Proposal for Practical Approach in Prenatal Diagnosis of Beckwith–Wiedemann Syndrome and Review of the Literature
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. DNA Extraction
2.3. Cytogenetic Analysis
2.4. Chromosome Microarray Analysis (CMA)
2.5. Methylation-Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA)
2.6. Literature Review
3. Results
3.1. Patients
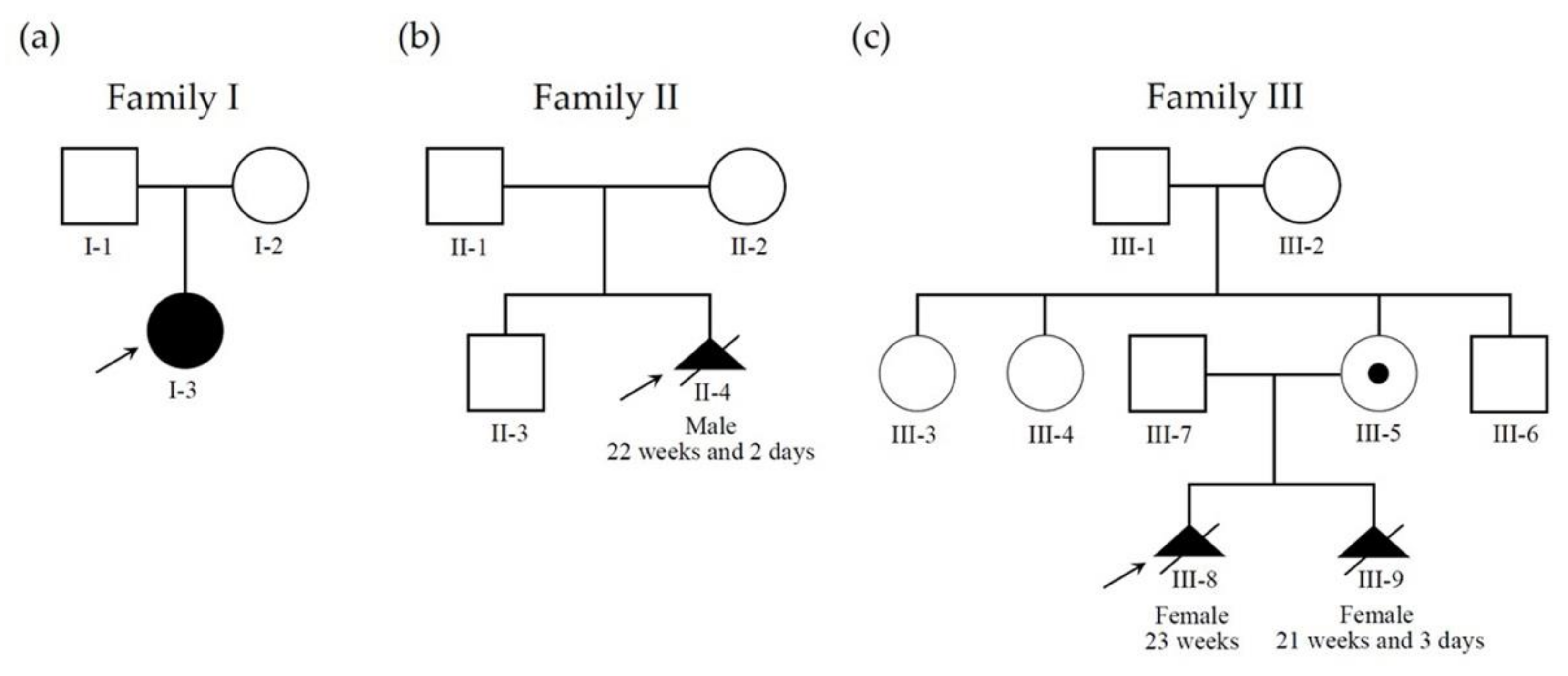
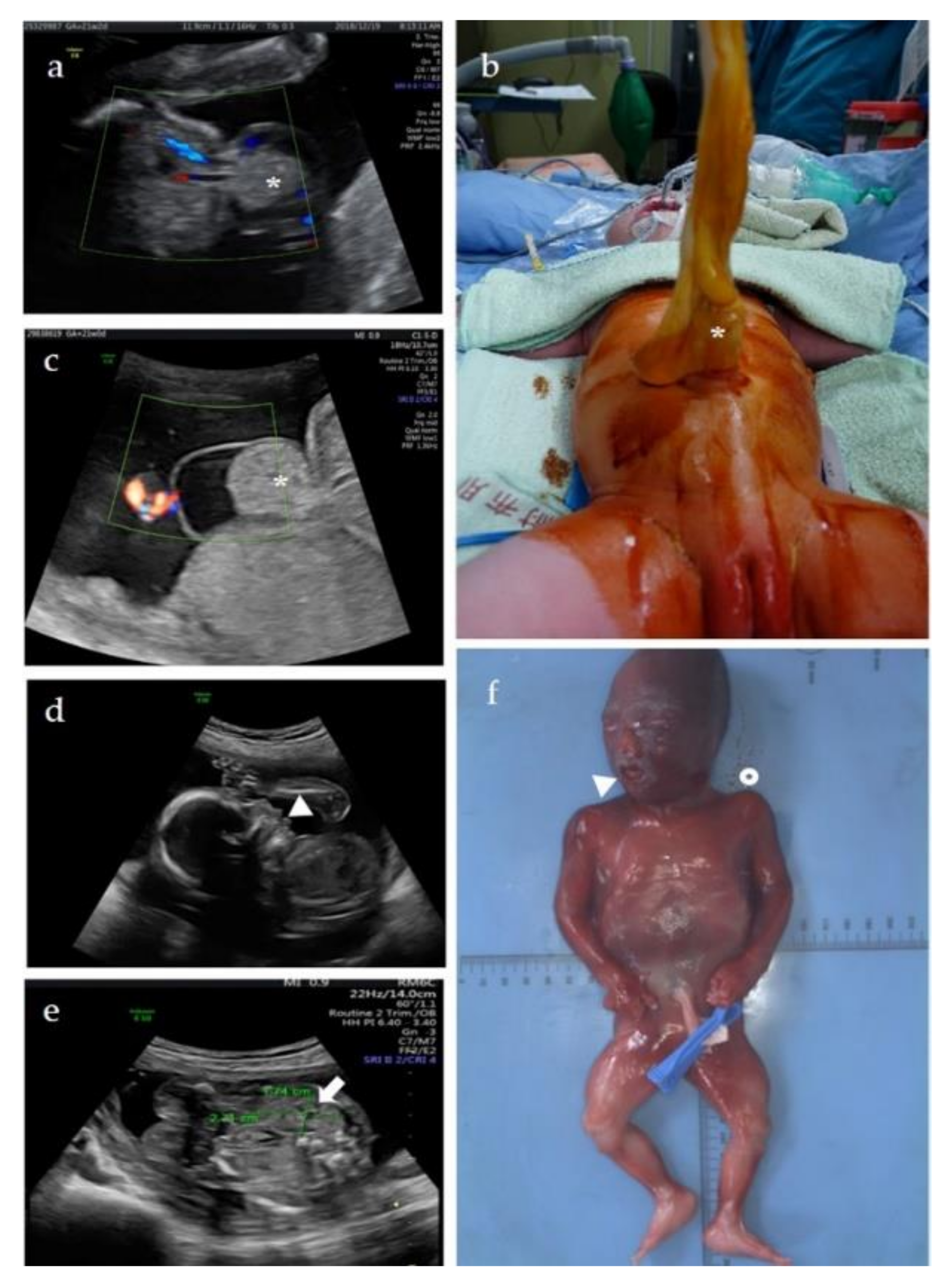
3.1.1. Patient 1
3.1.2. Patient 2
3.1.3. Patient 3
3.2. Literature Review
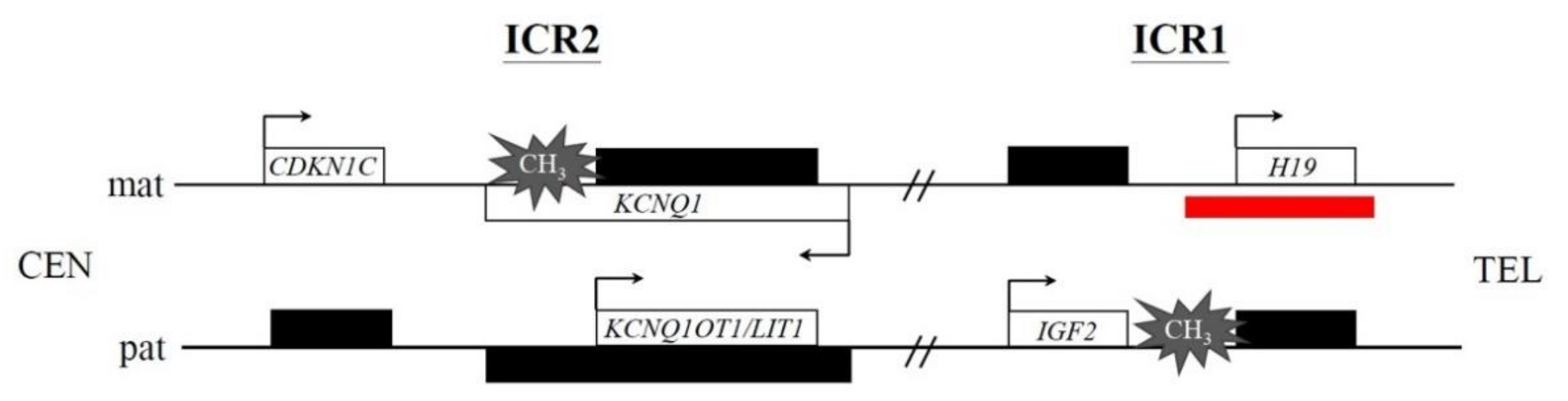
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, D.H.; Gauthier, D.W.; Maizels, M. Prenatal diagnosis of Beckwith-Wiedemann syndrome. Prenat. Diagn. 2005, 25, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Brioude, F.; Kalish, J.M.; Mussa, A.; Foster, A.C.; Bliek, J.; Ferrero, G.B.; Boonen, S.E.; Cole, T.; Baker, R.; Bertoletti, M.; et al. Expert consensus document: Clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: An international consensus statement. Nat. Rev. Endocrinol. 2018, 14, 229–249. [Google Scholar] [CrossRef] [PubMed]
- Mussa, A.; Molinatto, C.; Baldassarre, G.; Riberi, E.; Russo, S.; Larizza, L.; Riccio, A.; Ferrero, G.B. Cancer Risk in Beckwith-Wiedemann Syndrome: A Systematic Review and Meta-Analysis Outlining a Novel (Epi)Genotype Specific Histotype Targeted Screening Protocol. J. Pediatr. 2016, 176, 142–149.e1. [Google Scholar] [CrossRef] [PubMed]
