 | 0.51 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.82 | 0.76 | 0.79 | 0.78 |
| Grade 1 | 0.76 | 0.80 | 0.78 | ||
| Grade 2 | 0.78 | 0.78 | 0.78 | ||
| Number of epochs = 20 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.74 | 0.83 | 0.79 | 0.82 |
| Grade 1 | 0.83 | 0.83 | 0.83 | ||
| Grade 2 | 0.87 | 0.79 | 0.83 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.64 | 0.55 | 0.59 | 0.62 |
| Grade 1 | 0.66 | 0.72 | 0.69 | ||
| Grade 2 | 0.55 | 0.56 | 0.55 | ||
| Model 3: DenseNet201 | Grade 0 | 0.66 | 0.67 | 0.66 | 0.69 |
| Grade 1 | 0.66 | 0.80 | 0.72 | ||
| Grade 2 | 0.83 | 0.56 | 0.67 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.82 | 0.88 | 0.85 | 0.85 |
| Grade 1 | 0.85 | 0.87 | 0.86 | ||
| Grade 2 | 0.89 | 0.80 | 0.84 | ||
| Number of Epochs = 5 | |||||
|---|---|---|---|---|---|
| CNN Model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.64 | 0.94 | 0.76 | 0.79 |
| Grade 1 | 0.82 | 0.73 | 0.78 | ||
| Grade 2 | 0.95 | 0.74 | 0.83 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.38 | 0.72 | 0.49 | 0.45 |
| Grade 1 | 0.51 | 0.38 | 0.43 | ||
| Grade 2 | 0.52 | 0.33 | 0.40 | ||
| Model 3: DenseNet201 | Grade 0 | 0.48 | 0.51 | 0.49 | 0.37 |
| Grade 1 | 0.26 | 0.11 | 0.15 | ||
| Grade 2 | 0.35 | 0.56 | 0.43 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.89 | 0.80 | 0.84 | 0.81 |
| Grade 1 | 0.76 | 0.83 | 0.80 | ||
| Grade 2 | 0.80 | 0.78 | 0.79 | ||
| Number of epochs = 10 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.84 | 0.93 | 0.88 | 0.85 |
| Grade 1 | 0.88 | 0.80 | 0.84 | ||
| Grade 2 | 0.84 | 0.86 | 0.85 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.94 | 0.21 | 0.34 | 0.53 |
| Grade 1 | 0.45 | 0.81 | 0.58 | ||
| Grade 2 | 0.65 | 0.45 | 0.53 | ||
| Model 3: DenseNet201 | Grade 0 | 0.54 | 0.77 | 0.64 | 0.62 |
| Grade 1 | 0.85 | 0.36 | 0.50 | ||
| Grade 2 | 0.61 | 0.81 | 0.70 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.87 | 0.92 | 0.89 | 0.88 |
| Grade 1 | 0.86 | 0.84 | 0.85 | ||
| Grade 2 | 0.90 | 0.88 | 0.89 | ||
| Number of epochs = 15 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.96 | 0.93 | 0.94 | 0.88 |
| Grade 1 | 0.84 | 0.87 | 0.86 | ||
