Automated Identification of Coronary Arteries in Assisting Inexperienced Readers: Comparison between Two Commercial Vendors
Abstract
1. Introduction
2. Materials and Methods
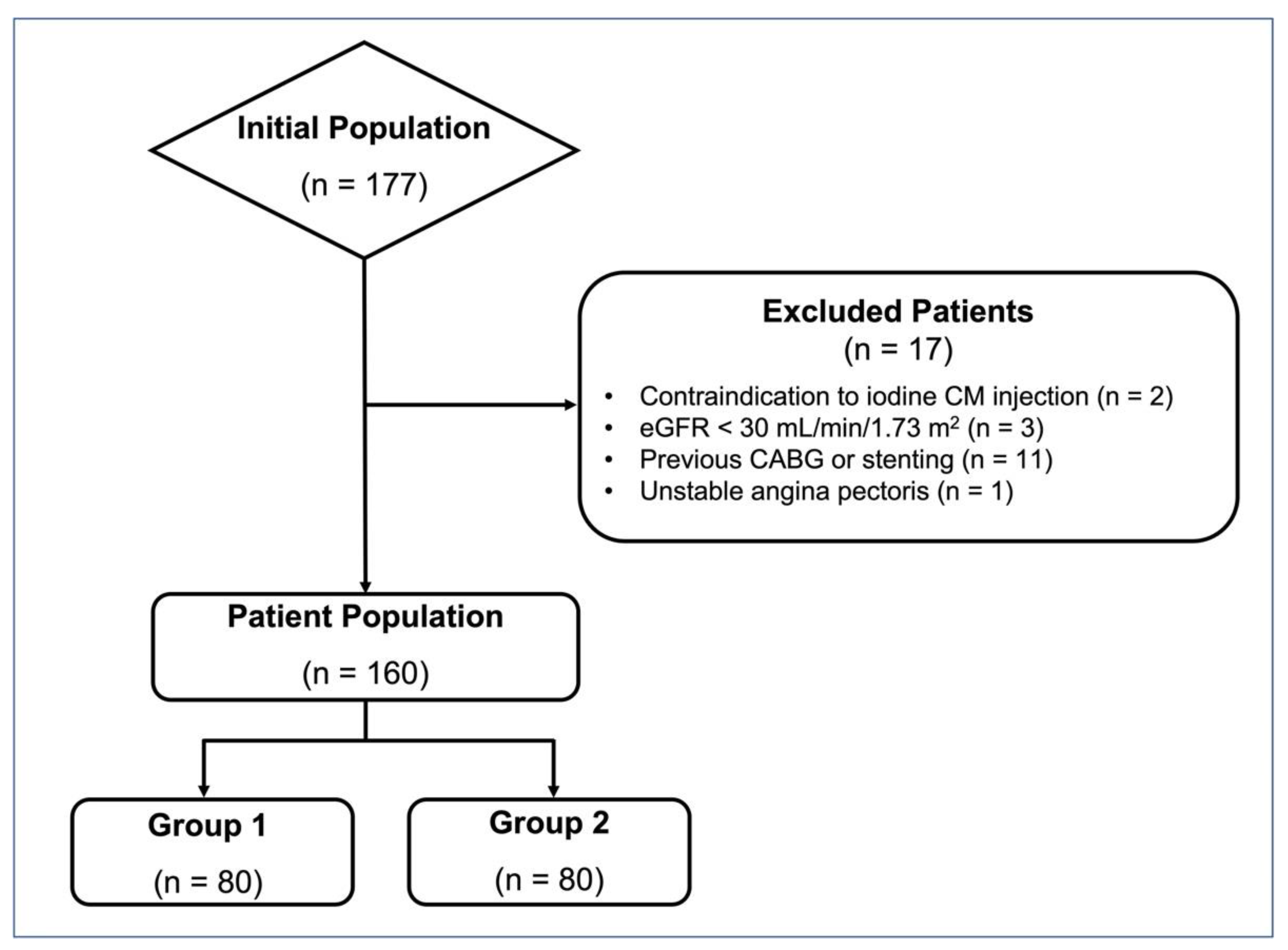
2.1. Patient Population
2.2. Image Acquisition—Group 1
2.3. Image Post Processing—Group 1
2.4. Image Acquisition—Group 2
2.5. Image Post Processing—Group 2
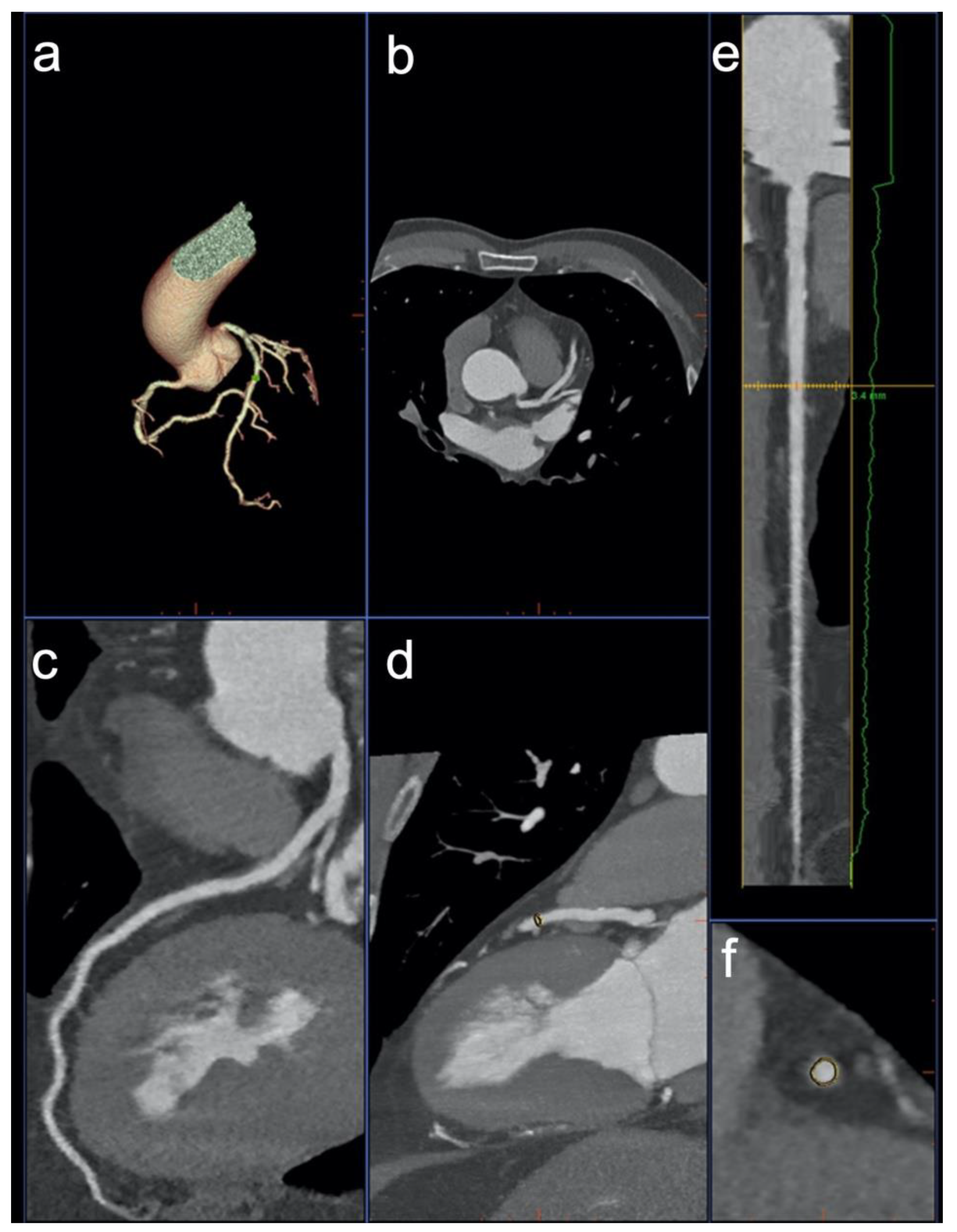
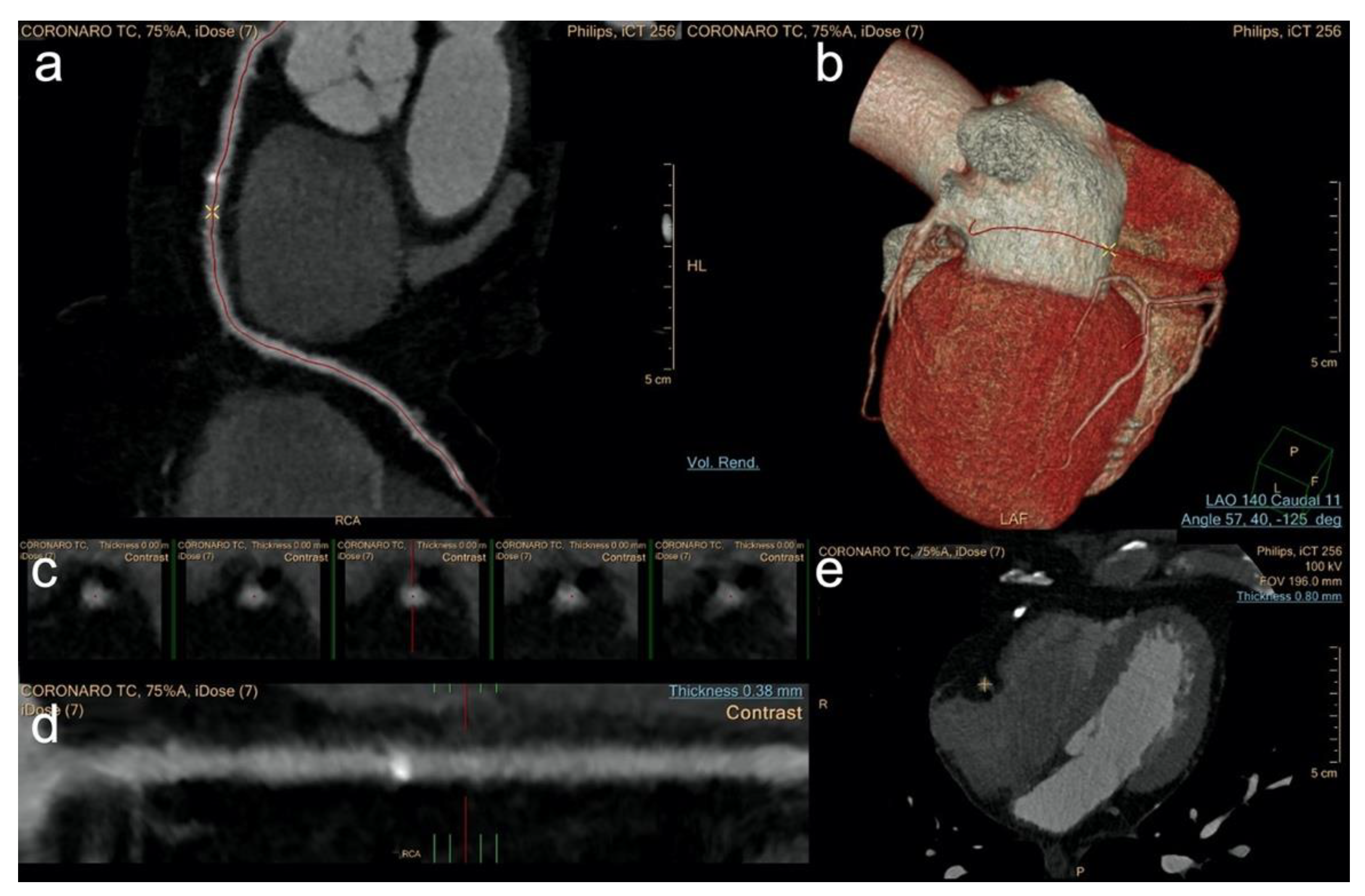
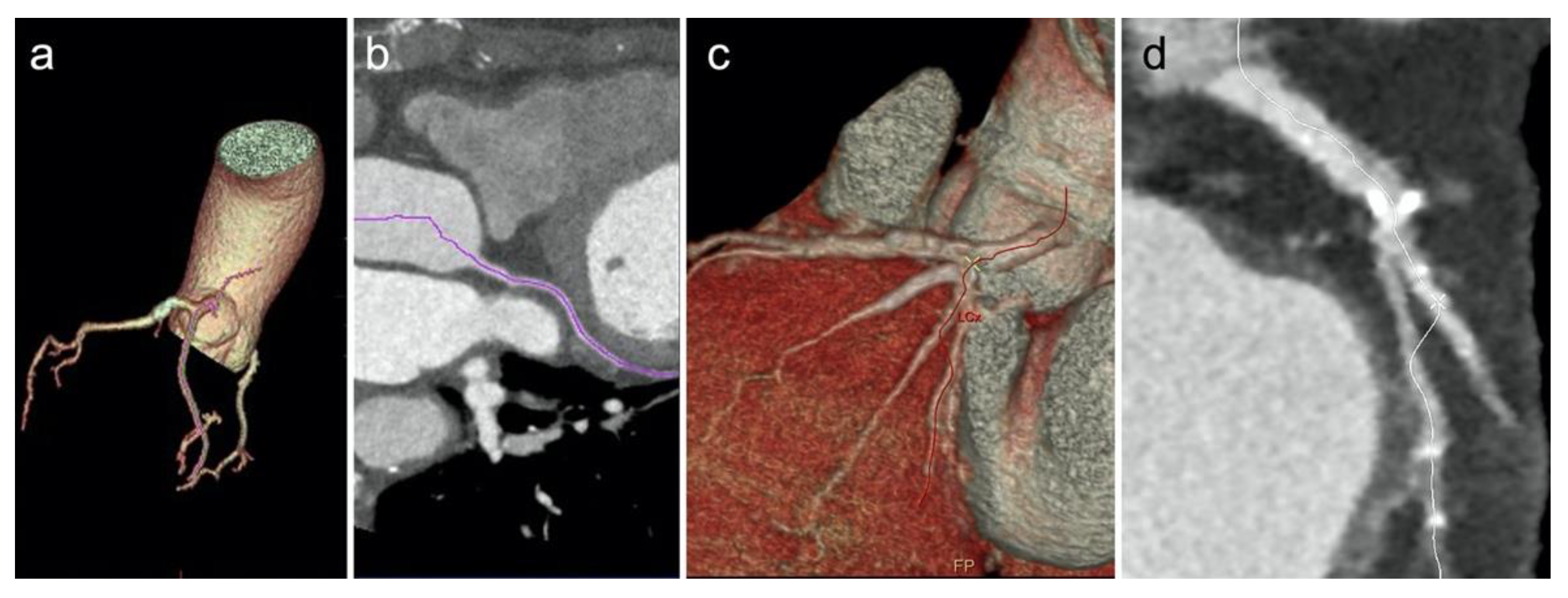
2.6. Image Analysis
2.7. Statistical Analysis
3. Results
3.1. Patient Population
3.2. Image Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, J.C.; Gerhardt, T.E.; Kwon, E. Risk Factors For Coronary Artery Disease. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Malakar, A.K.; Choudhury, D.; Halder, B.; Paul, P.; Uddin, A.; Chakraborty, S. A Review on Coronary Artery Disease, Its Risk Factors, and Therapeutics. J. Cell. Physiol. 2019, 234, 16812–16823. [Google Scholar] [CrossRef] [PubMed]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the Diagnosis and Management of Chronic Coronary Syndromes. Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef] [PubMed]
- Meijboom, W.B.; Meijs, M.F.L.; Schuijf, J.D.; Cramer, M.J.; Mollet, N.R.; van Mieghem, C.A.G.; Nieman, K.; van Werkhoven, J.M.; Pundziute, G.; Weustink, A.C.; et al. Diagnostic Accuracy of 64-Slice Computed Tomography Coronary Angiography: A Prospective, Multicenter, Multivendor Study. J. Am. Coll. Cardiol. 2008, 52, 2135–2144. [Google Scholar] [CrossRef] [PubMed]
