Applications of Liquid Biopsies in Non-Small-Cell Lung Cancer
Abstract
1. Introduction
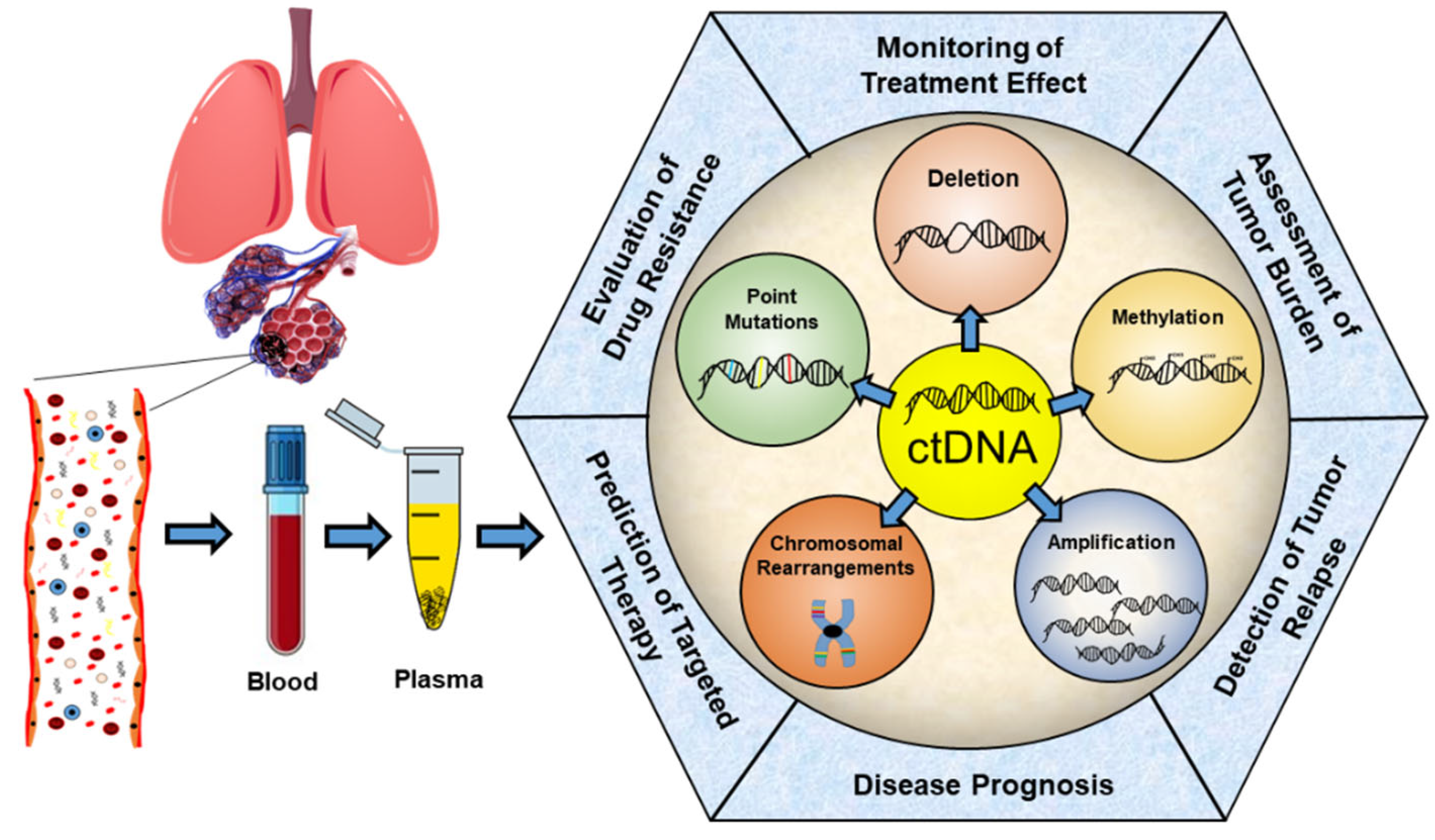
2. ctDNA—Properties and Analysis
3. Prediction of Response to Targeted Therapy
3.1. EGFR Inhibitors
3.2. ALK/ROS Inhibitors
3.3. RET Inhibitors
3.4. BRAF/MEK Inhibitors
3.5. MET Inhibitors
4. Prediction of Response to Immunotherapy
5. Prediction of Response to Chemotherapy
6. Monitoring of the Disease Course and Effect of the Therapy
6.1. EGFR Inhibitors
6.2. ALK Inhibitors
6.3. Immunotherapy (Anti-PD1/PDL1)
6.4. Chemotherapy
6.5. Radiotherapy
7. Further Use of ctDNA in NSCLC
7.1. NSCLC Diagnosis
7.2. Assessment of Tumor Burden
7.3. Estimation of Prognosis
7.4. Prediction and Detection of Recurrence after Tumor Resection
7.5. ctDNA Detection from Cerebrospinal Fluid
7.6. ctDNA Detection from Urine
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- European Lung White Book. Available online: https://www.erswhitebook.org/ (accessed on 24 February 2022).
