Abstract
Digital droplet PCR (ddPCR) is a recent version of quantitative PCR (QT-PCR), useful for measuring gene expression, doing clonality assays and detecting hot spot mutations. In respect of QT-PCR, ddPCR is more sensitive, does not need any reference curve and can quantify one quarter of samples already defined as “positive but not quantifiable”. In the IgH and TCR clonality assessment, ddPCR recapitulates the allele-specific oligonucleotide PCR (ASO-PCR), being not adapt for detecting clonal evolution, that, on the contrary, does not represent a pitfall for the next generation sequencing (NGS) technique. Differently from NGS, ddPCR is not able to sequence the whole gene, but it is useful, cheaper, and less time-consuming when hot spot mutations are the targets, such as occurs with IDH1, IDH2, NPM1 in acute leukemias or T315I mutation in Philadelphia-positive leukemias or JAK2 in chronic myeloproliferative neoplasms. Further versions of ddPCR, that combine different primers/probes fluorescences and concentrations, allow measuring up to four targets in the same PCR reaction, sparing material, time, and money. ddPCR is also useful for quantitating BCR-ABL1 fusion gene, WT1 expression, donor chimerism, and minimal residual disease, so helping physicians to realize that “patient-tailored therapy” that is the aim of the modern hematology.
Keywords:
digital PCR; quantitative PCR; multiplexing PCR; MRD; clonality; NGS; point mutations; hematology 1. Introduction
Digital Droplet Polymerase Chain Reaction (ddPCR) is a specific, accurate and time-saving technique capables of accurately quantifying gene expression or detecting point mutations applicable in several hematologic disorders, such as leukemias, lymphomas, myeloma, and chronic myeloproliferative neoplasms, and in transplant field. The ddPCR might provide useful informations in prognostic and therapeutic setting.
2. Digital PCR: General Features and Applications
Digital Droplet Polymerase Chain Reaction (ddPCR) technique is a recent “version” of quantitative PCR (QT-PCR), based on the partition of sample into several thousand droplets, so that—at least nominally—one single DNA/cDNA copy is partitioned into a single droplet. After the end-point amplification phase, an appropriate software counts and quantifies the numbers of droplets containing the amplified products, applying the Poisson’s correction (it would be possible that any DNA/cDNA molecule enters a droplet or that two or more DNA/cDNA copies would be co-present in a single droplet) (Figure 1) [1].
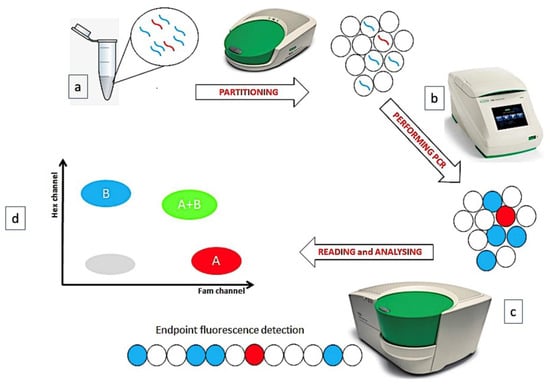
Figure 1.
The phases of ddPCR technique. (a) The sample is partitioned in many thousands of droplets. (b) In each droplet a target is amplified. (c) The endpoint amplification results are analyzed. (d) A plot is generated, reading 2 fluorescence channels.
The first mention of this technique was done by Vogelstein and Kinzler in 1999 [2], but its development effectively occurred after 2012, when instruments with greater than 10,000 partitions per reaction became commercially available, so increasing precision, dynamic range and analytical sensitivity of the new method. Since then, many different applications have been set and published: quantification of donor cell-free DNA in plasma of transplanted patients [3], rapid detection of the most common pathogens in patients with bloodstream infection [4], extracellular RNA [5] or circulating miRNAs quantitation [6].
More recently, ddPCR has been used with success during the Coronavirus pandemics: with a minimum cutoff of 0.04 copies/μL, ddPCR was able to quantify the Coronavirus genome with a sensitivity and specificity of 97.6% and 100%, respectively. Interestingly, in 12 out of 18 patients who converted back to Coronavirus positivity after a negative phase, only ddPCR—and not QT-PCR—still detected viral genome, so reducing the diagnostic error during the recovery phase from the SARS-CoV-2 infection [7]. Moreover, ddPCR accurately quantified Coronavirus genome from crude lysate, with high concordance with measures from purified RNA, thus making more rapid and simpler the viral genome detection [8].