- Tesche, C.; Caruso, D.; De Cecco, C.N.; Shuler, D.C.; Rames, J.D.; Albrecht, M.H.; Duguay, T.M.; Varga-Szemes, A.; Jochheim, D.; Baquet, M.; et al. Coronary Computed Tomography Angiography–Derived Plaque Quantification in Patients With Acute Coronary Syndrome. Am. J. Cardiol. 2017, 119, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Mangold, S.; Wichmann, J.L.; Schoepf, U.J.; Caruso, D.; Tesche, C.; Steinberg, D.H.; Varga-Szemes, A.; Stubenrauch, A.C.; Bayer, R.R.; Biancalana, M.; et al. Diagnostic Accuracy of Coronary CT Angiography Using 3rd-Generation Dual-Source CT and Automated Tube Voltage Selection: Clinical Application in a Non-Obese and Obese Patient Population. Eur. Radiol. 2017, 27, 2298–2308. [Google Scholar] [CrossRef]
- Du, H.; Shao, K.; Bao, F.; Zhang, Y.; Gao, C.; Wu, W.; Zhang, C. Automated Coronary Artery Tree Segmentation in Coronary CTA Using a Multiobjective Clustering and Toroidal Model-Guided Tracking Method. Comput. Methods Programs Biomed. 2021, 199, 105908. [Google Scholar] [CrossRef]
- Cheung, W.K.; Bell, R.; Nair, A.; Menezes, L.J.; Patel, R.; Wan, S.; Chou, K.; Chen, J.; Torii, R.; Davies, R.H.; et al. A Computationally Efficient Approach to Segmentation of the Aorta and Coronary Arteries Using Deep Learning. IEEE Access 2021, 9, 108873–108888. [Google Scholar] [CrossRef]
- Khan, M.F.; Wesarg, S.; Gurung, J.; Dogan, S.; Maataoui, A.; Brehmer, B.; Herzog, C.; Ackermann, H.; Assmus, B.; Vogl, T.J. Facilitating Coronary Artery Evaluation in MDCT Using a 3D Automatic Vessel Segmentation Tool. Eur. Radiol. 2006, 16, 1789–1795. [Google Scholar] [CrossRef]
- Dewey, M.; Schnapauff, D.; Laule, M.; Lembcke, A.; Borges, A.C.; Rutsch, W.; Hamm, B.; Rogalla, P. Multislice CT Coronary Angiography: Evaluation of an Automatic Vessel Detection Tool. Rofo 2004, 176, 478–483. [Google Scholar] [CrossRef]