- Sher, T.; Dy, G.K.; Adjei, A.A. Small Cell Lung Cancer. Mayo Clin. Proc. 2008, 83, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Uramoto, H.; Tanaka, F. Recurrence after Surgery in Patients with NSCLC. Transl. Lung Cancer Res. 2014, 3, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Martins, I.; Ribeiro, I.P.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Melo, J.B.; Carreira, I.M. Liquid Biopsies: Applications for Cancer Diagnosis and Monitoring. Genes 2021, 12, 349. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical Utility of Circulating Non-Coding RNAs—An Update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Beinert, T.; Ermitsch, M.; Seferi, D.; Possinger, K.; Engelmann, C.; Jandrig, B. Detection of Amplifiable Messenger RNA in the Serum of Patients with Lung Cancer. Ann. N. Y. Acad. Sci. 2001, 945, 179–188. [Google Scholar] [CrossRef]
- Wang, N.; Song, X.; Liu, L.; Niu, L.; Wang, X.; Song, X.; Xie, L. Circulating Exosomes Contain Protein Biomarkers of Metastatic Non-Small-Cell Lung Cancer. Cancer Sci. 2018, 109, 1701–1709. [Google Scholar] [CrossRef]
- Chen, X.; Bonnefoi, H.; Diebold-Berger, S.; Lyautey, J.; Lederrey, C.; Faltin-Traub, E.; Stroun, M.; Anker, P. Detecting Tumor-Related Alterations in Plasma or Serum DNA of Patients Diagnosed with Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1999, 5, 2297–2303. [Google Scholar]
- Goebel, G.; Zitt, M.; Zitt, M.; Müller, H.M. Circulating Nucleic Acids in Plasma or Serum (CNAPS) as Prognostic and Predictive Markers in Patients with Solid Neoplasias. Dis. Markers 2005, 21, 105–120. [Google Scholar] [CrossRef]
- Calabuig-Fariñas, S.; Jantus-Lewintre, E.; Herreros-Pomares, A.; Camps, C. Circulating Tumor Cells versus Circulating Tumor DNA in Lung Cancer—which One Will Win? Transl. Lung Cancer Res. 2016, 5, 466–482. [Google Scholar] [CrossRef]
- Pesta, M.; Kulda, V.; Narsanska, A.; Fichtl, J.; Topolcan, O. May CTC Technologies Promote Better Cancer Management? EPMA J. 2015, 6, 1. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network Comprehensive Molecular Profiling of Lung Adenocarcinoma. Nature 2014, 511, 543–550. [CrossRef]
- Bartels, S.; Persing, S.; Hasemeier, B.; Schipper, E.; Kreipe, H.; Lehmann, U. Molecular Analysis of Circulating Cell-Free DNA from Lung Cancer Patients in Routine Laboratory Practice. J. Mol. Diagn. 2017, 19, 722–732. [Google Scholar] [CrossRef]
- Ijzerman, M.J.; Berghuis, A.M.S.; de Bono, J.S.; Terstappen, L.W.M.M. Health Economic Impact of Liquid Biopsies in Cancer Management. Expert Rev. Pharmacoecon. Outcomes Res. 2018, 18, 593–599. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.H.; Chang, Y.S. Recent Advances in Diagnostic Technologies in Lung Cancer. Korean J. Intern. Med. 2020, 35, 257–268. [Google Scholar] [CrossRef]
- Normanno, N.; Denis, M.G.; Thress, K.S.; Ratcliffe, M.; Reck, M. Guide to Detecting Epidermal Growth Factor Receptor (EGFR) Mutations in ctDNA of Patients with Advanced Non-Small-Cell Lung Cancer. Oncotarget 2017, 8, 12501–12516. [Google Scholar] [CrossRef]
- Zhong, X.Y.; Ladewig, A.; Schmid, S.; Wight, E.; Hahn, S.; Holzgreve, W. Elevated Level of Cell-Free Plasma DNA Is Associated with Breast Cancer. Arch. Gynecol. Obstet. 2007, 276, 327–331. [Google Scholar] [CrossRef]
- Jaiswal, S.; Ebert, B.L. Clonal Hematopoiesis in Human Aging and Disease. Science 2019, 366, eaan4673. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, Y.; Xu, Y.; Li, L.; Gong, Y.; Zhang, K.; Zhang, M.; Guan, Y.; Chang, L.; Xia, X.; et al. Pan-Cancer Circulating Tumor DNA Detection in over 10,000 Chinese Patients. Nat. Commun. 2021, 12, 11. [Google Scholar] [CrossRef]
- Qiu, B.; Guo, W.; Zhang, F.; Lv, F.; Ji, Y.; Peng, Y.; Chen, X.; Bao, H.; Xu, Y.; Shao, Y.; et al. Dynamic Recurrence Risk and Adjuvant Chemotherapy Benefit Prediction by ctDNA in Resected NSCLC. Nat. Commun. 2021, 12, 6770. [Google Scholar] [CrossRef]
- Ashworth, T.R. A Case of Cancer in Which Cells Similar to Those in the Tumours Were Seen in the Blood after Death. Aust. Med. J. 1869, 14, 146–149. [Google Scholar]
- Mandel, P.; Metais, P. Nuclear Acids in Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble Normal and Mutated DNA Sequences from Single-Copy Genes in Human Blood. Cancer Epidemiol. Biomarkers Prev. 1994, 3, 67–71. [Google Scholar]
- Khier, S.; Lohan, L. Kinetics of Circulating Cell-Free DNA for Biomedical Applications: Critical Appraisal of the Literature. Future Sci. OA 2018, 4, FSO295. [Google Scholar] [CrossRef]
- Mouliere, F.; Robert, B.; Arnau Peyrotte, E.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, Structures, and Functions of Circulating DNA in Oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef]
- Bohers, E.; Viailly, P.-J.; Jardin, F. cfDNA Sequencing: Technological Approaches and Bioinformatic Issues. Pharmaceuticals 2021, 14, 596. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Chan, K.C.A.; Jiang, P.; Zheng, Y.W.L.; Liao, G.J.W.; Sun, H.; Wong, J.; Siu, S.S.N.; Chan, W.C.; Chan, S.L.; Chan, A.T.C.; et al. Cancer Genome Scanning in Plasma: Detection of Tumor-Associated Copy Number Aberrations, Single-Nucleotide Variants, and Tumoral Heterogeneity by Massively Parallel Sequencing. Clin. Chem. 2013, 59, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.-S.; Squire, J.; et al. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Thorac. Oncol. 2013, 8, 823–859. [Google Scholar] [CrossRef] [PubMed]