- Mussa, A.; Russo, S.; De Crescenzo, A.; Chiesa, N.; Molinatto, C.; Selicorni, A.; Richiardi, L.; Larizza, L.; Silengo, M.C.; Riccio, A.; et al. Prevalence of Beckwith-Wiedemann syndrome in North West of Italy. Am. J. Med. Genet. A 2013, 161A, 2481–2486. [Google Scholar] [CrossRef] [PubMed]
- Mussa, A.; Molinatto, C.; Cerrato, F.; Palumbo, O.; Carella, M.; Baldassarre, G.; Carli, D.; Peris, C.; Riccio, A.; Ferrero, G.B. Assisted Reproductive Techniques and Risk of Beckwith-Wiedemann Syndrome. Pediatrics 2017, 140, e20164311. [Google Scholar] [CrossRef] [PubMed]
- Lustig, O.; Ariel, I.; Ilan, J.; Lev-Lehman, E.; De-Groot, N.; Hochberg, A. Expression of the imprinted gene H19 in the human fetus. Mol. Reprod. Dev. 1994, 38, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.F.; Guo, N.H.; Zi, F.M.; Cheng, J. Long Noncoding RNA H19 Promotes Tumorigenesis of Multiple Myeloma by Activating BRD4 Signaling by Targeting MicroRNA 152-3p. Mol. Cell Biol. 2020, 40, e00382-19. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Pintar, J.; Efstratiadis, A. Pattern of the insulin-like growth factor II gene expression during early mouse embryogenesis. Development 1990, 110, 151–159. [Google Scholar] [CrossRef]
- Matsuoka, S.; Edwards, M.C.; Bai, C.; Parker, S.; Zhang, P.; Baldini, A.; Harper, J.W.; Elledge, S.J. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995, 9, 650–662. [Google Scholar] [CrossRef]
- Jespersen, T.; Grunnet, M.; Olesen, S.P. The KCNQ1 potassium channel: From gene to physiological function. Physiology 2005, 20, 408–416. [Google Scholar] [CrossRef]
- Mohammad, F.; Mondal, T.; Guseva, N.; Pandey, G.K.; Kanduri, C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010, 137, 2493–2499. [Google Scholar] [CrossRef]