- Pugliese, F.; Hunink, M.G.M.; Gruszczynska, K.; Alberghina, F.; Malagó, R.; van Pelt, N.; Mollet, N.R.; Cademartiri, F.; Weustink, A.C.; Meijboom, W.B.; et al. Learning Curve for Coronary CT Angiography: What Constitutes Sufficient Training? Radiology 2009, 251, 359–368. [Google Scholar] [CrossRef]
- Schroeder, S.; Achenbach, S.; Bengel, F.; Burgstahler, C.; Cademartiri, F.; de Feyter, P.; George, R.; Kaufmann, P.; Kopp, A.F.; Knuuti, J.; et al. Cardiac Computed Tomography: Indications, Applications, Limitations, and Training Requirements: Report of a Writing Group Deployed by the Working Group Nuclear Cardiology and Cardiac CT of the European Society of Cardiology and the European Council of Nuclear Cardiology. Eur. Heart J. 2008, 29, 531–556. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.; Kerl, J.M.; De Rosa, S.; Tekin, T.; Boehme, E.; Liem, S.; Scheuchenzuber, M.; Kim, H.-R.; Bauer, R.W.; Silverman, J.R.; et al. Influence of Observer Experience and Training on Proficiency in Coronary CT Angiography Interpretation. Eur. J. Radiol. 2013, 82, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Anders, K.; Achenbach, S.; Petit, I.; Daniel, W.G.; Uder, M.; Pflederer, T. Accuracy of Automated Software-Guided Detection of Significant Coronary Artery Stenosis by CT Angiography: Comparison with Invasive Catheterisation. Eur. Radiol. 2013, 23, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Pan, Y.; Duan, F.; Zhao, S.; Wang, Q.; Wang, W. Automated Segmentation of Coronary Arteries Based on Statistical Region Growing and Heuristic Decision Method. Biomed. Res. Int. 2016, 2016, 3530251. [Google Scholar] [CrossRef] [PubMed]
- Rief, M.; Kranz, A.; Hartmann, L.; Roehle, R.; Laule, M.; Dewey, M. Computer-Aided CT Coronary Artery Stenosis Detection: Comparison with Human Reading and Quantitative Coronary Angiography. Int. J. Cardiovasc. Imaging 2014, 30, 1621–1627. [Google Scholar] [CrossRef]
