SNP rs2920280 in PSCA Is Associated with Susceptibility to Gastric Mucosal Atrophy and Is a Promising Biomarker in Japanese Individuals with Helicobacter pylori Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. GMA Classification
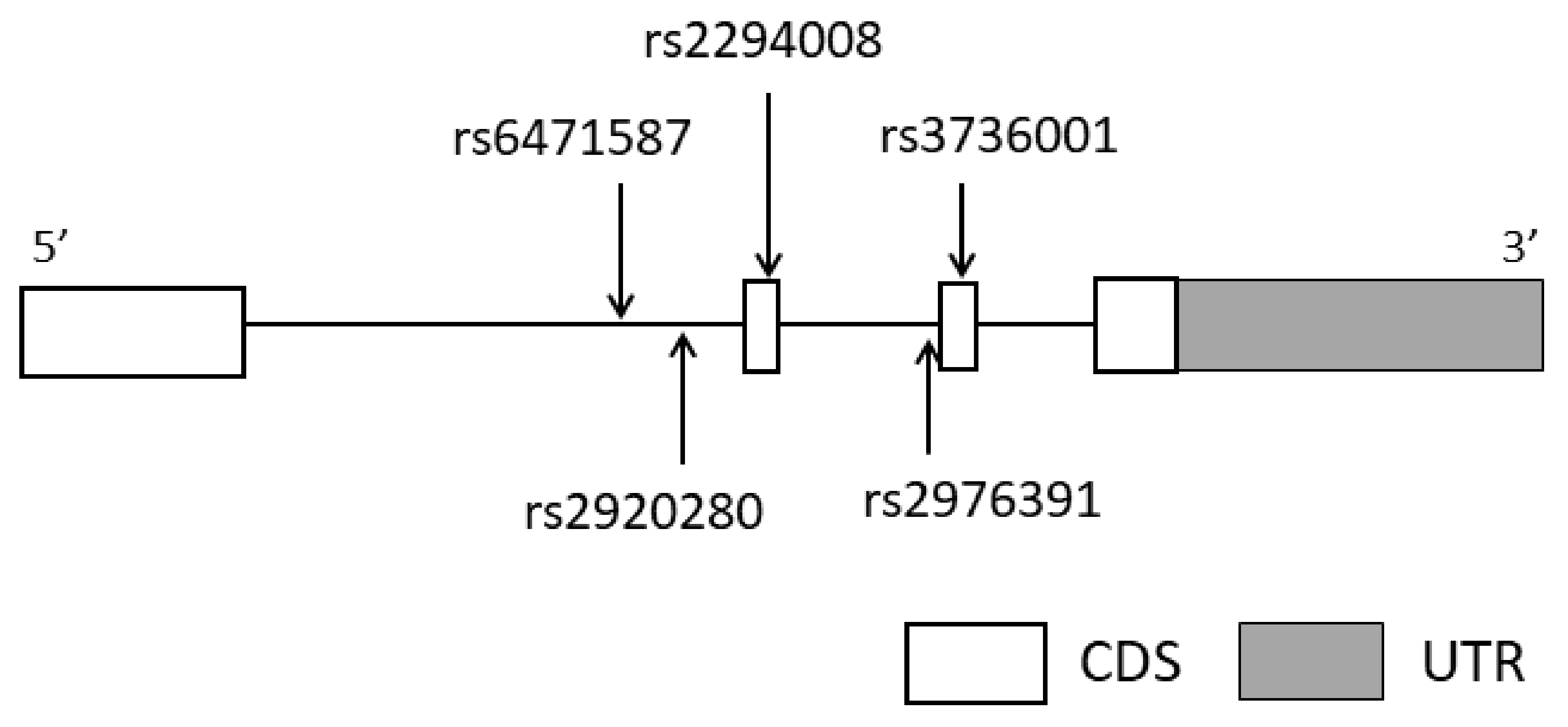
2.3. Selection of Tag SNPs in PSCA
2.4. SNP Genotyping
2.5. Statistical Analyses
3. Results
3.1. Association between PSCA Polymorphisms and GMA Susceptibility by the PG Method
3.2. Association between PSCA Polymorphisms and GMA Susceptibility Based on Endoscopic Findings According to the Kimura–Takemoto Classification
3.3. Comparison of Clinical Characteristics between GMA and Non-GMA Groups Based on Endoscopic Findings According to Kimura–Takemoto Classification
3.4. Contribution of Gene–Environment Interactions to GMA Susceptibility
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamaoka, Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 629–641. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Atherton, J.; Axon, A.T.; Bazzoli, F.; Gensini, G.F.; Gisbert, J.P.; Graham, D.Y.; Rokkas, T.; et al. Management of Helicobacter pylori infection—The Maastricht IV/Florence Consensus Report. Gut 2012, 61, 646–664. [Google Scholar] [CrossRef] [PubMed]
- Correa, P. Human gastric carcinogenesis: A multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar] [PubMed]
- Uemura, N.; Okamoto, S.; Yamamoto, S.; Matsumura, N.; Yamaguchi, S.; Yamakido, M.; Taniyama, K.; Sasaki, N.; Schlemper, R.J. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001, 345, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Saeki, N.; Ono, H.; Sakamoto, H.; Yoshida, T. Genetic factors related to gastric cancer susceptibility identified using a genome-wide association study. Cancer Sci. 2013, 104, 1–8. [Google Scholar] [CrossRef]
- Reiter, R.E.; Gu, Z.; Watabe, T.; Thomas, G.; Szigeti, K.; Davis, E.; Wahl, M.; Nisitani, S.; Yamashiro, J.; Le Beau, M.M.; et al. Prostate stem cell antigen: A cell surface marker overexpressed in prostate cancer. Proc. Natl. Acad. Sci. USA 1998, 95, 1735–1740. [Google Scholar] [CrossRef]
- Sakamoto, H.; Yoshimura, K.; Saeki, N.; Katai, H.; Shimoda, T.; Matsuno, Y.; Saito, D.; Sugimura, H.; Tanioka, F.; Kato, S.; et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat. Genet. 2008, 40, 730–740. [Google Scholar] [CrossRef]
- Tanikawa, C.; Urabe, Y.; Matsuo, K.; Kubo, M.; Takahashi, A.; Ito, H.; Tajima, K.; Kamatani, N.; Nakamura, Y.; Matsuda, K. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat. Genet. 2012, 44, 430–434. [Google Scholar] [CrossRef]
- Lochhead, P.; Frank, B.; Hold, G.L.; Rabkin, C.S.; Ng, M.T.; Vaughan, T.L.; Risch, H.A.; Gammon, M.D.; Lissowska, J.; Weck, M.N.; et al. Genetic variation in the prostate stem cell antigen gene and upper gastrointestinal cancer in white individuals. Gastroenterology 2011, 140, 435–441. [Google Scholar] [CrossRef]