- Pennell, N.A.; Arcila, M.E.; Gandara, D.R.; West, H. Biomarker Testing for Patients with Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 531–542. [Google Scholar] [CrossRef]
- Herbreteau, G.; Vallée, A.; Charpentier, S.; Normanno, N.; Hofman, P.; Denis, M.G. Circulating Free Tumor DNA in Non-Small Cell Lung Cancer (NSCLC): Clinical Application and Future Perspectives. J. Thorac. Dis. 2019, 11, S113–S126. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, S.; MacLennan, G.T.; Eble, J.N.; Lopez-Beltran, A.; Yang, X.J.; Pan, C.-X.; Zhou, H.; Montironi, R.; Cheng, L. Epidermal Growth Factor Receptor Protein Expression and Gene Amplification in Small Cell Carcinoma of the Urinary Bladder. Clin. Cancer Res. 2007, 13, 953–957. [Google Scholar] [CrossRef]
- Herbst, R.S.; Heymach, J.V.; Lippman, S.M. Lung Cancer. N. Engl. J. Med. 2008, 359, 1367–1380. [Google Scholar] [CrossRef]
- Sequist, L.V.; Bell, D.W.; Lynch, T.J.; Haber, D.A. Molecular Predictors of Response to Epidermal Growth Factor Receptor Antagonists in Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2007, 25, 587–595. [Google Scholar] [CrossRef]
- Ladanyi, M.; Pao, W. Lung Adenocarcinoma: Guiding EGFR-Targeted Therapy and beyond. Mod. Pathol. 2008, 21, S16–S22. [Google Scholar] [CrossRef]
- Yamamoto, H.; Toyooka, S.; Mitsudomi, T. Impact of EGFR Mutation Analysis in Non-Small Cell Lung Cancer. Lung Cancer 2009, 63, 315–321. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance Mechanisms to Osimertinib in EGFR-Mutated Non-Small Cell Lung Cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, J.; Liu, T.; Dang, J.; Li, G. First-Line Treatments in EGFR-Mutated Advanced Non-Small Cell Lung Cancer: A Network Meta-Analysis. PLoS ONE 2019, 14, e0223530. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Shen, L.; Zheng, D. Diagnostic Value of Circulating Free DNA for the Detection of EGFR Mutation Status in NSCLC: A Systematic Review and Meta-Analysis. Sci. Rep. 2014, 4, 6269. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.; Wu, Y.-L.; Lee, J.S.; Yu, C.-J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-Line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef]
- Ai, X.; Cui, J.; Zhang, J.; Chen, R.; Lin, W.; Xie, C.; Liu, A.; Zhang, J.; Yang, W.; Hu, X.; et al. Clonal Architecture of EGFR Mutation Predicts the Efficacy of EGFR-Tyrosine Kinase Inhibitors in Advanced NSCLC: A Prospective Multicenter Study (NCT03059641). Clin. Cancer Res. 2021, 27, 704–712. [Google Scholar] [CrossRef]
- Del Re, M.; Petrini, I.; Mazzoni, F.; Valleggi, S.; Gianfilippo, G.; Pozzessere, D.; Chella, A.; Crucitta, S.; Rofi, E.; Restante, G.; et al. Incidence of T790M in Patients with NSCLC Progressed to Gefitinib, Erlotinib, and Afatinib: A Study on Circulating Cell-Free DNA. Clin. Lung Cancer 2020, 21, 232–237. [Google Scholar] [CrossRef]
- Kitazono, S.; Sakai, K.; Yanagitani, N.; Ariyasu, R.; Yoshizawa, T.; Dotsu, Y.; Koyama, J.; Saiki, M.; Sonoda, T.; Nishikawa, S.; et al. Barcode Sequencing Identifies Resistant Mechanisms to Epidermal Growth Factor Receptor Inhibitors in Circulating Tumor DNA of Lung Cancer Patients. Cancer Sci. 2019, 110, 3350–3357. [Google Scholar] [CrossRef]
- Deng, Q.; Xie, B.; Wu, L.; Ji, X.; Li, C.; Feng, L.; Fang, Q.; Bao, Y.; Li, J.; Jin, S.; et al. Competitive Evolution of NSCLC Tumor Clones and the Drug Resistance Mechanism of First-Generation EGFR-TKIs in Chinese NSCLC Patients. Heliyon 2018, 4, e01031. [Google Scholar] [CrossRef]
- Romero, A.; Serna-Blasco, R.; Alfaro, C.; Sánchez-Herrero, E.; Barquín, M.; Turpin, M.C.; Chico, S.; Sanz-Moreno, S.; Rodrigez-Festa, A.; Laza-Briviesca, R.; et al. ctDNA Analysis Reveals Different Molecular Patterns upon Disease Progression in Patients Treated with Osimertinib. Transl. Lung Cancer Res. 2020, 9, 532–540. [Google Scholar] [CrossRef]
- Ishii, H.; Azuma, K.; Sakai, K.; Naito, Y.; Matsuo, N.; Tokito, T.; Yamada, K.; Hoshino, T.; Nishio, K. Determination of Somatic Mutations and Tumor Mutation Burden in Plasma by CAPP-Seq during Afatinib Treatment in NSCLC Patients Resistance to Osimertinib. Sci. Rep. 2020, 10, 691. [Google Scholar] [CrossRef]
- Shaw, A.T.; Kim, D.-W.; Nakagawa, K.; Seto, T.; Crinó, L.; Ahn, M.-J.; De Pas, T.; Besse, B.; Solomon, B.J.; Blackhall, F.; et al. Crizotinib versus Chemotherapy in Advanced ALK -Positive Lung Cancer. N. Engl. J. Med. 2013, 368, 2385–2394. [Google Scholar] [CrossRef]
- Camidge, D.R.; Bang, Y.-J.; Kwak, E.L.; Iafrate, A.J.; Varella-Garcia, M.; Fox, S.B.; Riely, G.J.; Solomon, B.; Ou, S.-H.I.; Kim, D.-W.; et al. Activity and Safety of Crizotinib in Patients with ALK-Positive Non-Small-Cell Lung Cancer: Updated Results from a Phase 1 Study. Lancet Oncol. 2012, 13, 1011–1019. [Google Scholar] [CrossRef]
- Shaw, A.T.; Yeap, B.Y.; Solomon, B.J.; Riely, G.J.; Gainor, J.; Engelman, J.A.; Shapiro, G.I.; Costa, D.B.; Ou, S.-H.I.; Butaney, M.; et al. Effect of Crizotinib on Overall Survival in Patients with Advanced Non-Small-Cell Lung Cancer Harbouring ALK Gene Rearrangement: A Retrospective Analysis. Lancet Oncol. 2011, 12, 1004–1012. [Google Scholar] [CrossRef]
- Devarakonda, S.; Morgensztern, D.; Govindan, R. Genomic Alterations in Lung Adenocarcinoma. Lancet Oncol. 2015, 16, e342–e351. [Google Scholar] [CrossRef]
- Bruno, R.; Fontanini, G. Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics 2020, 10, 521. [Google Scholar] [CrossRef]
- Gainor, J.F.; Dardaei, L.; Yoda, S.; Friboulet, L.; Leshchiner, I.; Katayama, R.; Dagogo-Jack, I.; Gadgeel, S.; Schultz, K.; Singh, M.; et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016, 6, 1118–1133. [Google Scholar] [CrossRef]