- Dankerl, P.; Hammon, M.; Tsymbal, A.; Cavallaro, A.; Achenbach, S.; Uder, M.; Janka, R. Evaluation of Novice Reader Diagnostic Performance in Coronary CT Angiography Using an Advanced Cardiac Software Package. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 609–615. [Google Scholar] [CrossRef][Green Version]
- Leipsic, J.; Abbara, S.; Achenbach, S.; Cury, R.; Earls, J.P.; Mancini, G.J.; Nieman, K.; Pontone, G.; Raff, G.L. SCCT Guidelines for the Interpretation and Reporting of Coronary CT Angiography: A Report of the Society of Cardiovascular Computed Tomography Guidelines Committee. J. Cardiovasc. Comput. Tomogr. 2014, 8, 342–358. [Google Scholar] [CrossRef]
- Richardson, J.T.E. The Analysis of 2 × 2 Contingency Tables-yet Again. Stat. Med. 2011, 30, 890, author reply 891–892. [Google Scholar] [CrossRef]
- Campbell, I. Chi-Squared and Fisher-Irwin Tests of Two-by-Two Tables with Small Sample Recommendations. Stat. Med. 2007, 26, 3661–3675. [Google Scholar] [CrossRef]
- Saur, S.C.; Alkadhi, H.; Stolzmann, P.; Baumüller, S.; Leschka, S.; Scheffel, H.; Desbiolles, L.; Fuchs, T.J.; Székely, G.; Cattin, P.C. Effect of Reader Experience on Variability, Evaluation Time and Accuracy of Coronary Plaque Detection with Computed Tomography Coronary Angiography. Eur. Radiol. 2010, 20, 1599–1606. [Google Scholar] [CrossRef][Green Version]
- Arnoldi, E.; Gebregziabher, M.; Schoepf, U.J.; Goldenberg, R.; Ramos-Duran, L.; Zwerner, P.L.; Nikolaou, K.; Reiser, M.F.; Costello, P.; Thilo, C. Automated Computer-Aided Stenosis Detection at Coronary CT Angiography: Initial Experience. Eur. Radiol. 