A Criterion of Colorectal Cancer Diagnosis Using Exosome Fluorescence-Lifetime Imaging
Abstract
:1. Introduction
- Finger rectal examination;
- Irrigoscopy (X-ray examination of the intestine);
- Test for hidden blood in feces;
- Capsule endoscopy (evaluation using an endocapsule with a digital video camera that passes through all parts of the intestine);
- Colonoscopy (endoscopic diagnostic method using a colonoscope with a video camera, if necessary with a biopsy);
- Cancer markers’ blood tests.
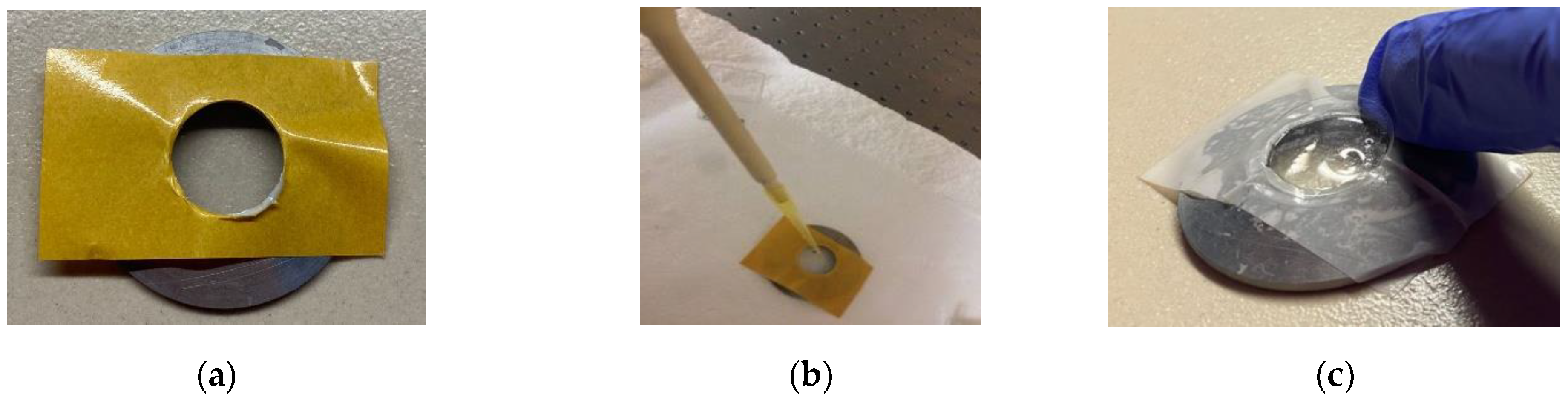
2. Materials and Methods
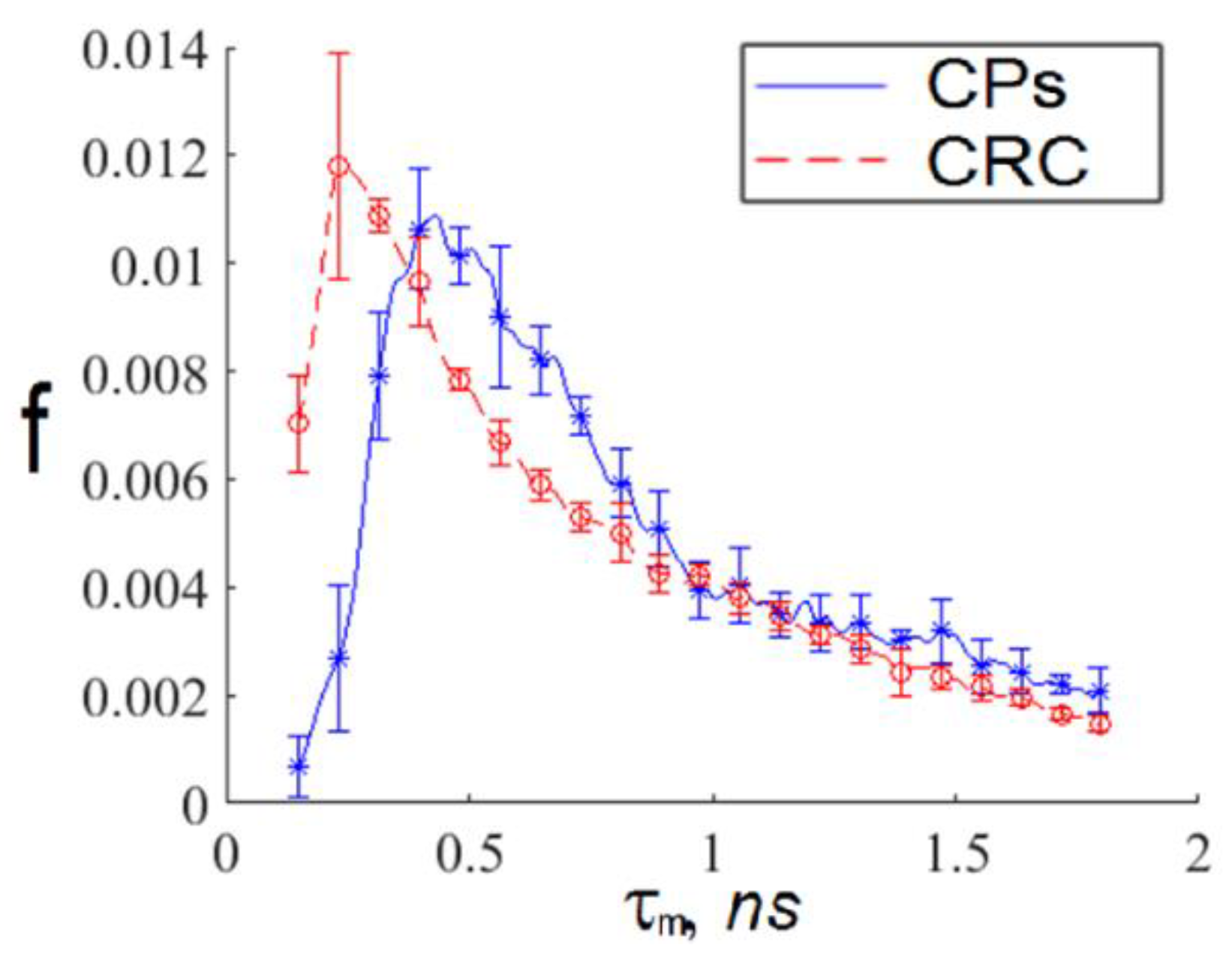
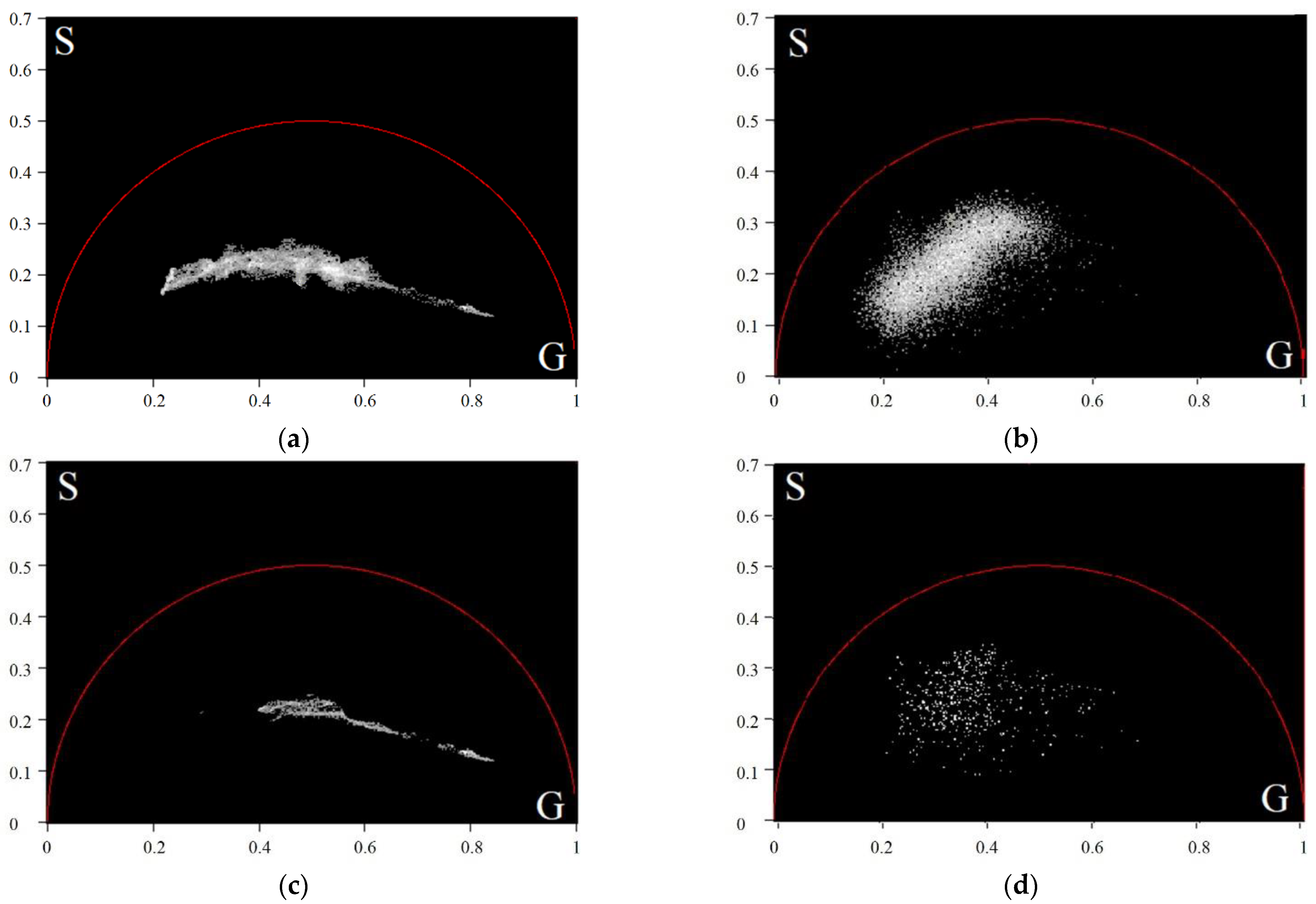
3. Results
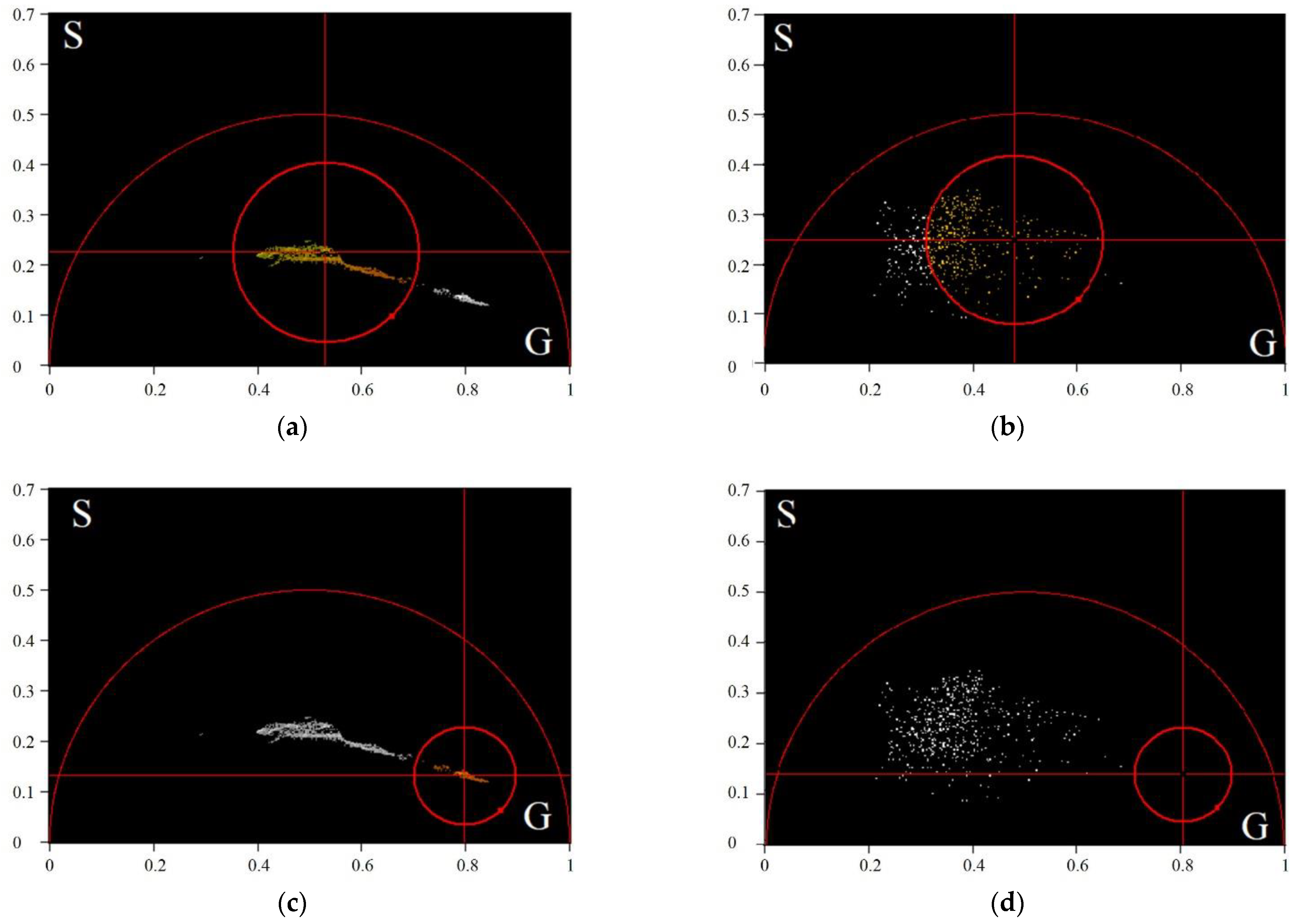
- For every exosome in an FLIM image, we calculated the TPAF average lifetime . If the value fell within the intersection of the intervals corresponding to short and long lifetime interval approximations, then this exosome was ignored. Otherwise, we assigned this exosome the appropriate index: “h”, corresponding to a CP-associated exosome, or “c”, corresponding to a CRC-associated exosome.
- For a whole FLIM dataset corresponding to a definite participant, we calculated the ratio as follows:where is the relative number of CRC-associated exosomes in the FLIM dataset, is the number of CRC-associated exosomes, and is the number of “non-cancerous” (CP-associated) exosomes. If the value is large, it is a sign of possible CRC presence.