- Matsuo, K.; Tajima, K.; Suzuki, T.; Kawase, T.; Watanabe, M.; Shitara, K.; Misawa, K.; Ito, S.; Sawaki, A.; Muro, K.; et al. Association of prostate stem cell antigen gene polymorphisms with the risk of stomach cancer in Japanese. Int. J. Cancer 2009, 125, 1961–1964. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Ding, Y.; Jin, G.; Wu, J.; Huang, H.; Deng, B.; Hua, Z.; Zhou, Y.; Shu, Y.; et al. Genetic variation of PSCA gene is associated with the risk of both diffuse- and intestinal-type gastric cancer in a Chinese population. Int. J. Cancer 2010, 127, 2183–2189. [Google Scholar] [CrossRef]
- Argani, P.; Rosty, C.; Reiter, R.E.; Wilentz, R.E.; Murugesan, S.R.; Leach, S.D.; Ryu, B.; Skinner, H.G.; Goggins, M.; Jaffee, E.M.; et al. Discovery of new markers of cancer through serial analysis of gene expression: Prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001, 61, 4320–4324. [Google Scholar]
- De Nooij-van Dalen, A.G.; van Dongen, G.A.; Smeets, S.J.; Nieuwenhuis, E.J.; Stigter-van Walsum, M.; Snow, G.B.; Brakenhoff, R.H. Characterization of the human Ly-6 antigens, the newly annotated member Ly-6K included, as molecular markers for head-and-neck squamous cell carcinoma. Int. J. Cancer 2003, 103, 768–774. [Google Scholar] [CrossRef]
- Wu, X.; Ye, Y.; Kiemeney, L.A.; Sulem, P.; Rafnar, T.; Matullo, G.; Seminara, D.; Yoshida, T.; Saeki, N.; Andrew, A.S.; et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009, 41, 991–995. [Google Scholar] [CrossRef]
- Marra, E.; Uva, P.; Viti, V.; Simonelli, V.; Dogliotti, E.; De Rinaldis, E.; Lahm, A.; La Monica, N.; Nicosia, A.; Ciliberto, G.; et al. Growth delay of human bladder cancer cells by Prostate Stem Cell Antigen downregulation is associated with activation of immune signaling pathways. BMC Cancer 2010, 10, 129. [Google Scholar] [CrossRef]
- Zhao, Z.; Ma, W.; Zeng, G.; Qi, D.; Ou, L.; Liang, Y. Small interference RNA-mediated silencing of prostate stem cell antigen attenuates growth, reduces migration and invasion of human prostate cancer PC-3M cells. Urol. Oncol. 2013, 31, 343–351. [Google Scholar] [CrossRef]
- Rizzato, C.; Kato, I.; Plummer, M.; Muñoz, N.; Canzian, F. Genetic variation in PSCA and risk of gastric advanced preneoplastic lesions and cancer in relation to Helicobacter pylori infection. PLoS ONE 2013, 8, e73100. [Google Scholar] [CrossRef]
- Mukoubayashi, C.; Yanaoka, K.; Ohata, H.; Arii, K.; Tamai, H.; Oka, M.; Ichinose, M. Serum pepsinogen and gastric cancer screening. Intern. Med. 2007, 46, 261–266. [Google Scholar] [CrossRef]
- Kimura, K. An endoscopic recognition of the atrophic border and its significance in chronic gastritis. Endoscopy 1969, 13, 87–97. [Google Scholar] [CrossRef]