- Shaw, A.T.; Friboulet, L.; Leshchiner, I.; Gainor, J.F.; Bergqvist, S.; Brooun, A.; Burke, B.J.; Deng, Y.-L.; Liu, W.; Dardaei, L.; et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N. Engl. J. Med. 2016, 374, 54–61. [Google Scholar] [CrossRef]
- Elliott, J.; Bai, Z.; Hsieh, S.-C.; Kelly, S.E.; Chen, L.; Skidmore, B.; Yousef, S.; Zheng, C.; Stewart, D.J.; Wells, G.A. ALK Inhibitors for Non-Small Cell Lung Cancer: A Systematic Review and Network Meta-Analysis. PLoS ONE 2020, 15, e0229179. [Google Scholar] [CrossRef]
- Solomon, B.J.; Mok, T.; Kim, D.-W.; Wu, Y.-L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef]
- Qiao, H.; Lovly, C.M. Cracking the Code of Resistance across Multiple Lines of ALK Inhibitor Therapy in Lung Cancer. Cancer Discov. 2016, 6, 1084–1086. [Google Scholar] [CrossRef]
- McCoach, C.E.; Le, A.T.; Gowan, K.; Jones, K.; Schubert, L.; Doak, A.; Estrada-Bernal, A.; Davies, K.D.; Merrick, D.T.; Bunn, P.A.; et al. Resistance Mechanisms to Targeted Therapies in ROS1+ and ALK+ Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 3334–3347. [Google Scholar] [CrossRef]
- Ali, S.M.; Hensing, T.; Schrock, A.B.; Allen, J.; Sanford, E.; Gowen, K.; Kulkarni, A.; He, J.; Suh, J.H.; Lipson, D.; et al. Comprehensive Genomic Profiling Identifies a Subset of Crizotinib-Responsive ALK-Rearranged Non-Small Cell Lung Cancer Not Detected by Fluorescence In Situ Hybridization. Oncologist 2016, 21, 762–770. [Google Scholar] [CrossRef]
- McCoach, C.E.; Blakely, C.M.; Banks, K.C.; Levy, B.; Chue, B.M.; Raymond, V.M.; Le, A.T.; Lee, C.E.; Diaz, J.; Waqar, S.N.; et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 2758–2770. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, P.-W.; Wang, W.-Y.; Wang, K.; Zhang, Z.; Chen, B.-J.; He, Y.-Q.; Li, L.; Liu, H.; Chuai, S.; et al. Noninvasive Genotyping and Monitoring of Anaplastic Lymphoma Kinase (ALK) Rearranged Non-Small Cell Lung Cancer by Capture-Based next-Generation Sequencing. Oncotarget 2016, 7, 65208–65217. [Google Scholar] [CrossRef][Green Version]
- Bordi, P.; Tiseo, M.; Rofi, E.; Petrini, I.; Restante, G.; Danesi, R.; Del Re, M. Detection of ALK and KRAS Mutations in Circulating Tumor DNA of Patients with Advanced ALK-Positive NSCLC with Disease Progression During Crizotinib Treatment. Clin. Lung Cancer 2017, 18, 692–697. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Rooney, M.; Nagy, R.J.; Lin, J.J.; Chin, E.; Ferris, L.A.; Ackil, J.; Lennerz, J.K.; Lanman, R.B.; Gainor, J.F.; et al. Molecular Analysis of Plasma from Patients with ROS1-Positive NSCLC. J. Thorac. Oncol. 2019, 14, 816–824. [Google Scholar] [CrossRef]
- Mezquita, L.; Swalduz, A.; Jovelet, C.; Ortiz-Cuaran, S.; Howarth, K.; Planchard, D.; Avrillon, V.; Recondo, G.; Marteau, S.; Benitez, J.C.; et al. Clinical Relevance of an Amplicon-Based Liquid Biopsy for Detecting ALK and ROS1 Fusion and Resistance Mutations in Patients with Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2020, 4, 272–282. [Google Scholar] [CrossRef]
- Horn, L.; Whisenant, J.G.; Wakelee, H.; Reckamp, K.L.; Qiao, H.; Leal, T.A.; Du, L.; Hernandez, J.; Huang, V.; Blumenschein, G.R.; et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients with ALK+ Lung Cancer. J. Thorac. Oncol. 2019, 14, 1901–1911. [Google Scholar] [CrossRef]
- Wang, R.; Hu, H.; Pan, Y.; Li, Y.; Ye, T.; Li, C.; Luo, X.; Wang, L.; Li, H.; Zhang, Y.; et al. RET Fusions Define a Unique Molecular and Clinicopathologic Subtype of Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 4352–4359. [Google Scholar] [CrossRef]
- Drilon, A.; Wang, L.; Arcila, M.E.; Balasubramanian, S.; Greenbowe, J.R.; Ross, J.S.; Stephens, P.; Lipson, D.; Miller, V.A.; Kris, M.G.; et al. Broad, Hybrid Capture–Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin. Cancer Res. 2015, 21, 3631–3639. [Google Scholar] [CrossRef]
- Piotrowska, Z.; Isozaki, H.; Lennerz, J.K.; Gainor, J.F.; Lennes, I.T.; Zhu, V.W.; Marcoux, N.; Banwait, M.K.; Digumarthy, S.R.; Su, W.; et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov. 2018, 8, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Rich, T.A.; Reckamp, K.L.; Chae, Y.K.; Doebele, R.C.; Iams, W.T.; Oh, M.; Raymond, V.M.; Lanman, R.B.; Riess, J.W.; Stinchcombe, T.E.; et al. Analysis of Cell-Free DNA from 32,989 Advanced Cancers Reveals Novel Co-Occurring Activating RET Alterations and Oncogenic Signaling Pathway Aberrations. Clin. Cancer Res. 2019, 25, 5832–5842. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Yang, D.; Velcheti, V.; Drilon, A.; Meric-Bernstam, F. State-of-the-Art Strategies for Targeting RET-Dependent Cancers. J. Clin. Oncol. 2020, 38, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Gautschi, O.; Milia, J.; Filleron, T.; Wolf, J.; Carbone, D.P.; Owen, D.; Camidge, R.; Narayanan, V.; Doebele, R.C.; Besse, B.; et al. Targeting RET in Patients with RET-Rearranged Lung Cancers: Results from the Global, Multicenter RET Registry. J. Clin. Oncol. 2017, 35, 1403–1410. [Google Scholar] [CrossRef]
- Sarfaty, M.; Moore, A.; Neiman, V.; Dudnik, E.; Ilouze, M.; Gottfried, M.; Katznelson, R.; Nechushtan, H.; Sorotsky, H.G.; Paz, K.; et al. RET Fusion Lung Carcinoma: Response to Therapy and Clinical Features in a Case Series of 14 Patients. Clin. Lung Cancer 2017, 18, e223–e232. [Google Scholar] [CrossRef]