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterology 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Haggar, F.A.; Boushey, R.P. Colorectal Cancer Epidemiology: Incidence, Mortality, Survival, and Risk Factors. Clin. Colon Rectal Surg. 2009, 22, 191–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebell, M.H.; Thai, T.N.; Royalty, K.J. Cancer screening recommendations: An international comparison of high income countries. Public Health Rev. 2018, 39, 7. [Google Scholar] [CrossRef]
- Blackmore, T.; Norman, K.; Kidd, J.; Cassim, S.; Chepulis, L.; Keenan, R.; Firth, M.; Jackson, C.; Stokes, T.; Weller, D.; et al. Barriers and facilitators to colorectal cancer diagnosis in New Zealand: A qualitative study. BMC Fam. Pract. 2020, 21, 1–206. [Google Scholar] [CrossRef]
- Garborg, K.; Holme, O.; Loberg, M.; Kalager, M.; Adami, H.O.; Bretthauer, M. Current status of screening for colorectal cancer. Ann. Oncol. 2013, 24, 1963–1972. [Google Scholar] [CrossRef]
- Beaulieu, J.-F. Colorectal Cancer. Methods and Protocols, 1st ed.; Humana New York: New York, NY, USA, 2018; pp. 1–353. [Google Scholar]
- Kolligs, F.T. Diagnostics and Epidemiology of Colorectal Cancer. Visc. Med. 2016, 32, 158–164. [Google Scholar] [CrossRef] [Green Version]