- Niemitz, E.L.; DeBaun, M.R.; Fallon, J.; Murakami, K.; Kugoh, H.; Oshimura, M.; Feinberg, A.P. Microdeletion of LIT1 in familial Beckwith-Wiedemann syndrome. Am. J. Hum. Genet. 2004, 75, 844–849. [Google Scholar] [CrossRef]
- Sparago, A.; Cerrato, F.; Vernucci, M.; Ferrero, G.B.; Silengo, M.C.; Riccio, A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat. Genet. 2004, 36, 958–960. [Google Scholar] [CrossRef]
- Choufani, S.; Shuman, C.; Weksberg, R. Molecular findings in Beckwith-Wiedemann syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163, 131–140. [Google Scholar] [CrossRef]
- Borjas Mendoza, P.A.; Mendez, M.D. Beckwith Wiedemann Syndrome. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Elliott, M.; Bayly, R.; Cole, T.; Temple, I.K.; Maher, E.R. Clinical features and natural history of Beckwith-Wiedemann syndrome: Presentation of 74 new cases. Clin. Genet. 1994, 46, 168–174. [Google Scholar] [CrossRef]
- DeBaun, M.R.; Tucker, M.A. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J. Pediatr. 1998, 132, 398–400. [Google Scholar] [CrossRef]
- Shieh, H.F.; Estroff, J.A.; Barnewolt, C.E.; Zurakowski, D.; Tan, W.H.; Buchmiller, T.L. Prenatal imaging throughout gestation in Beckwith-Wiedemann syndrome. Prenat. Diagn. 2019, 39, 792–795. [Google Scholar] [CrossRef]
- Shuman, C.; Beckwith, J.B.; Weksberg, R. Beckwith-Wiedemann Syndrome. 2000 Mar 3 [Updated 2016 Aug 11]. In GeneReviews® [Internet]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1394/ (accessed on 1 July 2022).