- Solomon, B.J.; Tan, L.; Lin, J.J.; Wong, S.Q.; Hollizeck, S.; Ebata, K.; Tuch, B.B.; Yoda, S.; Gainor, J.F.; Sequist, L.V.; et al. RET Solvent Front Mutations Mediate Acquired Resistance to Selective RET Inhibition in RET-Driven Malignancies. J. Thorac. Oncol. 2020, 15, 541–549. [Google Scholar] [CrossRef]
- Barlesi, F.; Mazieres, J.; Merlio, J.-P.; Debieuvre, D.; Mosser, J.; Lena, H.; Ouafik, L.; Besse, B.; Rouquette, I.; Westeel, V.; et al. Routine Molecular Profiling of Patients with Advanced Non-Small-Cell Lung Cancer: Results of a 1-Year Nationwide Programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016, 387, 1415–1426. [Google Scholar] [CrossRef]
- Paik, P.K.; Arcila, M.E.; Fara, M.; Sima, C.S.; Miller, V.A.; Kris, M.G.; Ladanyi, M.; Riely, G.J. Clinical Characteristics of Patients with Lung Adenocarcinomas Harboring BRAF Mutations. J. Clin. Oncol. 2011, 29, 2046–2051. [Google Scholar] [CrossRef]
- Alvarez, J.G.B.; Otterson, G.A. Agents to Treat BRAF-Mutant Lung Cancer. Drugs Context 2019, 8, 212566. [Google Scholar] [CrossRef]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or Chemotherapy for Non–Small-Cell Lung Cancer with Mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus Standard Chemotherapy as First-Line Treatment for European Patients with Advanced EGFR Mutation-Positive Non-Small-Cell Lung Cancer (EURTAC): A Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Chung, C. Tyrosine Kinase Inhibitors for Epidermal Growth Factor Receptor Gene Mutation–positive Non-Small Cell Lung Cancers: An Update for Recent Advances in Therapeutics. J. Oncol. Pharm. Pract. 2016, 22, 461–476. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, X.; Li, R.; Shen, J.; Zhang, H.; Yu, L.; Liu, B.; Wang, L. The Detection and Significance of EGFR and BRAF in Cell-Free DNA of Peripheral Blood in NSCLC. Oncotarget 2017, 8, 49773–49782. [Google Scholar] [CrossRef]
- Ortiz-Cuaran, S.; Mezquita, L.; Swalduz, A.; Aldea, M.; Mazieres, J.; Leonce, C.; Jovelet, C.; Pradines, A.; Avrillon, V.; Chumbi Flores, W.R.; et al. Circulating Tumor DNA Genomics Reveal Potential Mechanisms of Resistance to BRAF-Targeted Therapies in Patients with BRAF-Mutant Metastatic Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 6242–6253. [Google Scholar] [CrossRef]
- Blumenschein, G.R.; Mills, G.B.; Gonzalez-Angulo, A.M. Targeting the Hepatocyte Growth Factor-cMET Axis in Cancer Therapy. J. Clin. Oncol. 2012, 30, 3287–3296. [Google Scholar] [CrossRef]
- Sadiq, A.A.; Salgia, R. MET as a Possible Target for Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2013, 31, 1089–1096. [Google Scholar] [CrossRef]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.-M.; Zhao, X.; Christensen, J.; et al. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef]
- Paik, P.K.; Drilon, A.; Fan, P.-D.; Yu, H.; Rekhtman, N.; Ginsberg, M.S.; Borsu, L.; Schultz, N.; Berger, M.F.; Rudin, C.M.; et al. Response to MET Inhibitors in Patients with Stage IV Lung Adenocarcinomas Harboring MET Mutations Causing Exon 14 Skipping. Cancer Discov. 2015, 5, 842–849. [Google Scholar] [CrossRef]
- Li, A.; Yang, J.-J.; Zhang, X.-C.; Zhang, Z.; Su, J.; Gou, L.-Y.; Bai, Y.; Zhou, Q.; Yang, Z.; Han-Zhang, H.; et al. Acquired MET Y1248H and D1246N Mutations Mediate Resistance to MET Inhibitors in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 4929–4937. [Google Scholar] [CrossRef]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Luft, A.; Vicente, D.; Tafreshi, A.; Gümüş, M.; Mazières, J.; Hermes, B.; Çay Şenler, F.; Csőszi, T.; Fülöp, A.; et al. Pembrolizumab plus Chemotherapy for Squamous Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2040–2051. [Google Scholar] [CrossRef]
- Savill, K.M.Z.; Zettler, M.E.; Feinberg, B.A.; Jeune-Smith, Y.; Gajra, A. Awareness and Utilization of Tumor Mutation Burden (TMB) as a Biomarker for Administration of Immuno-Oncology (I-O) Therapeutics by Practicing Community Oncologists in the United States (U.S.). J. Clin. Oncol. 2021, 39, 2608. [Google Scholar] [CrossRef]
- Büttner, R.; Longshore, J.W.; López-Ríos, F.; Merkelbach-Bruse, S.; Normanno, N.; Rouleau, E.; Penault-Llorca, F. Implementing TMB Measurement in Clinical Practice: Considerations on Assay Requirements. ESMO Open 2019, 4, e000442. [Google Scholar] [CrossRef]
- Vilimas, T. Measuring Tumor Mutational Burden Using Whole-Exome Sequencing. Methods Mol. Biol. Clifton N. J. 2020, 2055, 63–91. [Google Scholar] [CrossRef]
- Fancello, L.; Gandini, S.; Pelicci, P.G.; Mazzarella, L. Tumor Mutational Burden Quantification from Targeted Gene Panels: Major Advancements and Challenges. J. Immunother. Cancer 2019, 7, 183. [Google Scholar] [CrossRef] [PubMed]
- Friedlaender, A.; Nouspikel, T.; Christinat, Y.; Ho, L.; McKee, T.; Addeo, A. Tissue-Plasma TMB Comparison and Plasma TMB Monitoring in Patients with Metastatic Non-Small Cell Lung Cancer Receiving Immune Checkpoint Inhibitors. Front. Oncol. 2020, 10, 142. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-Based Tumor Mutational Burden as a Predictor of Clinical Benefit in Non-Small-Cell Lung Cancer Patients Treated with Atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Chae, Y.K.; Davis, A.A.; Agte, S.; Pan, A.; Simon, N.I.; Iams, W.T.; Cruz, M.R.; Tamragouri, K.; Rhee, K.; Mohindra, N.; et al. Clinical Implications of Circulating Tumor DNA Tumor Mutational Burden (ctDNA TMB) in Non-Small Cell Lung Cancer. Oncologist 2019, 24, 820–828. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, J.; Wang, G.; Zhao, J.; Xu, J.; Han, J.; Zhao, Z.; Zhao, J.; Zhu, B.; Zhuo, M.; et al. Allele Frequency-Adjusted Blood-Based Tumor Mutational Burden as a Predictor of Overall Survival for Patients with NSCLC Treated with PD-(L)1 Inhibitors. J. Thorac. Oncol. 2020, 15, 556–567. [Google Scholar] [CrossRef]