| Grade 2 | 0.86 | 0.84 | 0.85 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.59 | 0.70 | 0.64 | 0.59 |
| Grade 1 | 0.67 | 0.45 | 0.54 | ||
| Grade 2 | 0.53 | 0.65 | 0.59 | ||
| Model 3: DenseNet201 | Grade 0 | 0.70 | 0.65 | 0.67 | 0.72 |
| Grade 1 | 0.65 | 0.84 | 0.74 | ||
| Grade 2 | 0.86 | 0.63 | 0.73 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.87 | 0.94 | 0.91 | 0.91 |
| Grade 1 | 0.91 | 0.88 | 0.89 | ||
| Grade 2 | 0.95 | 0.92 | 0.93 | ||
| Number of epochs = 20 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.29 | 0.39 | 0.34 | 0.32 |
| Grade 1 | 0.35 | 0.24 | 0.28 | ||
| Grade 2 | 0.33 | 0.36 | 0.34 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.00 | 0.00 | 0.00 | 0.36 |
| Grade 1 | 0.00 | 0.00 | 0.00 | ||
| Grade 2 | 0.36 | 1.00 | 0.53 | ||
| Model 3: DenseNet201 | Grade 0 | 0.23 | 0.58 | 0.33 | 0.27 |
| Grade 1 | 0.32 | 0.31 | 0.32 | ||
| Grade 2 | 0.00 | 0.00 | 0.00 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.97 | 0.93 | 0.95 | 0.94 |
| Grade 1 | 0.90 | 0.95 | 0.92 | ||
| Grade 2 | 0.97 | 0.93 | 0.95 | ||
| Number of Epochs = 5 | |||||
|---|---|---|---|---|---|
| CNN Model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.59 | 0.90 | 0.71 | 0.70 |
| Grade 1 | 0.92 | 0.50 | 0.64 | ||
| Grade 2 | 0.68 | 0.80 | 0.73 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.31 | 0.90 | 0.47 | 0.37 |
| Grade 1 | 0.49 | 0.29 | 0.36 | ||
| Grade 2 | 0.55 | 0.06 | 0.10 | ||
| Model 3: DenseNet201 | IDC Grade 0 | 0.38 | 0.82 | 0.52 | 0.44 |
| Grade 1 | 0.45 | 0.10 | 0.16 | ||
| Grade 2 | 0.53 | 0.60 | 0.56 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.78 | 0.72 | 0.75 | 0.79 |
| Grade 1 | 0.78 | 0.83 | 0.81 | ||
| Grade 2 | 0.82 | 0.80 | 0.81 | ||
| Number of epochs = 10 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.27 | 0.03 | 0.06 | 0.32 |
| Grade 1 | 0.39 | 0.09 | 0.15 | ||
| Grade 2 | 0.32 | 0.85 | 0.46 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.00 | 0.00 | 0.00 | 0.40 |
| Grade 1 | 0.40 | 1.00 | 0.57 | ||
| Grade 2 | 0.00 | 0.00 | 0.00 | ||
| Model 3: DenseNet201 | Grade 0 | 0.00 | 0.00 | 0.00 | 0.38 |
| Grade 1 | 0.39 | 0.93 | 0.55 | ||
| Grade 2 | 0.22 | 0.02 | 0.04 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.79 | 0.89 | 0.83 | 0.86 |