2010, 20, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Schoepf, U.J.; Fink, C.; Goldenberg, R.; Apfaltrer, P.; Gruettner, J.; Vajcs, D.; Schoenberg, S.O.; Henzler, T. Diagnostic Performance Evaluation of a Computer-Aided Simple Triage System for Coronary CT Angiography in Patients with Intermediate Risk for Acute Coronary Syndrome. Acad. Radiol. 2013, 20, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Karlo, C.A.; Leschka, S.; Stolzmann, P.; Glaser-Gallion, N.; Wildermuth, S.; Alkadhi, H. A Systematic Approach for Analysis, Interpretation, and Reporting of Coronary CTA Studies. Insights Imaging 2012, 3, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Husmann, L.; Leschka, S.; Desbiolles, L.; Schepis, T.; Gaemperli, O.; Seifert, B.; Cattin, P.; Frauenfelder, T.; Flohr, T.G.; Marincek, B.; et al. Coronary Artery Motion and Cardiac Phases: Dependency on Heart Rate—Implications for CT Image Reconstruction. Radiology 2007, 245, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Dodge, J.T.; Brown, B.G.; Bolson, E.L.; Dodge, H.T. Lumen Diameter of Normal Human Coronary Arteries. Influence of Age, Sex, Anatomic Variation, and Left Ventricular Hypertrophy or Dilation. Circulation 1992, 86, 232–246. [Google Scholar] [CrossRef]
- Kerl, J.M.; Schoepf, U.J.; Bauer, R.W.; Tekin, T.; Costello, P.; Vogl, T.J.; Herzog, C. 64-Slice Multidetector-Row Computed Tomography in the Diagnosis of Coronary Artery Disease: Interobserver Agreement among Radiologists with Varied Levels of Experience on a per-Patient and per-Segment Basis. J. Thorac. Imaging 2012, 27, 29–35. [Google Scholar] [CrossRef]
- van Assen, M.; Muscogiuri, G.; Caruso, D.; Lee, S.J.; Laghi, A.; De Cecco, C.N. Artificial Intelligence in Cardiac Radiology. Radiol. Med. 2020, 125, 1186–1199. [Google Scholar] [CrossRef]