- Rosell, R.; Gomez-Codina, J.; Camps, C.; Maestre, J.; Padille, J.; Canto, A.; Mate, J.L.; Li, S.; Roig, J.; Olazabal, A.; et al. A Randomized Trial Comparing Preoperative Chemotherapy Plus Surgery with Surgery Alone in Patients with Non-Small-Cell Lung Cancer. N. Engl. J. Med. 1994, 330, 153–158. [Google Scholar] [CrossRef]
- Roth, J.A.; Fossella, F.; Komaki, R.; Ryan, M.B.; Putnam, J.B.; Lee, J.S.; Dhingra, H.; De Caro, L.; Chasen, M.; McGavran, M.; et al. A Randomized Trial Comparing Perioperative Chemotherapy and Surgery with Surgery Alone in Resectable Stage IIIA Non-Small-Cell Lung Cancer. JNCI J. Natl. Cancer Inst. 1994, 86, 673–680. [Google Scholar] [CrossRef]
- Coco, S.; Alama, A.; Vanni, I.; Fontana, V.; Genova, C.; Dal Bello, M.G.; Truini, A.; Rijavec, E.; Biello, F.; Sini, C.; et al. Circulating Cell-Free DNA and Circulating Tumor Cells as Prognostic and Predictive Biomarkers in Advanced Non-Small Cell Lung Cancer Patients Treated with First-Line Chemotherapy. Int. J. Mol. Sci. 2017, 18, 1035. [Google Scholar] [CrossRef]
- Han, X.; Han, Y.; Tan, Q.; Huang, Y.; Yang, J.; Yang, S.; He, X.; Zhou, S.; Song, Y.; Pi, J.; et al. Tracking Longitudinal Genetic Changes of Circulating Tumor DNA (ctDNA) in Advanced Lung Adenocarcinoma Treated with Chemotherapy. J. Transl. Med. 2019, 17, 339. [Google Scholar] [CrossRef]
- Xu, R.; Zhong, G.; Huang, T.; He, W.; Kong, C.; Zhang, X.; Wang, Y.; Liu, M.; Xu, M.; Chen, S. Sequencing of Circulating Tumor DNA for Dynamic Monitoring of Gene Mutations in Advanced Non-Small Cell Lung Cancer. Oncol. Lett. 2018, 15, 3726–3734. [Google Scholar] [CrossRef]
- Chiou, C.-C.; Wang, C.-L.; Luo, J.-D.; Liu, C.-Y.; Ko, H.-W.; Yang, C.-T. Targeted Sequencing of Circulating Cell Free DNA Can Be Used to Monitor Therapeutic Efficacy of Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer Patients. Cancer Genom.-Proteom. 2020, 17, 417–423. [Google Scholar] [CrossRef]
- Sakai, K.; Takahama, T.; Shimokawa, M.; Azuma, K.; Takeda, M.; Kato, T.; Daga, H.; Okamoto, I.; Akamatsu, H.; Teraoka, S.; et al. Predicting Osimertinib-treatment Outcomes through EGFR Mutant-fraction Monitoring in the Circulating Tumor DNA of EGFR T790M-positive Patients with Non-small Cell Lung Cancer (WJOG8815L). Mol. Oncol. 2021, 15, 126–137. [Google Scholar] [CrossRef]
- Iwama, E.; Sakai, K.; Azuma, K.; Harada, T.; Harada, D.; Nosaki, K.; Hotta, K.; Ohyanagi, F.; Kurata, T.; Fukuhara, T.; et al. Monitoring of Somatic Mutations in Circulating Cell-Free DNA by Digital PCR and next-Generation Sequencing during Afatinib Treatment in Patients with Lung Adenocarcinoma Positive for EGFR Activating Mutations. Ann. Oncol. 2017, 28, 136–141. [Google Scholar] [CrossRef]
- Dietz, S.; Christopoulos, P.; Yuan, Z.; Angeles, A.K.; Gu, L.; Volckmar, A.-L.; Ogrodnik, S.J.; Janke, F.; Fratte, C.D.; Zemojtel, T.; et al. Longitudinal Therapy Monitoring of ALK-Positive Lung Cancer by Combined Copy Number and Targeted Mutation Profiling of Cell-Free DNA. EBioMedicine 2020, 62, 103103. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; Sznol, M.; Kluger, H.M.; McDermott, D.F.; Carvajal, R.D.; Lawrence, D.P.; Topalian, S.L.; Atkins, M.B.; Powderly, J.D.; Sharfman, W.H.; et al. Long-Term Survival of Ipilimumab-Naive Patients (pts) with Advanced Melanoma (MEL) Treated with Nivolumab (anti-PD-1, BMS-936558, ONO-4538) in a Phase I Trial. J. Clin. Oncol. 2014, 32, 9002. [Google Scholar] [CrossRef]
- Guibert, N.; Mazières, J. Nivolumab for Treating Non-Small Cell Lung Cancer. Expert Opin. Biol. Ther. 2015, 15, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S.B.; Narayan, A.; Kole, A.J.; Decker, R.H.; Teysir, J.; Carriero, N.J.; Lee, A.; Nemati, R.; Nath, S.K.; Mane, S.M.; et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018, 24, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Guibert, N.; Mazieres, J.; Delaunay, M.; Casanova, A.; Farella, M.; Keller, L.; Favre, G.; Pradines, A. Monitoring of KRAS-Mutated ctDNA to Discriminate Pseudo-Progression from True Progression during Anti-PD-1 Treatment of Lung Adenocarcinoma. Oncotarget 2017, 8, 38056–38060. [Google Scholar] [CrossRef]
- Anagnostou, V.; Forde, P.M.; White, J.R.; Niknafs, N.; Hruban, C.; Naidoo, J.; Marrone, K.; Sivakumar, I.K.A.; Bruhm, D.C.; Rosner, S.; et al. Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Res. 2019, 79, 1214–1225. [Google Scholar] [CrossRef]
- Jiang, T.; Li, X.; Wang, J.; Su, C.; Han, W.; Zhao, C.; Wu, F.; Gao, G.; Li, W.; Chen, X.; et al. Mutational Landscape of cfDNA Identifies Distinct Molecular Features Associated with Therapeutic Response to First-Line Platinum-Based Doublet Chemotherapy in Patients with Advanced NSCLC. Theranostics 2017, 7, 4753–4762. [Google Scholar] [CrossRef]
- Guibert, N.; Pradines, A.; Farella, M.; Casanova, A.; Gouin, S.; Keller, L.; Favre, G.; Mazieres, J. Monitoring KRAS Mutations in Circulating DNA and Tumor Cells Using Digital Droplet PCR during Treatment of KRAS-Mutated Lung Adenocarcinoma. Lung Cancer 2016, 100, 1–4. [Google Scholar] [CrossRef]
- Kageyama, S.-I.; Nihei, K.; Karasawa, K.; Sawada, T.; Koizumi, F.; Yamaguchi, S.; Kato, S.; Hojo, H.; Motegi, A.; Tsuchihara, K.; et al. Radiotherapy Increases Plasma Levels of Tumoral Cell-Free DNA in Non-Small Cell Lung Cancer Patients. Oncotarget 2018, 9, 19368–19378. [Google Scholar] [CrossRef]