- Rao, J.; Wan, X.; Tou, F.; He, Q.; Xiong, A.; Chen, X.; Cui, W.; Zheng, Z. Molecular Characterization of Advanced Colorectal Cancer Using Serum Proteomics and Metabolomics. Front. Mol. Biosci. 2021, 8, 1–11. [Google Scholar] [CrossRef]
- Sharma, S. Tumor markers in clinical practice: General principles and guidelines. Indian J. Med. Paediatr. Oncol. 2009, 30, 1–8. [Google Scholar] [CrossRef] [Green Version]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidu, J.; Chang, B.H.; Kurre, P. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci. Rep. 2015, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yuan, X.; Shi, H.; Wu, L.; Qian, H.; Xu, W. Exosomes in cancer: Small particle, big player. J. Hematol. Oncol. 2015, 8, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Tamkovich, S.N.; Yunusova, N.V.; Stakheeva, M.N.; Somov, A.K.; Frolova, A.Y.; Kirushina, N.A.; Afanasyev, S.G.; Grigoryeva, A.E.; Laktionov, P.P.; Kondakova, I.V. Isolation and characterization of exosomes from blood plasma of breast cancer and colorectal cancer patients. Biomeditsinskaya khimiya 2017, 63, 165–169. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhong, J.; Zhong, B.; Huang, J.; Jiang, L.; Jiang, Y.; Yuan, J.; Sun, J.; Dai, L.; Yang, C.; et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 476, 13–22. [Google Scholar] [CrossRef]