- Nanashima, K.; Mawatari, T.; Tahara, N.; Higuchi, N.; Nakaura, A.; Inamine, T.; Kondo, S.; Yanagihara, K.; Fukushima, K.; Suyama, N.; et al. Genetic variants in antioxidant pathway: Risk factors for hepatotoxicity in tuberculosis patients. Tuberculosis 2012, 92, 253–259. [Google Scholar] [CrossRef]
- Urabe, S.; Isomoto, H.; Ishida, T.; Maeda, K.; Inamine, T.; Kondo, S.; Higuchi, N.; Sato, K.; Uehara, R.; Yajima, H.; et al. Genetic Polymorphisms of IL-17F and TRAF3IP2 Could Be Predictive Factors of the Long-Term Effect of Infliximab against Crohn’s Disease. Biomed. Res. Int. 2015, 2015, 416838. [Google Scholar] [CrossRef] [PubMed]
- Araki, C.; Yoshimura, M.; Fukumitsu, Y.; Ma, S.; Ishida, T.; Urabe, S.; Matsushima, K.; Honda, T.; Uehara, R.; Fukuda, Y.; et al. The evidence of genetic polymorphisms of genes involved in the P2RX7 signaling pathway as predictive biomarkers for response and loss of response to infliximab against Crohn’s disease. Integr. Mol. Med. 2016, 3, 1–15. [Google Scholar] [CrossRef]
- Yamashita, A.; Inamine, T.; Suzuki, S.; Fukuda, S.; Unoike, M.; Kawafuchi, Y.; Machida, H.; Isomoto, H.; Nakao, K.; Tsukamoto, K. Genetic variants of SMAD2/3/4/7 are associated with susceptibility to ulcerative colitis in a Japanese genetic background. Immunol. Lett. 2019, 207, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Chooi, E.Y.; Chen, H.M.; Miao, Q.; Weng, Y.R.; Chen, X.Y.; Ge, Z.Z.; Xiao, S.D.; Fang, J.Y. Chronic atrophic gastritis is a progressive disease: Analysis of medical reports from Shanghai (1985–2009). Singap. Med. J. 2012, 53, 318–324. [Google Scholar]
- Toyoshima, O.; Tanikawa, C.; Yamamoto, R.; Watanabe, H.; Yamashita, H.; Sakitani, K.; Yoshida, S.; Kubo, M.; Matsuo, K.; Ito, H.; et al. Decrease in PSCA expression caused by Helicobacter pylori infection may promote progression to severe gastritis. Oncotarget 2018, 9, 3936–3945. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Wu, J.L.; Qiu, H.B.; Dong, S.S.; Zhu, Y.H.; Lee, V.H.; Qin, Y.R.; Li, Y.; Chen, J.; Liu, H.B.; et al. PSCA acts as a tumor suppressor by facilitating the nuclear translocation of RB1CC1 in esophageal squamous cell carcinoma. Carcinogenesis 2016, 37, 320–332. [Google Scholar] [CrossRef]
- Xu, L.P.; Qiu, H.B.; Yuan, S.Q.; Chen, Y.M.; Zhou, Z.W.; Chen, Y.B. Downregulation of PSCA promotes gastric cancer proliferation and is related to poor prognosis. J. Cancer 2020, 11, 2708–2715. [Google Scholar] [CrossRef]
- Saeki, N.; Gu, J.; Yoshida, T.; Wu, X. Prostate stem cell antigen: A Jekyll and Hyde molecule? Clin. Cancer Res. 2010, 16, 3533–3538. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.; Zhang, Z.; Shen, J.; Zhang, N.; Yang, M.; Yang, M.; Yu, Y. PSCArs2294008 T polymorphism increases the risk of bladder cancer in Bai, Dai, and Han ethnicity in China and a potential mechanism. Genes Genom. 2018, 40, 531–541. [Google Scholar] [CrossRef]