- Walls, G.M.; McConnell, L.; McAleese, J.; Murray, P.; Lynch, T.B.; Savage, K.; Hanna, G.G.; de Castro, D.G. Early Circulating Tumour DNA Kinetics Measured by Ultra-Deep next-Generation Sequencing during Radical Radiotherapy for Non-Small Cell Lung Cancer: A Feasibility Study. Radiat. Oncol. 2020, 15, 132. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and Localization of Surgically Resectable Cancers with a Multi-Analyte Blood Test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.C.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef]
- Yang, Z.; Qi, W.; Sun, L.; Zhou, H.; Zhou, B.; Hu, Y. DNA Methylation Analysis of Selected Genes for the Detection of Early-Stage Lung Cancer Using Circulating Cell-Free DNA. Adv. Clin. Exp. Med. 2019, 28, 355–360. [Google Scholar] [CrossRef]
- Ooki, A.; Maleki, Z.; Tsay, J.-C.J.; Goparaju, C.; Brait, M.; Turaga, N.; Nam, H.-S.; Rom, W.N.; Pass, H.I.; Sidransky, D.; et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non–Small Cell Lung Cancer and Serum DNA. Clin. Cancer Res. 2017, 23, 7141–7152. [Google Scholar] [CrossRef]
- Passiglia, F.; Rizzo, S.; Di Maio, M.; Galvano, A.; Badalamenti, G.; Listì, A.; Gulotta, L.; Castiglia, M.; Fulfaro, F.; Bazan, V.; et al. The Diagnostic Accuracy of Circulating Tumor DNA for the Detection of EGFR-T790M Mutation in NSCLC: A Systematic Review and Meta-Analysis. Sci. Rep. 2018, 8, 13379. [Google Scholar] [CrossRef]
- Czarnecka, A.M.; Brodziak, A.; Sobczuk, P.; Dendek, C.; Labochka, D.; Korniluk, J.; Bartnik, E.; Szczylik, C. Metastatic Tumor Burden and Loci as Predictors of First Line Sunitinib Treatment Efficacy in Patients with Renal Cell Carcinoma. Sci. Rep. 2019, 9, 7754. [Google Scholar] [CrossRef]
- Tateishi, U.; Tatsumi, M.; Terauchi, T.; Ando, K.; Niitsu, N.; Kim, W.S.; Suh, C.; Ogura, M.; Tobinai, K. Prognostic Significance of Metabolic Tumor Burden by Positron Emission Tomography/computed Tomography in Patients with Relapsed/refractory Diffuse Large B-cell Lymphoma. Cancer Sci. 2015, 106, 186–193. [Google Scholar] [CrossRef]
- Gobbi, P.G.; Broglia, C.; Di Giulio, G.; Mantelli, M.; Anselmo, P.; Merli, F.; Zinzani, P.L.; Rossi, G.; Callea, V.; Iannitto, E.; et al. The Clinical Value of Tumor Burden at Diagnosis in Hodgkin Lymphoma. Cancer 2004, 101, 1824–1834. [Google Scholar] [CrossRef]
- Leek, R.D.; Landers, R.J.; Harris, A.L.; Lewis, C.E. Necrosis Correlates with High Vascular Density and Focal Macrophage Infiltration in Invasive Carcinoma of the Breast. Br. J. Cancer 1999, 79, 991–995. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and Quantification of Mutations in the Plasma of Patients with Colorectal Tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef]
- Cho, M.-S.; Park, C.H.; Lee, S.; Park, H.S. Clinicopathological Parameters for Circulating Tumor DNA Shedding in Surgically Resected Non-Small Cell Lung Cancer with EGFR or KRAS Mutation. PLoS ONE 2020, 15, e0230622. [Google Scholar] [CrossRef]
- Zhu, Y.-J.; Zhang, H.-B.; Liu, Y.-H.; Zhang, F.-L.; Zhu, Y.-Z.; Li, Y.; Bai, J.-P.; Liu, L.-R.; Qu, Y.-C.; Qu, X.; et al. Quantitative Cell-Free Circulating EGFR Mutation Concentration Is Correlated with Tumor Burden in Advanced NSCLC Patients. Lung Cancer 2017, 109, 124–127. [Google Scholar] [CrossRef]
- Yanagita, M.; Redig, A.J.; Paweletz, C.P.; Dahlberg, S.E.; O’Connell, A.; Feeney, N.; Taibi, M.; Boucher, D.; Oxnard, G.R.; Johnson, B.E.; et al. A Prospective Evaluation of Circulating Tumor Cells and Cell-Free DNA in EGFR-Mutant Non–Small Cell Lung Cancer Patients Treated with Erlotinib on a Phase II Trial. Clin. Cancer Res. 2016, 22, 6010–6020. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Fan, G.; Zhang, K.; Yang, X.; Ding, J.; Wang, Z.; Li, J. Prognostic Value of Circulating Tumor DNA in Patients with Colon Cancer: Systematic Review. PLoS ONE 2017, 12, e0171991. [Google Scholar] [CrossRef]
- Jia, J.; Huang, B.; Zhuang, Z.; Chen, S. Circulating Tumor DNA as Prognostic Markers for Late Stage NSCLC with Bone Metastasis. Int. J. Biol. Markers 2018, 33, 222–230. [Google Scholar] [CrossRef]
- Michaelidou, K.; Koutoulaki, C.; Mavridis, K.; Vorrias, E.; Papadaki, M.A.; Koutsopoulos, A.V.; Mavroudis, D.; Agelaki, S. Detection of KRAS G12/G13 Mutations in Cell Free-DNA by Droplet Digital PCR, Offers Prognostic Information for Patients with Advanced Non-Small Cell Lung Cancer. Cells 2020, 9, 2514. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Z.; Zhao, S.; Ye, D.; Cai, X.; Cheng, B.; Li, C.; Xiong, S.; Li, J.; Liang, H.; et al. Presence of Allele Frequency Heterogeneity Defined by ctDNA Profiling Predicts Unfavorable Overall Survival of NSCLC. Transl. Lung Cancer Res. 2019, 8, 1045–1050. [Google Scholar] [CrossRef]
- Song, Y.; Hu, C.; Xie, Z.; Wu, L.; Zhu, Z.; Rao, C.; Liu, L.; Chen, Y.; Liang, N.; Chen, J.; et al. Circulating Tumor DNA Clearance Predicts Prognosis across Treatment Regimen in a Large Real-World Longitudinally Monitored Advanced Non-Small Cell Lung Cancer Cohort. Transl. Lung Cancer Res. 2020, 9, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating Mutant DNA to Assess Tumor Dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhao, H.; Shi, Y.; Yang, F.; Wang, L.T.; Kang, G.; Nie, Y.; Wang, J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin. Cancer Res. 2019, 25, 7058–7067. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Lou, F.; Ma, Y.; Li, J.; Yang, B.; Chen, W.; Ye, H.; Zhang, J.-B.; Zhao, M.-Y.; Wu, W.-J.; et al. Circulating Tumor DNA Detection in Lung Cancer Patients before and after Surgery. Sci. Rep. 2016, 6, 33519. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, X.; Huang, Z.; Wang, F.; Lin, Y.; Wen, Y.; Liu, L.; Li, J.; Liu, X.; Xie, W.; et al. Development and Validation of a Preoperative Noninvasive Predictive Model Based on Circular Tumor DNA for Lymph Node Metastasis in Resectable Non-Small Cell Lung Cancer. Transl. Lung Cancer Res. 2020, 9, 722–730. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.; Chirieac, L.R.; D’Amico, T.A.; DeCamp, M.M.; Dilling, T.J.; Dobelbower, M.; et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2017, 15, 504–535. [Google Scholar] [CrossRef]