- Fischer, A.M.; Eid, M.; De Cecco, C.N.; Gulsun, M.A.; van Assen, M.; Nance, J.W.; Sahbaee, P.; De Santis, D.; Bauer, M.J.; Jacobs, B.E.; et al. Accuracy of an Artificial Intelligence Deep Learning Algorithm Implementing a Recurrent Neural Network With Long Short-Term Memory for the Automated Detection of Calcified Plaques From Coronary Computed Tomography Angiography. J. Thorac. Imaging 2020, 35, S49. [Google Scholar] [CrossRef]
- Duguay, T.M.; Tesche, C.; Vliegenthart, R.; De Cecco, C.N.; Lin, H.; Albrecht, M.H.; Varga-Szemes, A.; De Santis, D.; Ebersberger, U.; Bayer 2nd, R.R. Coronary Computed Tomographic Angiography-Derived Fractional Flow Reserve Based on Machine Learning for Risk Stratification of Non-Culprit Coronary Narrowings in Patients with Acute Coronary Syndrome. Am. J. Cardiol. 2017, 120, 1260–1266. [Google Scholar] [CrossRef]
- Mastrodicasa, D.; Albrecht, M.H.; Schoepf, U.J.; Varga-Szemes, A.; Jacobs, B.E.; Gassenmaier, S.; De Santis, D.; Eid, M.H.; van Assen, M.; Tesche, C.; et al. Artificial Intelligence Machine Learning-Based Coronary CT Fractional Flow Reserve (CT-FFRML): Impact of Iterative and Filtered Back Projection Reconstruction Techniques. J. Cardiovasc. Comput. Tomogr. 2019, 13, 331–335. [Google Scholar] [CrossRef]
- von Knebel Doeberitz, P.L.; De Cecco, C.N.; Schoepf, U.J.; Duguay, T.M.; Albrecht, M.H.; van Assen, M.; Bauer, M.J.; Savage, R.H.; Pannell, J.T.; De Santis, D. Coronary CT Angiography–Derived Plaque Quantification with Artificial Intelligence CT Fractional Flow Reserve for the Identification of Lesion-Specific Ischemia. Eur. Radiol. 2019, 29, 2378–2387. [Google Scholar] [CrossRef] [PubMed]