- Lucchetti, D.; Litta, F.; Ratto, C.; Sgambato, A. The role of extracellular vesicles as biomarkers in colorectal cancer. Tech. Coloproctol. 2018, 22, 989–990. [Google Scholar] [CrossRef]
- Borisov, A.V.; Vrazhnov, D.A. Breathomics for Lung Cancer Diagnosis. In Multimodal Optical Diagnostics of Cancer, 1st ed.; Tuchin, V.V., Popp, J., Zakharov, V.P., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2020; Volume 1, Chapter 6; pp. 209–244. [Google Scholar]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef]
- Malloci, M.; Perdomo, L.; Veerasamy, M.; Andriantsitohaina, R.; Simard, G.; Martínez, M.C. Extracellular Vesicles: Mechanisms in Human Health and Disease. Antioxid. Redox Signal. 2018, 30, 813–856. [Google Scholar] [CrossRef]
- Meleppat, R.K.; Ronning, K.E.; Karlen, S.J.; Burns, M.E.; Pugh, E.N.; Zawadzki, R.J. In vivo multimodal retinal imaging of disease-related pigmentary changes in retinal pigment epithelium. Sci. Rep. 2021, 11, 16252. [Google Scholar] [CrossRef] [PubMed]
- Meleppat, R.K.; Prabhathan, P.; Keey, S.-L.; Matham, M.-V. Plasmon Resonant Silica-Coated Silver Nanoplates as Contrast Agents for Optical Coherence Tomography. J. Biomed. Nanotechnol. 2016, 12, 1929–1937. [Google Scholar] [CrossRef] [PubMed]
- Sorrells, J.E.; Martin, E.M.; Aksamitiene, E.; Mukherjee, P.; Alex, A.; Chaney, E.J.; Marjanovic, M.; Boppart, S.A. Label-free characterization of single extracellular vesicles using two-photon fluorescence lifetime imaging microscopy of NAD(P)H. Sci. Rep. 2021, 11, 3308. [Google Scholar] [CrossRef] [PubMed]
