Rapid Extraction of Viral Nucleic Acids Using Rotating Blade Lysis and Magnetic Beads
Abstract
:1. Introduction
2. Materials and Methods
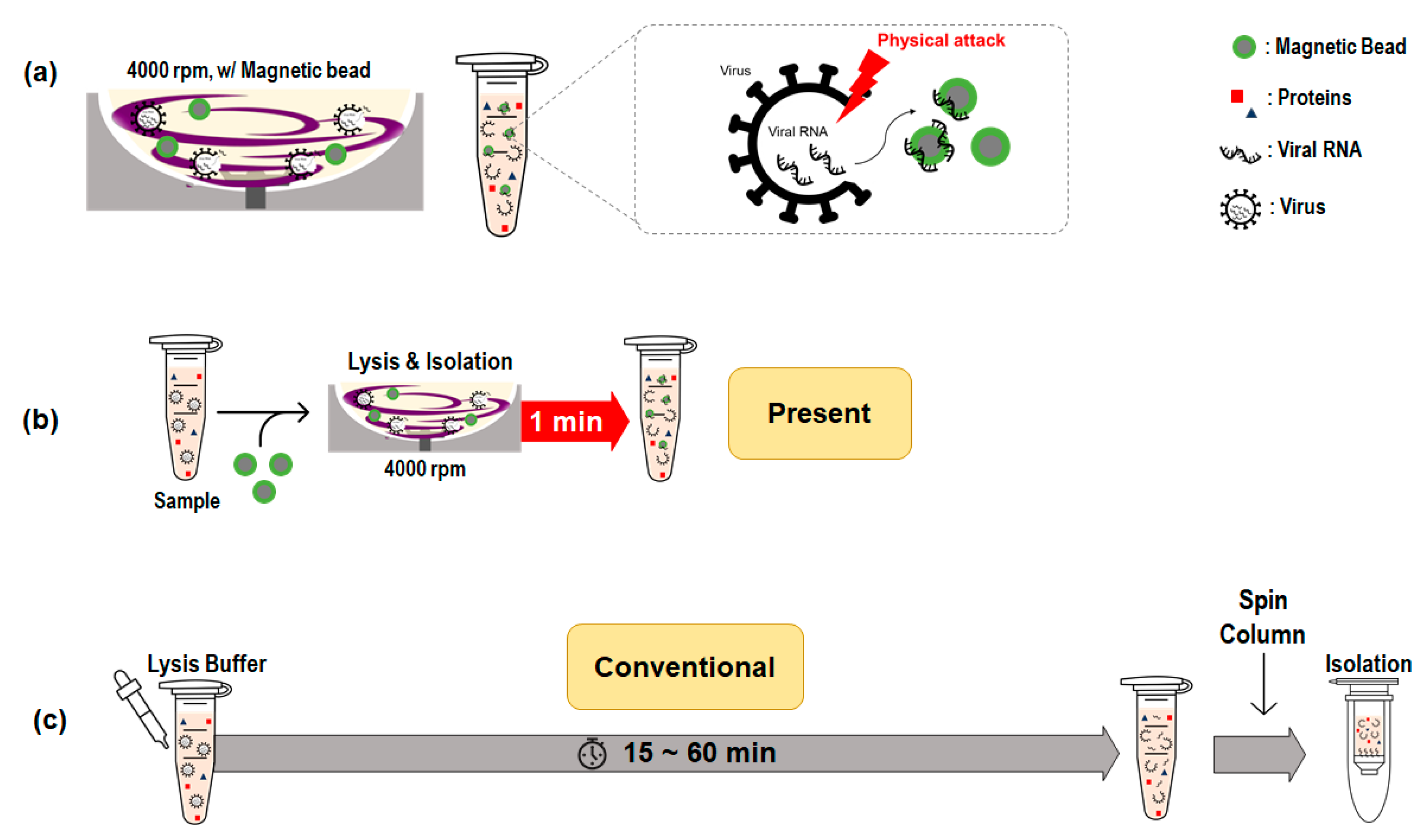
2.1. Design of Rotating Blade System
2.2. Sample Collection
2.3. Viral RNA Extraction Using Spin Column
2.4. Analysis of Extracted Viral RNA
2.5. Agarose Gelelectrophoresis
3. Results
3.1. Effect of Rotational Speed of Stirrer
3.2. Effect of Rotation Time
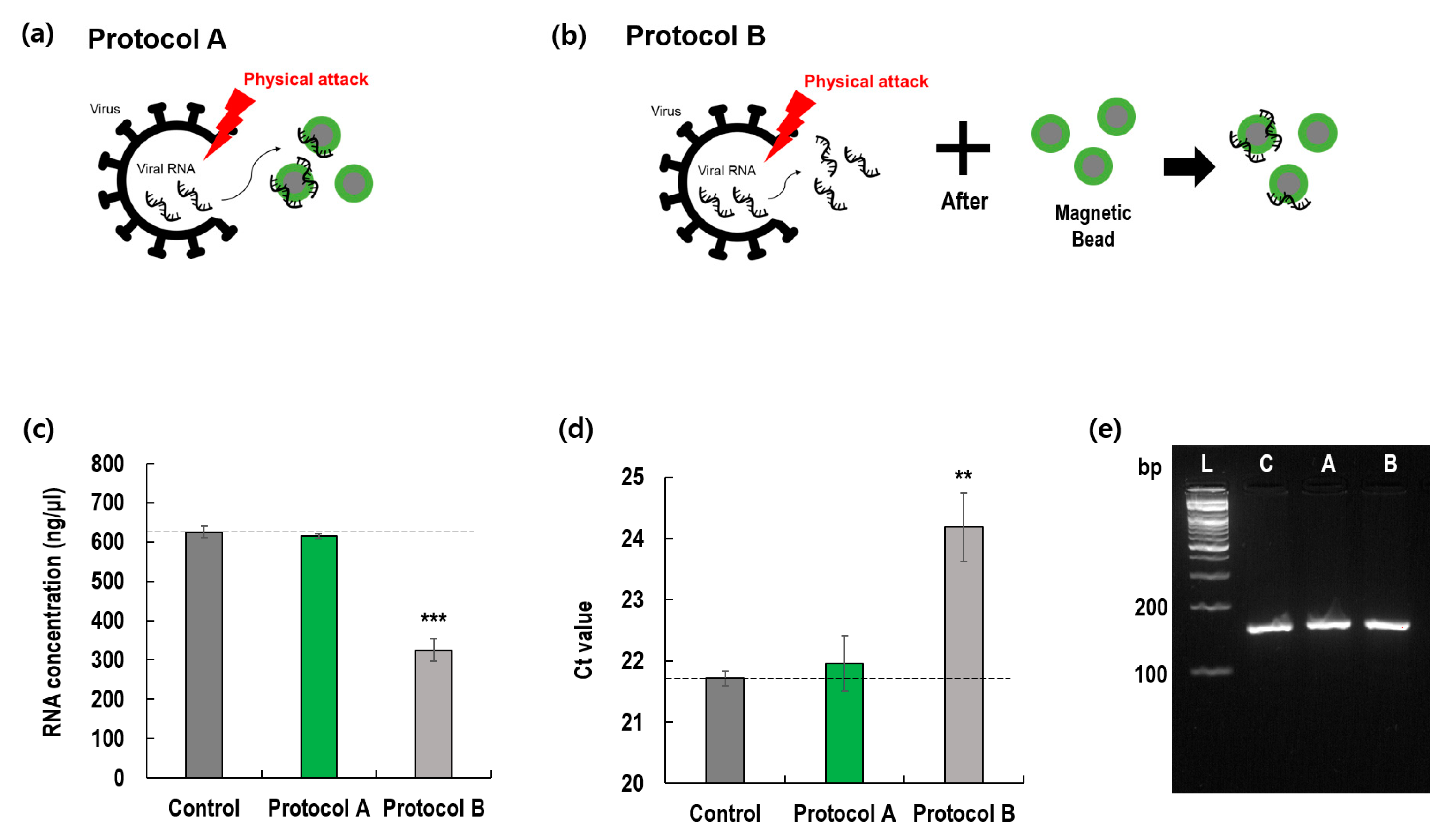
3.3. Comparison of Two Protocols
3.4. Application to Clinical Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fink, G.; Tediosi, F.; Felder, S. Burden of COVID-19 restrictions: National, regional and global estimates. eClinicalMedicine 2022, 45, 101305. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yue, F.; Lee, L.P. Integrated Point-of-Care Molecular Diagnostic Devices for Infectious Diseases. Acc. Chem. Res. 2021, 54, 4107–4119. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.; Lee, S.H.; Song, J.; Bhattacharjee, S.; Feng, J.; Hong, S.; Song, M.; Kim, W.; Lee, J.; Bang, D.; et al. Nanophotonic Cell Lysis and Polymerase Chain Reaction with Gravity-Driven Cell Enrichment for Rapid Detection of Pathogens. ACS Nano 2019, 13, 13866–13874. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Degani, I.; Cheong, J.; Weissleder, R.; Lee, J.-H.; Cheon, J.; Lee, H. Development of Integrated Systems for On-Site Infection Detection. Acc. Chem. Res. 2021, 54, 3991–4000. [Google Scholar] [CrossRef] [PubMed]
- Butler-Wu, S.M.; Wald-Dickler, N.; Holtom, P.; Zangwill, K.M.; Van, T.T. Under-allocation: Critical supply chain hurdles negatively impact the ability of community hospitals to perform repeat SARS-CoV-2 testing. J. Clin. Microbiol. 2020, 58, e01160-20. [Google Scholar] [CrossRef] [PubMed]