- von Knebel Doeberitz, P.L.; De Cecco, C.N.; Schoepf, U.J.; Albrecht, M.H.; van Assen, M.; De Santis, D.; Gaskins, J.; Martin, S.; Bauer, M.J.; Ebersberger, U. Impact of Coronary Computerized Tomography Angiography-Derived Plaque Quantification and Machine-Learning Computerized Tomography Fractional Flow Reserve on Adverse Cardiac Outcome. Am. J. Cardiol. 2019, 124, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.-S.; Li, C.-W.; Su, S.-F.; Tay, S.-Y.; Tran, Q.-V.; Chan, W.P. Coronary Artery Segmentation under Class Imbalance Using a U-Net Based Architecture on Computed Tomography Angiography Images. Sci. Rep. 2021, 11, 14493. [Google Scholar] [CrossRef]
- Thilo, C.; Gebregziabher, M.; Meinel, F.G.; Goldenberg, R.; Nance, J.W.; Arnoldi, E.M.; Soma, L.D.; Ebersberger, U.; Blanke, P.; Coursey, R.L.; et al. Computer-Aided Stenosis Detection at Coronary CT Angiography: Effect on Performance of Readers with Different Experience Levels. Eur. Radiol. 2015, 25, 694–702. [Google Scholar] [CrossRef] [PubMed]
| Group 1 | Group 2 | p Value | |
|---|---|---|---|
| Patient Characteristics | |||
| Age, y * | 63 ± 11 | 64 ± 12 | 0.583 |
| BMI * | 28.3 ± 5.5 | 27.7 ± 4.4 | 0.675 |
| HR * | 58 ± 7 | 59 ± 7 | 0.367 |
| Sex: Male † | 50 (63) | 46 (58) | 0.519 |
| Sex: Female † | 30 (38) | 34 (43) | 0.520 |
| Cardiovascular Risk Factors † | |||
| Family history of CAD | 57 (71.3) | 56 (70) | 0.857 |
| Hypertension | 53 (66.3) | 56 (70) | 0.616 |
| Hypercholesterolemia | 33 (41.3) | 40 (50) | 0.270 |
| Diabetes Mellitus | 24 (30) | 16(20) | 0.145 |
| Current of former smoking | 29 (36.3) | 45 (56.3) | 0.011 |
| Medications † | |||
| Beta-blockers | 24 (30) | 19 (23.8) | 0.378 |
| Nitrates | 33 (41.3) | 46 (57.5) | 0.004 |
| Group 1 | Group 2 | p Value | |
|---|---|---|---|
| RCA | 65/80 (81.2) | 67/80 (83.7) | 0.679 |