- Becker, W. Fluorescence lifetime imaging–techniques and applications. J. Microsc. 2012, 247, 119–136. [Google Scholar] [CrossRef]
- Zhuo, G.-Y.; Kistenev, Y.V.; Kao, F.-J.; Nikolaev, V.V.; Zuhayri, H.; Krivova, N.A.; Mazumder, N. Label-free multimodal nonlinear optical microscopy for biomedical applications. J. Appl. Phys. 2021, 129, 214901. [Google Scholar] [CrossRef]
- Yunusova, N.; Kolegova, E.; Sereda, E.; Kolomiets, L.; Villert, A.; Patysheva, M.; Rekeda, I.; Grigor’eva, A.; Tarabanovskaya, N.; Kondakova, I.; et al. Plasma Exosomes of Patients with Breast and Ovarian Tumors Contain an Inactive 20S Proteasome. Molecules 2021, 26, 6965. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef]
- Pena, A.M.; Decencière, E.; Brizion, S.; Sextius, P.; Koudoro, S.; Baldeweck, T.; Tancrède-Bohin, E. In vivo melanin 3D quantification and z-epidermal distribution by multiphoton FLIM, phasor and Pseudo-FLIM analyses. Sci. Rep. 2022, 12, 1642. [Google Scholar] [CrossRef]
- Zakharova, O.A.; Zhambalova, H.A.; Yunusova, N.I.; Nikolaev, V.V.; Borisov, A.V. Visualization of biological nano-objects with the help of multiphoton microscopy. In Proceedings of the 25th International Symposium on Atmospheric and Ocean Optics: Atmospheric Physics, 112080G, Novosibirsk, Russia, 18 December 2019; SPIE: Bellingham, DC, USA, 2019. [Google Scholar] [CrossRef]