- Van Vuuren, S.H.; Damen-Elias, H.A.; Stigter, R.H.; van der Doef, R.; Goldschmeding, R.; de Jong, T.P.; Westers, P.; Visser, G.H.; Pistorius, L.R. Size and volume charts of fetal kidney, renal pelvis and adrenal gland. Ultrasound Obstet. Gynecol. 2012, 40, 659–664. [Google Scholar] [CrossRef]
- Hamada, H.; Fujiki, Y.; Obata-Yasuoka, M.; Watanabe, H.; Yamada, N.; Kubo, T. Prenatal sonographic diagnosis of Beckwith-Wiedemann syndrome in association with a single umbilical artery. J. Clin. Ultrasound 2001, 29, 535–538. [Google Scholar] [CrossRef]
- Pelizzo, G.; Conoscenti, G.; Kalache, K.D.; Vesce, F.; Guerrini, P.; Cavazzini, L. Antenatal manifestation of congenital pancreatoblastoma in a fetus with Beckwith-Wiedemann syndrome. Prenat. Diagn. 2003, 23, 292–294. [Google Scholar] [CrossRef]
- Le Caignec, C.; Gicquel, C.; Gubler, M.C.; Guyot, C.; You, M.C.; Laurent, A.; Joubert, M.; Winer, N.; David, A.; Rival, J.M. Sonographic findings in Beckwith-Wiedemann syndrome related to H19 hypermethylation. Prenat. Diagn. 2004, 24, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Mulik, V.; Wellesley, D.; Sawdy, R.; Howe, D.T. Unusual prenatal presentation of Beckwith-Wiedemann syndrome. Prenat. Diagn. 2004, 24, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Sinico, M.; Touboul, C.; Haddad, B.; Encha-Razavi, F.; Paniel, J.B.; Gicquel, C.; Gérard-Blanluet, M. Giant omphalocele and "prune belly" sequence as components of the Beckwith-Wiedemann syndrome. Am. J. Med. Genet. Part A 2004, 129A, 198–200. [Google Scholar] [CrossRef] [PubMed]
- Gocmen, R.; Basaran, C.; Karcaaltincaba, M.; Cinar, A.; Yurdakok, M.; Akata, D.; Haliloglu, M. Bilateral hemorrhagic adrenal cysts in an incomplete form of Beckwith-Wiedemann syndrome: MRI and prenatal US findings. Abdom. Imaging 2005, 30, 786–789. [Google Scholar] [CrossRef]
- Aagaard-Tillery, K.M.; Buchbinder, A.; Boente, M.P.; Ramin, K.D. Beckwith-Wiedemann syndrome presenting with an elevated triple screen in the second trimester of pregnancy. Fetal Diagn. Ther. 2007, 22, 18–22. [Google Scholar] [CrossRef]
- Grati, F.R.; Turolla, L.; D’Ajello, P.; Ruggeri, A.; Miozzo, M.; Bracalente, G.; Baldo, D.; Laurino, L.; Boldorini, R.; Frate, E.; et al. Chromosome 11 segmental paternal isodisomy in amniocytes from two fetuses with omphalocoele: New highlights on phenotype-genotype correlations in Beckwith-Wiedemann syndrome. J. Med. Genet. 2007, 44, 257–263. [Google Scholar] [CrossRef]
- Gomes, M.V.; Gomes, C.C.; Pinto, W., Jr.; Ramos, E.S. Methylation pattern at the KvDMR in a child with Beckwith-Wiedemann syndrome conceived by ICSI. Am. J. Med. Genet. A 2007, 143A, 625–629. [Google Scholar] [CrossRef]
- Ma, G.C.; Chang, S.D.; Chang, Y.; Chang, S.P.; Yang, C.W.; Lee, M.J.; Lee, T.H.; Chen, M. Rapid prenatal confirmation of LIT1 hypomethylation using a novel quantitative method (E-Q-PCR) in fetuses with Beckwith-Wiedemann syndrome impressed with ultrasonography. Fertil. Steril. 2008, 90, 1279–1282. [Google Scholar] [CrossRef]
- Percesepe, A.; Bertucci, E.; Ferrari, P.; Lugli, L.; Ferrari, F.; Mazza, V.; Forabosco, A. Familial Beckwith-Wiedemann syndrome due to CDKN1C mutation manifesting with recurring omphalocele. Prenat. Diagn. 2008, 28, 447–449. [Google Scholar] [CrossRef]