| Grade 1 | 0.92 | 0.84 | 0.87 | ||
| Grade 2 | 0.86 | 0.86 | 0.86 | ||
| Number of epochs = 15 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.36 | 0.04 | 0.07 | 0.34 |
| Grade 1 | 0.35 | 0.91 | 0.51 | ||
| Grade 2 | 0.21 | 0.05 | 0.09 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.32 | 1.00 | 0.48 | 0.32 |
| Grade 1 | 0.00 | 0.00. | 0.00 | ||
| Grade 2 | 0.00 | 0.00 | 0.00 | ||
| Model 3: DenseNet201 | Grade 0 | 0.26 | 0.56 | 0.36 | 0.25 |
| Grade 1 | 0.26 | 0.22 | 0.24 | ||
| Grade 2 | 0.00 | 0.00 | 0.00 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.92 | 0.81 | 0.86 | 0.86 |
| Grade 1 | 0.81 | 0.94 | 0.87 | ||
| Grade 2 | 0.88 | 0.83 | 0.85 | ||
| Number of epochs = 20 | |||||
| CNN model | Grade | Precision | Recall | F1-Score | Accuracy |
| Model 1: EfficientNetV2L | Grade 0 | 0.24 | 0.64 | 0.35 | 0.24 |
| Grade 1 | 0.45 | 0.04 | 0.07 | ||
| Grade 2 | 0.19 | 0.15 | 0.16 | ||
| Model 2: ResNet152V2 | Grade 0 | 0.23 | 0.14 | 0.17 | 0.39 |
| Grade 1 | 0.42 | 0.88 | 0.57 | ||
| Grade 2 | 0.00 | 0.00 | 0.00 | ||
| Model 3: DenseNet201 | Grade 0 | 0.43 | 0.28 | 0.34 | 0.47 |
| Grade 1 | 0.48 | 0.67 | 0.56 | ||
| Grade 2 | 0.48 | 0.39 | 0.43 | ||
| Ensemble of CNN model (Model 1 + Model 2 + Model 3) | Grade 0 | 0.86 | 0.85 | 0.86 | 0.87 |
| Grade 1 | 0.85 | 0.89 | 0.87 | ||
| Grade 2 | 0.89 | 0.84 | 0.87 | ||
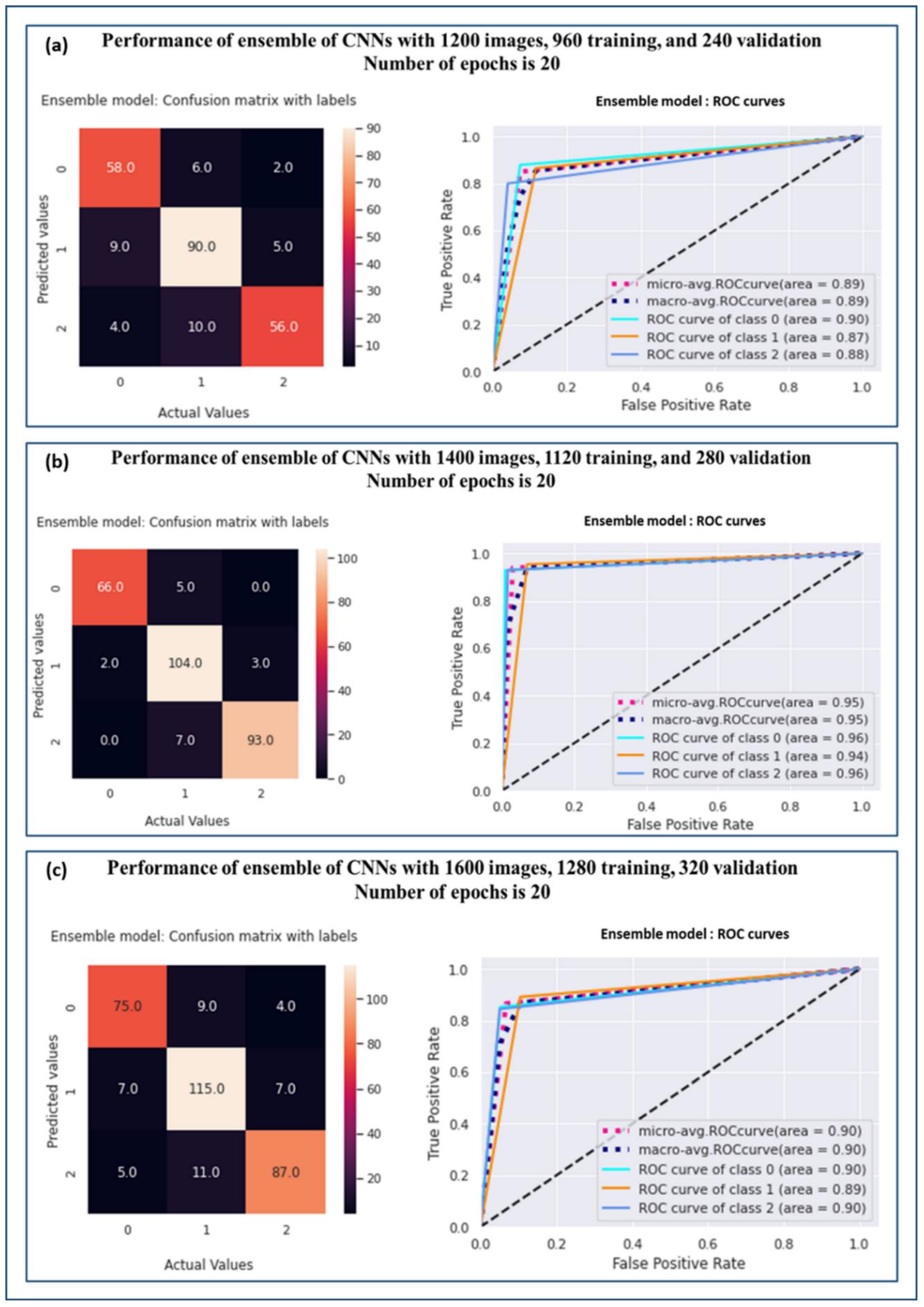
| Sample Size | Grade | Precision | Recall | F1-Score | AUC | Accuracy |
|---|---|---|---|---|---|---|
| 1200 | Grade 0 | 0.82 | 0.88 | 0.85 | 0.90 | 0.85 |
| Grade 1 | 0.85 | 0.87 | 0.86 | 0.87 | ||
| Grade 2 | 0.89 | 0.80 | 0.84 | 0.88 | ||
| 1400 | Grade 0 | 0.97 | 0.93 | 0.95 | 0.96 | 0.94 |
| Grade 1 | 0.90 | 0.95 | 0.92 | 0.94 | ||
| Grade 2 | 0.97 | 0.93 | 0.95 | 0.96 | ||
| 1600 | Grade 0 | 0.86 | 0.85 | 0.86 | 0.90 | 0.87 |
| Grade 1 | 0.85 | 0.89 | 0.87 | 0.89 | ||
| Grade 2 | 0.89 | 0.84 | 0.87 | 0.90 |
| Samples | Epochs | Time for Data Augmentation (In Minutes) | Time for Training and Validation (In Minutes) | Total Time (In Minutes) |
|---|---|---|---|---|
| 1200 | 5 | 21 | 27 | 48 |
| 10 | 21 | 33 | 54 | |
| 15 | 21 | 46 | 67 | |