- Datta, R.; Heaster, T.M.; Sharick, J.T.; Gillette, A.A.; Skala, M.C. Fluorescence lifetime imaging microscopy: Fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 2020, 25, 1–44. [Google Scholar] [CrossRef]
- Ma, N.; Mochel, N.R.; Pham, P.D.; Yoo, T.Y.; Cho, K.W.Y.; Digman, M.A. Label-free assessment of pre-implantation embryo quality by the Fluorescence Lifetime Imaging Microscopy (FLIM)-phasor approach. Sci. Rep. 2019, 9, 13206. [Google Scholar] [CrossRef] [Green Version]
- Redford, G.I.; Clegg, R.M. Polar Plot Representation for Frequency-Domain Analysis of Fluorescence Lifetimes. J. Fluoresc. 2005, 15, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Becker, W. Advanced Time-Correlated Single Photon Counting Applications; Springer International Publishing: Cham, Switzerland, 2015; Volume 111, p. 642. [Google Scholar]
- Birnbaum, Z.W. On a use of Mann-Whitney statistics. In Proceeding of the Third Berkley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 26–31 December 1956. [Google Scholar]
- David, A.; Vassilvitskii, S. K-means++: The advantages of careful seeding. In Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms (SODA ‘07), New Orleans Louisiana, LA, USA, 7–9 January 2007; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2007; pp. 1027–1035. [Google Scholar]