- Descartes, M.; Romp, R.; Franklin, J.; Biggio, J.R.; Zehnbauer, B. Constitutional H19 hypermethylation in a patient with isolated cardiac tumor. Am. J. Med. Genet. Part A 2008, 146A, 2126–2129. [Google Scholar] [CrossRef]
- Ramadan, G.I.; Kennea, N.L. Beckwith-Wiedemann syndrome associated with congenital hypothyroidism in a preterm neonate: A case report and literature review. J. Perinatol. 2009, 29, 455–457. [Google Scholar] [CrossRef][Green Version]
- Bui, C.; Picone, O.; Mas, A.E.; Levaillant, J.M.; Ami, O.; Netchine, I.; Frydman, R.; Senat, M.V. Beckwith-Wiedemann syndrome in association with posterior hypoplasia of the cerebellar vermis. Prenat. Diagn. 2009, 29, 906–907. [Google Scholar] [CrossRef]
- Sorrentino, S.; Conte, M.; Nozza, P.; Granata, C.; Capra, V.; Avanzini, S.; Garaventa, A. Simultaneous occurrence of pancreatoblastoma and neuroblastoma in a newborn with Beckwith-Wiedemann syndrome. J. Pediatr. Hematol. Oncol. 2010, 32, e207–e209. [Google Scholar] [CrossRef]
- Storm, D.W.; Hirselj, D.A.; Rink, B.; O’Shaughnessy, R.; Alpert, S.A. The prenatal diagnosis of Beckwith-Wiedemann syndrome using ultrasound and magnetic resonance imaging. Urology 2011, 77, 208–210. [Google Scholar] [CrossRef]
- Aoki, A.; Shiozaki, A.; Sameshima, A.; Higashimoto, K.; Soejima, H.; Saito, S. Beckwith-Wiedemann syndrome with placental chorangioma due to H19-differentially methylated region hypermethylation: A case report. J. Obstet. Gynaecol. Res. 2011, 37, 1872–1876. [Google Scholar] [CrossRef]
- Eckmann-Scholz, C.; Jonat, W. 3-D ultrasound imaging of a prenatally diagnosed Beckwith-Wiedemann syndrome. Arch. Gynecol. Obstet. 2011, 284, 1051–1052. [Google Scholar] [CrossRef]
- Guanciali-Franchi, P.; Di Luzio, L.; Iezzi, I.; Celentano, C.; Matarrelli, B.; Liberati, M.; Palka, G. Elevated maternal serum α-fetoprotein level in a fetus with Beckwith-Wiedemann syndrome in the second trimester of pregnancy. J. Prenat. Med. 2012, 6, 7–9. [Google Scholar]
- Moreira-Pinto, J.; Pereira, J.; Osório, A.; Enes, C.; Mota, C.R. Beckwith-Wiedemann syndrome, delayed abdominal wall closure, and neonatal intussusception--case report and literature review. Fetal. Pediatr. Pathol. 2012, 31, 448–452. [Google Scholar] [CrossRef]
- Longardt, A.C.; Nonnenmacher, A.; Graul-Neumann, L.; Opgen-Rhein, B.; Henrich, W.; Bührer, C.; Hüseman, D. Fetal intracardiac rhabdomyoma in Beckwith-Wiedemann syndrome. J. Clin. Ultrasound 2014, 42, 569–573. [Google Scholar] [CrossRef]
- Chen, C.P.; Su, Y.N.; Chen, S.U.; Chang, T.Y.; Wu, P.C.; Chern, S.R.; Wu, P.S.; Kuo, Y.L.; Wang, W. Prenatal diagnosis of hypomethylation at KvDMR1 and Beckwith-Wiedemann syndrome in a pregnancy conceived by intracytoplasmic sperm injection and in vitro fertilization and embryo transfer. Taiwan. J. Obstet. Gynecol. 2014, 53, 90–94. [Google Scholar] [CrossRef][Green Version]