- Sung, H.; Hu, N.; Yang, H.H.; Giffen, C.A.; Zhu, B.; Song, L.; Su, H.; Wang, C.; Parisi, D.M.; Goldstein, A.M.; et al. Association of high-evidence gastric cancer susceptibility loci and somatic gene expression levels with survival. Carcinogenesis 2017, 38, 1119–1128. [Google Scholar] [CrossRef]
- Choi, I.J.; Kook, M.C.; Kim, Y.I.; Cho, S.J.; Lee, J.Y.; Kim, C.G.; Park, B.; Nam, B.H. Helicobacter pylori Therapy for the Prevention of Metachronous Gastric Cancer. N. Engl. J. Med. 2018, 378, 1085–1095. [Google Scholar] [CrossRef]
- Wong, B.C.; Lam, S.K.; Wong, W.M.; Chen, J.S.; Zheng, T.T.; Feng, R.E.; Lai, K.C.; Hu, W.H.; Yuen, S.T.; Leung, S.Y.; et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: A randomized controlled trial. JAMA 2004, 291, 187–194. [Google Scholar] [CrossRef]
- Leung, W.K.; Lin, S.R.; Ching, J.Y.; To, K.F.; Ng, E.K.; Chan, F.K.; Lau, J.Y.; Sung, J.J. Factors predicting progression of gastric intestinal metaplasia: Results of a randomised trial on Helicobacter pylori eradication. Gut 2004, 53, 1244–1249. [Google Scholar] [CrossRef]
- Hishida, A.; Ugai, T.; Fujii, R.; Nakatochi, M.; Wu, M.C.; Ito, H.; Oze, I.; Tajika, M.; Niwa, Y.; Nishiyama, T.; et al. GWAS analysis reveals a significant contribution of PSCA to the risk of Heliobacter pylori-induced gastric atrophy. Carcinogenesis 2019, 40, 661–668. [Google Scholar] [CrossRef]
- Heinrichs, S.K.M.; Hess, T.; Becker, J.; Hamann, L.; Vashist, Y.K.; Butterbach, K.; Schmidt, T.; Alakus, H.; Krasniuk, I.; Höblinger, A.; et al. Evidence for PTGER4, PSCA, and MBOAT7 as risk genes for gastric cancer on the genome and transcriptome level. Cancer Med. 2018, 7, 5057–5065. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Nagata, Y.; Hiratsuka, R.; Kawase, Y.; Tominaga, T.; Takeuchi, S.; Sakagami, S.; Ishida, S. Gastric Cancer Screening by Combined Assay for Serum Anti-Helicobacter pylori IgG Antibody and Serum Pepsinogen Levels—The ABC Method. Digestion 2016, 93, 13–18. [Google Scholar] [CrossRef]
- Hatakeyama, M. Helicobacter pylori CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis. Cell Host Microbe 2014, 15, 306–316. [Google Scholar] [CrossRef]
| SNP | Primer Sequences (5′ to 3′) | Annealing Temperature (°C) | |
|---|---|---|---|
| forward | TCAGTGCACAGCTTGATGGT | ||
| rs6471587 | reverse | CCCCGTATAACCCAGACATT | 56 |
| probe | CTGGTTGGCAATCGCTTGCAAGAGTTATC | ||
| forward | GGAACAGCCAGCGTTAACAT | ||
| rs2920280 | reverse | CATCTTCTGGTTGGCAATCG | 55 |
| probe | GTCTCCACAACCCGGTATAACCCAGA | ||
| forward | CCTCACTGGCTCCAGGAAAC | ||
| rs2294008 | reverse | AAGCCTGCCATCAACAGGG | 58 |
| probe | CCACCAGTGACCATGAAGGCTGTGCT | ||
| forward | GGAGGAGACATTGAGGGTGA | ||
| rs2976391 | reverse | TTTGCAGGAGTAGCACAGCA | 57 |
| probe | CAGCCTCTGAGGCCCCTCCCACTCCA | ||
| forward | TGCTGTGCTACTCCTGCAAA | ||
| rs3736001 | reverse | AGGTGGAAGGAAAACAGCAC | 56 |
| probe | TGCCTGCAGGTGGAGAACTGCACCC | ||