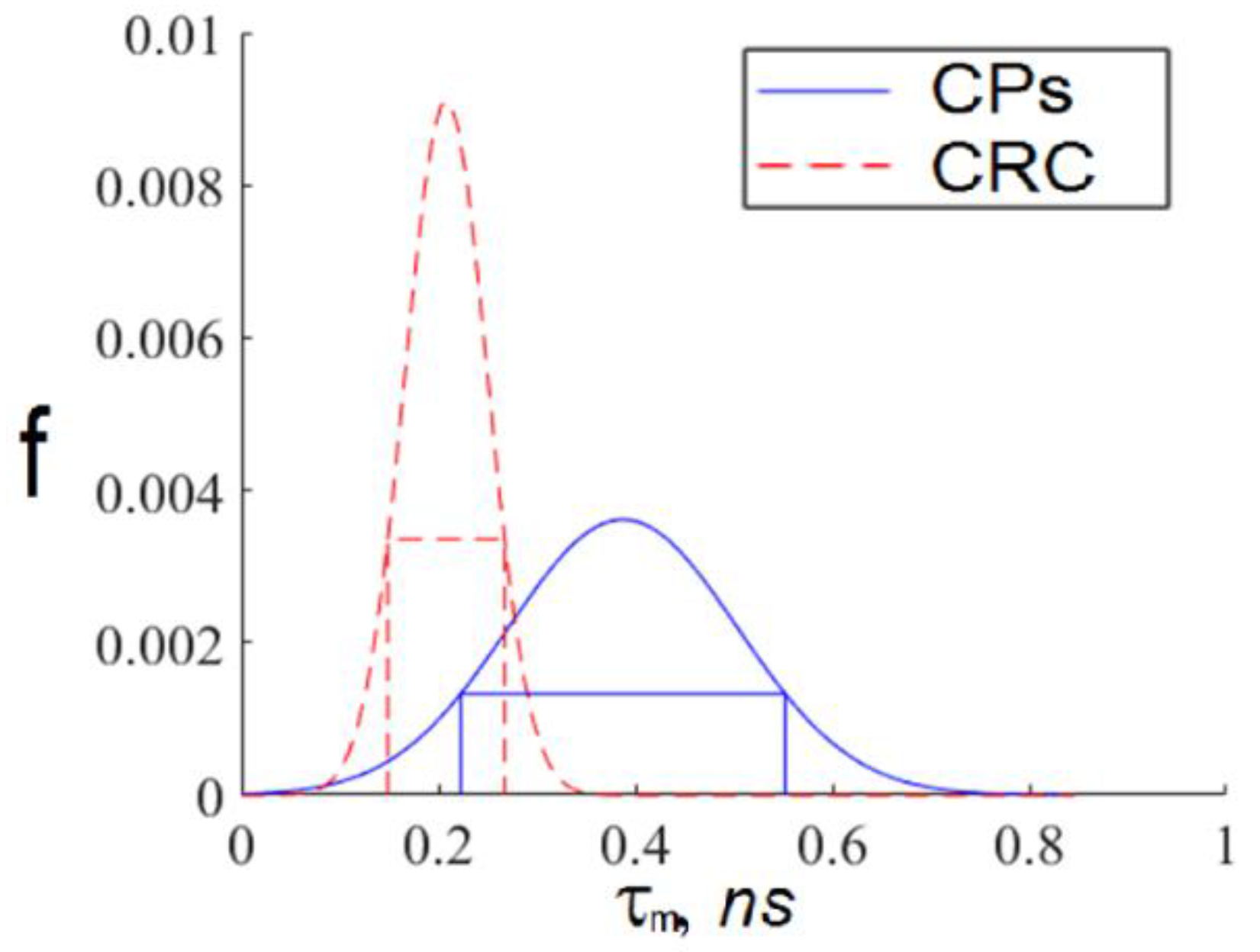

| Mean Value, ns | Standard Deviation, ns | |
|---|---|---|
| The short average TPAF lifetime distribution | 0.21 | 0.06 |
| The long average TPAF lifetime distribution | 0.43 | 0.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borisov, A.V.; Zakharova, O.A.; Samarinova, A.A.; Yunusova, N.V.; Cheremisina, O.V.; Kistenev, Y.V. A Criterion of Colorectal Cancer Diagnosis Using Exosome Fluorescence-Lifetime Imaging. Diagnostics 2022, 12, 1792. https://doi.org/10.3390/diagnostics12081792
Borisov AV, Zakharova OA, Samarinova AA, Yunusova NV, Cheremisina OV, Kistenev YV. A Criterion of Colorectal Cancer Diagnosis Using Exosome Fluorescence-Lifetime Imaging. Diagnostics. 2022; 12(8):1792. https://doi.org/10.3390/diagnostics12081792
Chicago/Turabian StyleBorisov, Alexey V., Olga A. Zakharova, Alisa A. Samarinova, Natalia V. Yunusova, Olga V. Cheremisina, and Yury V. Kistenev. 2022. "A Criterion of Colorectal Cancer Diagnosis Using Exosome Fluorescence-Lifetime Imaging" Diagnostics 12, no. 8: 1792. https://doi.org/10.3390/diagnostics12081792
APA StyleBorisov, A. V., Zakharova, O. A., Samarinova, A. A., Yunusova, N. V., Cheremisina, O. V., & Kistenev, Y. V. (2022). A Criterion of Colorectal Cancer Diagnosis Using Exosome Fluorescence-Lifetime Imaging. Diagnostics, 12(8), 1792. https://doi.org/10.3390/diagnostics12081792