- Jurkiewicz, D.; Kugaudo, M.; Tańska, A.; Wawrzkiewicz-Witkowska, A.; Tomaszewska, A.; Kucharczyk, M.; Cieślikowska, A.; Ciara, E.; Krajewska-Walasek, M. 11p15 duplication and 13q34 deletion with Beckwith-Wiedemann syndrome and factor VII deficiency. Pediatr. Int. 2015, 57, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.J.; Liu, Y.M.; Li, C.H.; Chang, Y.L.; Chang, S.D. Prenatal diagnosis of paternal duplication of 11p15.5→14.3, Its implication of Beckwith-Wiedemann syndrome. Taiwan. J. Obstet. Gynecol. 2016, 55, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Geng, Q.; Zhou, Q.; Luo, F.; Li, P.; Xie, J. De novo paternal origin duplication of chromosome 11p15.5, report of two Chinese cases with Beckwith-Wiedemann syndrome. Mol. Cytogenet. 2017, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Beaufrère, A.; Bonnière, M.; Tantau, J.; Roth, P.; Schaerer, E.; Brioude, F.; Netchine, I.; Bessières, B.; Gelot, A.; Vekemans, M.; et al. Corpus Callosum Abnormalities and Short Femurs in Beckwith-Wiedemann Syndrome: A Report of Two Fetal Cases. Fetal Pediatr. Pathol. 2018, 37, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Moore, A.; Chiu, P.; Ryan, G.; Weksberg, R.; Shuman, C.; Steele, L.; Chitayat, D. Prenatally diagnosed omphaloceles: Report of 92 cases and association with Beckwith-Wiedemann syndrome. Prenat. Diagn. 2021, 41, 798–816. [Google Scholar] [CrossRef]
- Baker, S.W.; Ryan, E.; Kalish, J.M.; Ganguly, A. Prenatal molecular testing and diagnosis of Beckwith-Wiedemann syndrome. Prenat. Diagn. 2021, 41, 817–822. [Google Scholar] [CrossRef]
- Carli, D.; Bertola, C.; Cardaropoli, S.; Ciuffreda, V.P.; Pieretto, M.; Ferrero, G.B.; Mussa, A. Prenatal features in Beckwith-Wiedemann syndrome and indications for prenatal testing. J. Med. Genet. 2021, 58, 842–849. [Google Scholar] [CrossRef]
- Wang, K.H.; Kupa, J.; Duffy, K.A.; Kalish, J.M. Diagnosis and Management of Beckwith-Wiedemann Syndrome. Front. Pediatr. 2020, 7, 562. [Google Scholar] [CrossRef]
- Queremel Milani, D.A.; Chauhan, P.R. Genetics, Mosaicism. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Moutou, C.; Junien, C.; Henry, I.; Bonaïti-Pellié, C. Beckwith-Wiedemann syndrome: A demonstration of the mechanisms responsible for the excess of transmitting females. J. Med. Genet. 1992, 29, 217–220. [Google Scholar] [CrossRef][Green Version]
- Evans, M.I.; Evans, S.M.; Bennett, T.A.; Wapner, R.J. The price of abandoning diagnostic testing for cell-free DNA screening. Prenat. Diagn. 2018, 38, 243–245. [Google Scholar] [CrossRef]
- Evans, M.I.; Andrioles, S.; Curtis, J.; Evans, S.M.; Kessler, A.A.; Rubenstein, A.F. The epidemic of abnormal copy number variant cases missed because of reliance upon noninvasive prenatal screening. Prenat. Diagn. 2018, 38, 730–734. [Google Scholar] [CrossRef]
- Benn, P.; Cuckle, H.; Pergament, E. Non-invasive prenatal testing for aneuploidy: Current status and future prospects. Ultrasound Obstet. Gynecol. 2013, 42, 15–33. [Google Scholar] [CrossRef]
- Cheng, H.H.; Ma, G.C.; Tsai, C.C.; Wu, W.J.; Lan, K.C.; Hsu, T.Y.; Yang, C.W.; Chen, M. Confined placental mosaicism of double trisomies 9 and 21: Discrepancy between non-invasive prenatal testing, chorionic villus sampling and postnatal confirmation. Ultrasound. Obstet. Gynecol. 2016, 48, 251–253. [Google Scholar] [CrossRef]