| LAD | 72/80 (90) | 70/80 (87.5) | 0.618 |
| LCx | 65/80 (81.2) | 54/80 (67.5) | 0.048 |
| RCA–LCx–LAD | 202/240 (84.2) | 191/240 (79.6) | 0.191 |
| Coronary Segments | 942/1062 (88.7) | 797/1078 (73.9) | <0.001 |
| Time of analysis | 13.8 ± 2 s | 21.9 ± 3 s | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Santis, D.; Tremamunno, G.; Rucci, C.; Polidori, T.; Zerunian, M.; Piccinni, G.; Pugliese, L.; Masci, B.; Ubaldi, N.; Laghi, A.; et al. Automated Identification of Coronary Arteries in Assisting Inexperienced Readers: Comparison between Two Commercial Vendors. Diagnostics 2022, 12, 1987. https://doi.org/10.3390/diagnostics12081987
De Santis D, Tremamunno G, Rucci C, Polidori T, Zerunian M, Piccinni G, Pugliese L, Masci B, Ubaldi N, Laghi A, et al. Automated Identification of Coronary Arteries in Assisting Inexperienced Readers: Comparison between Two Commercial Vendors. Diagnostics. 2022; 12(8):1987. https://doi.org/10.3390/diagnostics12081987
Chicago/Turabian StyleDe Santis, Domenico, Giuseppe Tremamunno, Carlotta Rucci, Tiziano Polidori, Marta Zerunian, Giulia Piccinni, Luca Pugliese, Benedetta Masci, Nicolò Ubaldi, Andrea Laghi, and et al. 2022. "Automated Identification of Coronary Arteries in Assisting Inexperienced Readers: Comparison between Two Commercial Vendors" Diagnostics 12, no. 8: 1987. https://doi.org/10.3390/diagnostics12081987
APA StyleDe Santis, D., Tremamunno, G., Rucci, C., Polidori, T., Zerunian, M., Piccinni, G., Pugliese, L., Masci, B., Ubaldi, N., Laghi, A., & Caruso, D. (2022). Automated Identification of Coronary Arteries in Assisting Inexperienced Readers: Comparison between Two Commercial Vendors. Diagnostics, 12(8), 1987. https://doi.org/10.3390/diagnostics12081987