- Liang, H.; Huang, J.; Wang, B.; Liu, Z.; He, J.; Liang, W. The Role of Liquid Biopsy in Predicting Post-Operative Recurrence of Non-Small Cell Lung Cancer. J. Thorac. Dis. 2018, 10, S838–S845. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Mayor, R.; Ng, C.K.Y.; Weigelt, B.; Martínez-Ricarte, F.; Torrejon, D.; Oliveira, M.; Arias, A.; Raventos, C.; Tang, J.; et al. Cerebrospinal Fluid-Derived Circulating Tumour DNA Better Represents the Genomic Alterations of Brain Tumours than Plasma. Nat. Commun. 2015, 6, 8839. [Google Scholar] [CrossRef]
- Paik, P.K.; Shen, R.; Won, H.; Rekhtman, N.; Wang, L.; Sima, C.S.; Arora, A.; Seshan, V.; Ladanyi, M.; Berger, M.F.; et al. Next-Generation Sequencing of Stage IV Squamous Cell Lung Cancers Reveals an Association of PI3K Aberrations and Evidence of Clonal Heterogeneity in Patients with Brain Metastases. Cancer Discov. 2015, 5, 610–621. [Google Scholar] [CrossRef]
- Schuette, W. Treatment of Brain Metastases from Lung Cancer: Chemotherapy. Lung Cancer 2004, 45, S253–S257. [Google Scholar] [CrossRef]
- Ma, C.; Yang, X.; Xing, W.; Yu, H.; Si, T.; Guo, Z. Detection of Circulating Tumor DNA from Non-Small Cell Lung Cancer Brain Metastasis in Cerebrospinal Fluid Samples. Thorac. Cancer 2020, 11, 588–593. [Google Scholar] [CrossRef]
- Huang, R.; Xu, X.; Li, D.; Chen, K.; Zhan, Q.; Ge, M.; Zhou, X.; Liang, X.; Guan, M. Digital PCR-Based Detection of EGFR Mutations in Paired Plasma and CSF Samples of Lung Adenocarcinoma Patients with Central Nervous System Metastases. Target. Oncol. 2019, 14, 343–350. [Google Scholar] [CrossRef]
- Reungwetwattana, T.; Nakagawa, K.; Cho, B.C.; Cobo, M.; Cho, E.K.; Bertolini, A.; Bohnet, S.; Zhou, C.; Lee, K.H.; Nogami, N.; et al. CNS Response to Osimertinib Versus Standard Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients with Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 3290–3297. [Google Scholar] [CrossRef]
- Zheng, M.-M.; Li, Y.-S.; Tu, H.-Y.; Jiang, B.-Y.; Yang, J.-J.; Zhou, Q.; Xu, C.-R.; Yang, X.-R.; Wu, Y.-L. Genotyping of Cerebrospinal Fluid Associated with Osimertinib Response and Resistance for Leptomeningeal Metastases in EGFR-Mutated NSCLC. J. Thorac. Oncol. 2021, 16, 250–258. [Google Scholar] [CrossRef]
- Botezatu, I.; Serdyuk, O.; Potapova, G.; Shelepov, V.; Alechina, R.; Molyaka, Y.; Ananév, V.; Bazin, I.; Garin, A.; Narimanov, M.; et al. Genetic Analysis of DNA Excreted in Urine: A New Approach for Detecting Specific Genomic DNA Sequences from Cells Dying in an Organism. Clin. Chem. 2000, 46, 1078–1084. [Google Scholar] [CrossRef]
- Reckamp, K.L.; Melnikova, V.O.; Karlovich, C.; Sequist, L.V.; Camidge, D.R.; Wakelee, H.; Perol, M.; Oxnard, G.R.; Kosco, K.; Croucher, P.; et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2016, 11, 1690–1700. [Google Scholar] [CrossRef]
- Zhang, H.; He, B.; Cui, J.; Zhao, M.; Zhang, Z. Comparison of Circulating DNA from Plasma and Urine for EGFR Mutations in NSCLC Patients. Cancer Biomark. Sect. Dis. Markers 2018, 23, 427–436. [Google Scholar] [CrossRef]
- Hu, T.; Shen, H.; Huang, H.; Song, M.; Yang, Z.; Zhou, Y.; Zhao, G. Urinary Circulating DNA Profiling in Non-Small Cell Lung Cancer Patients Following Treatment Shows Prognostic Potential. J. Thorac. Dis. 2018, 10, 4137–4146. [Google Scholar] [CrossRef]
- Husain, H.; Melnikova, V.O.; Kosco, K.; Woodward, B.; More, S.; Pingle, S.C.; Weihe, E.; Park, B.H.; Tewari, M.; Erlander, M.G.; et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 4716–4723. [Google Scholar] [CrossRef]
- Tchekmedyian, N.; Mudad, R.; Blanco, F.F.; Raymond, V.M.; Garst, J.; Erlander, M.G.; Haura, E.; Berz, D. Longitudinal Monitoring of ctDNA EGFR Mutation Burden from Urine Correlates with Patient Response to EGFR TKIs: A Case Series. Lung Cancer 2017, 108, 22–28. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pesta, M.; Shetti, D.; Kulda, V.; Knizkova, T.; Houfkova, K.; Sharif Bagheri, M.; Svaton, M.; Polivka, J. Applications of Liquid Biopsies in Non-Small-Cell Lung Cancer. Diagnostics 2022, 12, 1799. https://doi.org/10.3390/diagnostics12081799
Pesta M, Shetti D, Kulda V, Knizkova T, Houfkova K, Sharif Bagheri M, Svaton M, Polivka J. Applications of Liquid Biopsies in Non-Small-Cell Lung Cancer. Diagnostics. 2022; 12(8):1799. https://doi.org/10.3390/diagnostics12081799
Chicago/Turabian StylePesta, Martin, Dattatrya Shetti, Vlastimil Kulda, Tereza Knizkova, Katerina Houfkova, Mahyar Sharif Bagheri, Martin Svaton, and Jiri Polivka. 2022. "Applications of Liquid Biopsies in Non-Small-Cell Lung Cancer" Diagnostics 12, no. 8: 1799. https://doi.org/10.3390/diagnostics12081799
APA StylePesta, M., Shetti, D., Kulda, V., Knizkova, T., Houfkova, K., Sharif Bagheri, M., Svaton, M., & Polivka, J. (2022). Applications of Liquid Biopsies in Non-Small-Cell Lung Cancer. Diagnostics, 12(8), 1799. https://doi.org/10.3390/diagnostics12081799