- Liao, C.H.; Chang, M.Y.; Ma, G.C.; Chang, S.P.; Lin, C.F.; Lin, W.H.; Chen, H.F.; Chen, S.U.; Lee, Y.C.; Chao, C.C.; et al. Preimplantation Genetic Diagnosis of Neurodegenerative Diseases: Review of Methodologies and Report of Our Experience as a Regional Reference Laboratory. Diagnostics 2019, 9, 44. [Google Scholar] [CrossRef]
- Yang, I.J.; Tu, Y.A.; Pan, S.P.; Huang, T.C.; Chen, C.L.; Lin, M.W.; Tsai, Y.Y.; Yao, Y.L.; Su, Y.N.; Chen, S.U. First report of a successful pregnancy by preimplantation genetic testing for Beckwith-Wiedemann syndrome. Taiwan. J. Obstet. Gynecol. 2022, 61, 174–179. [Google Scholar] [CrossRef]
- Adams, A.D.; Stover, S.; Rac, M.W. Omphalocele-What should we tell the prospective parents? Prenat. Diagn. 2021, 41, 486–496. [Google Scholar] [CrossRef]
- Van den Veyver, I.B. Improving the prenatal diagnosis of Beckwith-Wiedemann syndrome. Prenat. Diagn. 2021, 41, 795–797. [Google Scholar] [CrossRef]
- Gicquel, C.; Rossignol, S.; Le Bouc, Y. Beckwith-Wiedemann Syndrome. Orphanet. Encyclopedia 2005. Available online: http://www.orpha.net/data/patho/GB/uk-BWS05.pdf (accessed on 26 February 2020).
- DeBaun, M.R.; Niemitz, E.L.; McNeil, D.E.; Brandenburg, S.A.; Lee, M.P.; Feinberg, A.P. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am. J. Hum. Genet. 2002, 70, 604–611. [Google Scholar] [CrossRef]
- Rump, P.; Zeegers, M.P.; van Essen, A.J. Tumor risk in Beckwith-Wiedemann syndrome: A review and meta-analysis. Am. J. Med. Genet. Part A 2005, 136, 95–104. [Google Scholar] [CrossRef]
- DeBaun, M.R.; Niemitz, E.L.; Feinberg, A.P. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am. J. Hum. Genet. 2003, 72, 156–160. [Google Scholar] [CrossRef]
- Gicquel, C.; Gaston, V.; Mandelbaum, J.; Siffroi, J.P.; Flahault, A.; Le Bouc, Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am. J. Hum. Genet. 2003, 72, 1338–1341. [Google Scholar] [CrossRef] [PubMed]
- Maher, E.R.; Brueton, L.A.; Bowdin, S.C.; Luharia, A.; Cooper, W.; Cole, T.R.; Macdonald, F.; Sampson, J.R.; Barratt, C.L.; Reik, W.; et al. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART). J. Med. Genet. 2003, 40, 62–64. [Google Scholar] [CrossRef]
- Johnson, J.P.; Beischel, L.; Schwanke, C.; Styren, K.; Crunk, A.; Schoof, J.; Elias, A.F. Overrepresentation of pregnancies conceived by artificial reproductive technology in prenatally identified fetuses with Beckwith-Wiedemann syndrome. J. Assist. Reprod. Genet. 2018, 35, 985–992. [Google Scholar] [CrossRef]
- Halliday, J.; Oke, K.; Breheny, S.; Algar, E.; Amor, D.J. Beckwith-Wiedemann syndrome and IVF: A case-control study. Am. J. Hum. Genet. 2004, 75, 526–528. [Google Scholar] [CrossRef]
- Weksberg, R.; Shuman, C.; Caluseriu, O.; Smith, A.C.; Fei, Y.L.; Nishikawa, J.; Stockley, T.L.; Best, L.; Chitayat, D.; Olney, A.; et al. Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum. Mol. Genet. 2002, 11, 1317–1325. [Google Scholar] [CrossRef]
- Fontana, L.; Bedeschi, M.F.; Cagnoli, G.A.; Costanza, J.; Persico, N.; Gangi, S.; Porro, M.; Ajmone, P.F.; Colapietro, P.; Santaniello, C.; et al. (Epi)genetic profiling of extraembryonic and postnatal tissues from female monozygotic twins discordant for Beckwith-Wiedemann syndrome. Mol. Genet. Genomic. Med. 2020, 8, e1386. [Google Scholar] [CrossRef]