| 20 | 21 | 50 | 71 | |
| 1400 | 5 | 24 | 30 | 54 |
| 10 | 24 | 39 | 63 | |
| 15 | 24 | 48 | 72 | |
| 20 | 24 | 53 | 77 | |
| 1600 | 5 | 28 | 34 | 62 |
| 10 | 28 | 42 | 70 | |
| 15 | 28 | 49 | 77 | |
| 20 | 28 | 58 | 86 |
| Reference | Year | Approach | Performance Metric |
|---|---|---|---|
| Zavareh et al. [22] | 2021 | Transfer learning approach (VGG16 used as feature extractor) | Accuracy of 72% |
| Kumaraswamy et al. [25] | 2021 | Transfer learning approach pre-trained CNNs: DensNet201 and NASNetMobile used as feature extractors) | Accuracy of 72%. AUC for Grade 1, and Grade 2 is 98% and 75%, respectively with DensNet201 AUC for Grade 3 is 69% with NASNetMobile |
| Sujatha et al. [24] | 2022 | Transfer learning approaches (Utilized VGG16, VGG19, InceptionReNetV2, DenseNet121, and DenseNet201) | DenseNet121 produced the highest accuracy of 92.64% |
| Talpur et al. [23] | 2022 | A sequential convolutional neural network is utilised | Accuracy of 92.81% |
| Present Work | 2023 | Proposed Ensemble Model(EfficientNetV2L + ResNet152V2 + DensNet201) | Accuracy of 94%. AUC of 96%, 94% and 96% for Grades 0, 1, and 2, respectively. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumaraswamy, E.; Kumar, S.; Sharma, M. An Invasive Ductal Carcinomas Breast Cancer Grade Classification Using an Ensemble of Convolutional Neural Networks. Diagnostics 2023, 13, 1977. https://doi.org/10.3390/diagnostics13111977
Kumaraswamy E, Kumar S, Sharma M. An Invasive Ductal Carcinomas Breast Cancer Grade Classification Using an Ensemble of Convolutional Neural Networks. Diagnostics. 2023; 13(11):1977. https://doi.org/10.3390/diagnostics13111977
Chicago/Turabian StyleKumaraswamy, Eelandula, Sumit Kumar, and Manoj Sharma. 2023. "An Invasive Ductal Carcinomas Breast Cancer Grade Classification Using an Ensemble of Convolutional Neural Networks" Diagnostics 13, no. 11: 1977. https://doi.org/10.3390/diagnostics13111977
APA StyleKumaraswamy, E., Kumar, S., & Sharma, M. (2023). An Invasive Ductal Carcinomas Breast Cancer Grade Classification Using an Ensemble of Convolutional Neural Networks. Diagnostics, 13(11), 1977. https://doi.org/10.3390/diagnostics13111977