- Ñique, A.M.; Coronado-Marquina, F.; Mendez Rico, J.A.; García Mendoza, M.P.; Rojas-Serrano, N.; Simas, P.V.M.; Cabezas Sanchez, C.; Drexler, J.F. A faster and less costly alternative for RNA extraction of SARS-CoV-2 using proteinase k treatment followed by thermal shock. PLoS ONE 2021, 16, e0248885. [Google Scholar] [CrossRef] [PubMed]
- Claas, E.C.J.; Smit, P.W.; van Bussel, M.J.A.W.M.; Verbakel, H.; Taouil, M.; Verweij, J.J.; Thijsen, S.F.T. A two minute liquid based sample preparation for rapid SARS-CoV2 real-time PCR screening: A multicentre evaluation. J. Clin. Virol. 2021, 135, 104720. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, G.; Lim, C.; Lee, B.; Shin, S. A simple method for activating the platelets used in microfluidic platelet aggregation tests: Stirring-induced platelet activation. Biomicrofluidics 2016, 10, 064118. [Google Scholar] [CrossRef] [PubMed]
- Macosko, C.W.; Larson, R.G. Rheology: Principles, Measurements, and Applications; Wiley-VCH: New York, NY, USA, 1994. [Google Scholar]
- Song, J.Y.; Cheong, H.J.; Choi, S.H.; Baek, J.H.; Han, S.B.; Wie, S.-H.; So, B.H.; Kim, H.Y.; Kim, Y.K.; Choi, W.S.; et al. Hospital-based influenza surveillance in Korea: Hospital-based influenza morbidity and mortality study group. J. Med. Virol. 2013, 85, 910–917. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Information for the Molecular Detection of Influenza Viruses; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Yu, Y.J.; Majumdar, A.P.; Nechvatal, J.M.; Ram, J.L.; Basson, M.D.; Heilbrun, L.K.; Kato, I. Exfoliated cells in stool: A source for Reverse Transcription-PCR–based analysis of biomarkers of gastrointestinal cancer. Cancer Epidemiol. Biomark. Prev. 2008, 17, 455–458. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, J.; Martínez, M.J.F. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 2002, 50 (Suppl. S3), 59–74. [Google Scholar] [CrossRef] [PubMed]
- Rossen, R.D.; Schade, A.L.; Butler, W.T.; Kasel, J.A. The proteins in nasal secretion: A longitudinal study of the gammaA-globulin, gammaG-globulin, albumin, siderophilin, and total protein concentrations in nasal washings from adult male volunteers. J. Clin. Investig. 1966, 45, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: The past and the present. J. Biomed. Biotechnol. 2009, 2009, 574398. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, M.; Park, J.; Seong, H.; Lee, H.; Choi, W.; Noh, J.; Kim, W.; Shin, S. Rapid Extraction of Viral Nucleic Acids Using Rotating Blade Lysis and Magnetic Beads. Diagnostics 2022, 12, 1995. https://doi.org/10.3390/diagnostics12081995
Bae M, Park J, Seong H, Lee H, Choi W, Noh J, Kim W, Shin S. Rapid Extraction of Viral Nucleic Acids Using Rotating Blade Lysis and Magnetic Beads. Diagnostics. 2022; 12(8):1995. https://doi.org/10.3390/diagnostics12081995
Chicago/Turabian StyleBae, Minju, Junsoo Park, Hyeonah Seong, Hansol Lee, Wonsuk Choi, Jiyun Noh, Woojoo Kim, and Sehyun Shin. 2022. "Rapid Extraction of Viral Nucleic Acids Using Rotating Blade Lysis and Magnetic Beads" Diagnostics 12, no. 8: 1995. https://doi.org/10.3390/diagnostics12081995
APA StyleBae, M., Park, J., Seong, H., Lee, H., Choi, W., Noh, J., Kim, W., & Shin, S. (2022). Rapid Extraction of Viral Nucleic Acids Using Rotating Blade Lysis and Magnetic Beads. Diagnostics, 12(8), 1995. https://doi.org/10.3390/diagnostics12081995