- Cohen, J.L.; Duffy, K.A.; Sajorda, B.J.; Hathaway, E.R.; Gonzalez-Gandolfi, C.X.; Richards-Yutz, J.; Gunter, A.T.; Ganguly, A.; Kaplan, J.; Deardorff, M.A.; et al. Diagnosis and management of the phenotypic spectrum of twins with Beckwith-Wiedemann syndrome. Am. J. Med. Genet. A 2019, 179, 1139–1147. [Google Scholar] [CrossRef]
- Chen, C.H.; Hsieh, H.C.; Tsai, H.D.; Chen, T.H.; Chen, M. Cardiac tamponade: An alternative procedure for late feticide. Taiwan. J. Obstet. Gynecol. 2009, 48, 159–162. [Google Scholar] [CrossRef][Green Version]
- Paganini, L.; Carlessi, N.; Fontana, L.; Silipigni, R.; Motta, S.; Fiori, S.; Guerneri, S.; Lalatta, F.; Cereda, A.; Sirchia, S.; et al. Beckwith-Wiedemann syndrome prenatal diagnosis by methylation analysis in chorionic villi. Epigenetics 2015, 10, 643–649. [Google Scholar] [CrossRef]
- Mussa, A.; Di Candia, S.; Russo, S.; Catania, S.; De Pellegrin, M.; Di Luzio, L.; Ferrari, M.; Tortora, C.; Meazzini, M.C.; Brusati, R.; et al. Recommendations of the Scientific Committee of the Italian Beckwith-Wiedemann Syndrome Association on the diagnosis, management and follow-up of the syndrome. Eur. J. Med. Genet. 2016, 59, 52–64. [Google Scholar] [CrossRef]
| ICR2 Hypomethylation † | ICR1 Hypermethylation † | patUPD11p15.5 † | Others †, ‡ | Total † | |
|---|---|---|---|---|---|
| Number of cases | 99 | 14 | 32 | 21 | 166 |
| Abdominal wall defects (e.g., umbilical hernia/omphalocele) | 57 (57.6%) | 0 (0) | 6 (18.8%) | 8 (38.1%) | 71 (42.8%) |
| Macroglossia | 20 (20.2%) | 6 (42.9%) | 2 (6.3%) | 2 (9.5%) | 30 (18.1%) |
| Macrosomia | 26 (26.3%) | 7 (50.0%) | 11 (34.4%) | 10 (47.6%) | 54 (32.5%) |
| Organomegaly (e.g., nephromegaly, hepatomegaly, cardiomegaly) | 14 (14.1%) | 8 (57.1%) | 6 (18.8%) | 4 (19.3%) | 29 (17.5%) |
| Polyhydramnios | 33 (33.3%) | 7 (50.0%) | 7 (21.9%) | 8 (38.1%) | 55 (33.1%) |
| Placentomegaly | 9 (9.1%) | 0 (0) | 2 (6.3%) | 2 (9.5%) | 13 (7.8%) |
| Corpus callosum anomaly | 1 (1.0%) | 0 (0) | 1 (3.1%) | 0 (0) | 2 (1.2%) |
| Tumor (e.g., placental tumor, macroglossia, intracardiac rhabdomyoma) | 1 (1.0%) | 3 (21.4%) | 1 (3.1%) | 1 (4.8%) | 6 (3.6%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, G.-C.; Chen, T.-H.; Wu, W.-J.; Lee, D.-J.; Lin, W.-H.; Chen, M. Proposal for Practical Approach in Prenatal Diagnosis of Beckwith–Wiedemann Syndrome and Review of the Literature. Diagnostics 2022, 12, 1709. https://doi.org/10.3390/diagnostics12071709
Ma G-C, Chen T-H, Wu W-J, Lee D-J, Lin W-H, Chen M. Proposal for Practical Approach in Prenatal Diagnosis of Beckwith–Wiedemann Syndrome and Review of the Literature. Diagnostics. 2022; 12(7):1709. https://doi.org/10.3390/diagnostics12071709
Chicago/Turabian StyleMa, Gwo-Chin, Tze-Ho Chen, Wan-Ju Wu, Dong-Jay Lee, Wen-Hsiang Lin, and Ming Chen. 2022. "Proposal for Practical Approach in Prenatal Diagnosis of Beckwith–Wiedemann Syndrome and Review of the Literature" Diagnostics 12, no. 7: 1709. https://doi.org/10.3390/diagnostics12071709
APA StyleMa, G.-C., Chen, T.-H., Wu, W.-J., Lee, D.-J., Lin, W.-H., & Chen, M. (2022). Proposal for Practical Approach in Prenatal Diagnosis of Beckwith–Wiedemann Syndrome and Review of the Literature. Diagnostics, 12(7), 1709. https://doi.org/10.3390/diagnostics12071709