| SNP | Genotype | GMA (%) | Non-GMA (%) | Inheritance Model | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| PG I ≤ 70 and PG I/II ≤ 3 | Others | |||||
| Clinical characteristics | Number | 92 | 103 | |||
| Age, mean ± SD | 59.0 ± 9.6 | 55.0 ± 11.0 | 0.0055 | |||
| Male/female | 37/55 | 49/54 | 0.302 | |||
| rs6471587 | G allele * | 24 (13.0) | 32 (15.5) | Allele | 0.816 (0.456–1.467) | 0.484 |
| C/C | 70 (76.1) | 74 (71.9) | ||||
| C/G | 20 (21.7) | 26 (25.2) | Dominant | 0.802 (0.432–1.520) | 0.501 | |
| G/G | 2 (2.2) | 3 (2.9) | Recessive | 0.741 (0.129–3.700) | 1.000 | |
| rs2920280 | G allele * | 92 (50.0) | 87 (42.2) | Allele | 1.368 (0.922–2.037) | 0.124 |
| C/C | 24 (26.1) | 33 (32.0) | ||||
| C/G | 44 (47.8) | 53 (51.5) | Dominant | 1.336 (0.732–2.502) | 0.362 | |
| G/G | 24 (26.1) | 17 (16.5) | Recessive | 1.785 (0.900–3.481) | 0.101 | |
| rs2294008 | C allele * | 69 (37.5) | 86 (41.7) | Allele | 0.837 (0.553–1.259) | 0.392 |
| T/T | 34 (37.0) | 31 (30.1) | ||||
| T/C | 47 (51.1) | 58 (56.3) | Dominant | 0.735 (0.398–1.342) | 0.310 | |
| C/C | 11 (11.9) | 14 (13.6) | Recessive | 0.863 (0.3588–1.997) | 0.733 | |
| rs2976391 | A allele * | 33 (17.9) | 39 (18.9) | Allele | 0.936 (0.551–1.563) | 0.800 |
| C/C | 61 (65.2) | 67 (65.1) | ||||
| C/A | 29 (32.6) | 33 (32.0) | Dominant | 0.946 (0.517–1.711) | 0.854 | |
| A/A | 2 (2.2) | 3 (2.9) | Recessive | 0.7461 (0.129–3.700) | 1.000 | |
| rs3736001 | A allele * | 15 (8.2) | 24 (11.7) | Allele | 0.673 (0.350–1.346) | 0.250 |
| G/G | 77 (83.7) | 80 (77.7) | ||||
| G/A | 15 (16.3) | 22 (21.3) | Dominant | 0.678 (0.326–1.400) | 0.289 | |
| A/A | 0 (0) | 1 (1.0) | Recessive | 0.000 (0.099–10.080) | 1.000 |
| SNP | Genotype | GMA (%) | Non-GMA (%) | Inheritance Model | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| C3 or O1-3 | C1 or C2 | |||||
| Clinical characteristics | Number | 123 | 72 | |||
| Age, mean ± SD | 59.1 ± 9.1 | 53.2 ± 11.6 | 0.0002 | |||
| Male/female | 55/68 | 31/41 | 0.822 | |||
| rs6471587 | G allele * | 33 (13.4) | 23 (16.0) | Allele | 0.815 (0.466–1.485) | 0.487 |
| C/C | 93 (75.6) | 51 (70.8) | ||||
| C/G | 27 (22.0) | 19 (26.4) | Dominant | 0.783 (0.406–1.480) | 0.464 | |
| G/G | 3 (2.4) | 2 (2.8) | Recessive | 0.875 (0.175–5.028) | 1.000 | |
| rs2920280 | G allele * | 125 (50.8) | 54 (37.5) | Allele | 1.722 (1.122–2.629) | 0.011 |
| C/C | 32 (26.0) | 25 (34.7) | ||||
| C/G | 57 (46.3) | 40 (55.6) | Dominant | 1.513 (0.801–2.808) | 0.197 | |
| G/G | 34 (27.7) | 7 (9.7) | Recessive | 3.547 (1.545–9.006) | 0.003 | |
| rs2294008 | C allele * | 88 (35.8) | 67 (46.5) | Allele | 0.640 (0.424–0.966) | 0.036 |
| T/T | 48 (39.0) | 17 (23.6) | ||||
| T/C | 62 (50.4) | 43 (59.7) | Dominant | 0.483 (0.246–0.905) | 0.028 | |
| C/C | 13 (10.6) | 12 (16.7) | Recessive | 0.591 (0.249–1.375) | 0.219 | |
| rs2976391 | A allele * | 39 (15.9) | 33 (22.9) | Allele | 0.634 (0.376–1.046) | 0.083 |
| C/C | 86 (69.9) | 42 (58.3) | ||||
| C/A | 35 (28.5) | 27 (37.5) | Dominant | 0.602 (0.322–1.131) | 0.100 | |
| A/A | 2 (1.6) | 3 (4.2) | Recessive | 0.380 (0.067–1.909) | 0.360 | |
| rs3736001 | A allele * | 22 (8.9) | 17 (11.8) | Allele | 0.734 (0.388–1.4361) | 0.363 |
| G/G | 102 (82.9) | 55 (76.4) | ||||
| G/A | 20 (16.3) | 17 (23.6) | Dominant | 0.666 (0.318–1.344) | 0.266 | |
| A/A | 1 (0.8) | 0 (0) | Recessive | 0.000 (0.065–15.380) | 1.000 |
| Factor | Factor Comparison | |
|---|---|---|
| OR (95% CI) | p-Value | |
| Age | 1.060 (1.028–1.093) | <0.001 |
| G/G genotype of rs2920280 in PSCA | 3.665 (1.499–8.911) | 0.004 |
| Factor | Factor Comparison | |
|---|---|---|
| OR (95% CI) | p-Value | |
| Age | 1.058 (1.026–1.091) | <0.001 |
| T/T genotype of rs2294008 in PSCA | 1.992 (1.016–3.906) | 0.045 |
| Biomarker | OR (95% CI) | p-Value | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| G/G genotype of rs2920280 | 3.547 (1.545–9.006) | 0.003 | 27.6 | 90.3 | 82.9 | 42.2 |
| T/T genotype of rs2294008 | 2.070 (1.105–4.065) | 0.028 | 39 | 76.4 | 73.8 | 42.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Isomoto, H.; Sakaguchi, T.; Inamine, T.; Takeshita, S.; Fukuda, D.; Ohnita, K.; Kanda, T.; Matsushima, K.; Honda, T.; Sugihara, T.; et al. SNP rs2920280 in PSCA Is Associated with Susceptibility to Gastric Mucosal Atrophy and Is a Promising Biomarker in Japanese Individuals with Helicobacter pylori Infection. Diagnostics 2022, 12, 1988. https://doi.org/10.3390/diagnostics12081988
Isomoto H, Sakaguchi T, Inamine T, Takeshita S, Fukuda D, Ohnita K, Kanda T, Matsushima K, Honda T, Sugihara T, et al. SNP rs2920280 in PSCA Is Associated with Susceptibility to Gastric Mucosal Atrophy and Is a Promising Biomarker in Japanese Individuals with Helicobacter pylori Infection. Diagnostics. 2022; 12(8):1988. https://doi.org/10.3390/diagnostics12081988
Chicago/Turabian StyleIsomoto, Hajime, Takuki Sakaguchi, Tatsuo Inamine, Shintaro Takeshita, Daisuke Fukuda, Ken Ohnita, Tsutomu Kanda, Kayoko Matsushima, Tetsuro Honda, Takaaki Sugihara, and et al. 2022. "SNP rs2920280 in PSCA Is Associated with Susceptibility to Gastric Mucosal Atrophy and Is a Promising Biomarker in Japanese Individuals with Helicobacter pylori Infection" Diagnostics 12, no. 8: 1988. https://doi.org/10.3390/diagnostics12081988
APA StyleIsomoto, H., Sakaguchi, T., Inamine, T., Takeshita, S., Fukuda, D., Ohnita, K., Kanda, T., Matsushima, K., Honda, T., Sugihara, T., Hirayama, T., Nakao, K., & Tsukamoto, K. (2022). SNP rs2920280 in PSCA Is Associated with Susceptibility to Gastric Mucosal Atrophy and Is a Promising Biomarker in Japanese Individuals with Helicobacter pylori Infection. Diagnostics, 12(8), 1988. https://doi.org/10.3390/diagnostics12081988