The principal distinctive feature of ddPCR in respect of QT-PCR is that the former does not require a reference standard curve, because the number of “amplified” droplets are divided by the total number of droplets giving an absolute percentage (for example: if 1000 droplets are “positive” among 20,000 total droplets: 1000/20,000 = 0.05%). This is relevant, making possible to quantitate new genes or mutations without need of cloning sequences into “ad hoc” plasmids whose presence in a laboratory significantly increases the probability of environmental contamination. Moreover, the “absolute quantitation” avoids the need of comparing a sample with itself in a different phase of disease, for example before and after a specific treatment, that might overcome the problem of the material consumption.
In terms of sensitivity, ddPCR is at least comparable to QT-PCR, and probably even higher, as shown in several different contexts. In non-Hodgkin’s lymphomas (NHLs) Dr. Drandi and coworkers showed a higher sensitivity for ddPCR (up to one log), especially in samples with very low tumor infiltration [9]. In Waldenstrom’s Macroglobulinemia (WM), it has been shown that ddPCR reached a sensitivity of 5 × 10−5, 1.5 log higher than that offered by the Allele-Specific Oligonucleotide PCR (ASO-PCR), the technique classically used for quantitating the rearrangement of the immunoglobulins heavy chain (IgH). In a series of 148 patients affected by WM, lymphoplasmacytic lymphoma (LPL) or IgM monoclonal gammopathy of undetermined significance (MGUS), 95% of cases showed the MYD88L265P mutation; the concordance with QT-PCR was 74%, and the discordance was always in favor of ddPCR [10]. In patients affected by acute B lymphoblastic leukemia (ALL), ddPCR has been compared to QT-PCR for assessment of minimal residual measurable disease (MRD): rearrangements of IgH or of immunoglobulins light chains (Ig, Ig), in addition to those of T cell receptors (TCRs) have been analyzed and concordant results were observed in 88% of cases, without significant prevalence of one or the two techniques in the discordant cases. On the contrary, 28% of samples defined as “positive but not quantifiable” by QT-PCR resulted quantifiable by ddPCR, so suggesting its higher sensitivity and accuracy [11]. Finally, in multiple myeloma (MM), ddPCR has been shown to have a comparable sensitivity of ASO-PCR in the MRD assessment [12].
Unfortunately, no guidelines about ddPCR setting are today available; nevertheless, the progressive dissemination of this technique in many laboratories prompted the scientific community to produce two useful documents for producing high-quality assays: the ISO 20395:2019 rules (available at the website https://www.iso.org/obp/ui#iso:std:iso:20395:ed-1:v1:en, accessed on 28 February 2022) and another work that summarized the minimum information for publication of ddPCR experiments (dMIQE guidelines) [13]. In this paper, several technical aspects of ddPCR are discussed: amplicons <150 bp are preferred, the fundamental role of perfectly setting annealing temperature and probe concentrations, the pre-amplification step for low-level targets or the dilution step for too concentrated samples. Finally, the rules for adequately placing the threshold that allows distinguishing positive events from the background and the best number of replicates to do. Moreover, a further application of ddPCR also includes the multiplexing ddPCR [13]. In this case, different fluorescent probes are simultaneously detected in different channels or, in the “higher order multiplexing” version, it is possible to evaluate different targets by varying the concentrations of different probes using the same fluorophore. This most recent version of ddPCR can be also performed by combination of more than one fluorophore with different probe concentrations, to detect up to 4 targets within a single reaction [1,14,15] (Figure 2). During the recent pandemics, the multiplexing ddPCR has been used to simultaneously detect the Coronavirus envelope, the viral RNA polymerase and the nucleocapsid genes, so avoiding the possible mismatch of primers and probes which could follow the virus changes that might lead to false negative results [16].
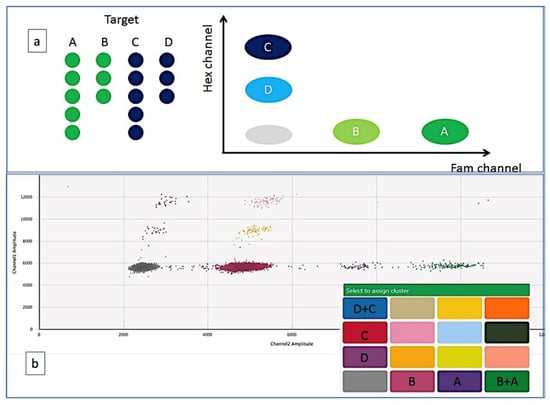
Figure 2.
Multiplex assay based on the amplitude of the amplifiers. (a) the different targets are detected by probes labeled with the same fluorochrome (FAM or HEX) but used at different concentrations. This strategy allows to quantify four targets within a single reaction (A, B, C, D). (b) Targets A and B have relative concentrations of 100% and 50% of FAM-labeled probe, respectively, while C and D have relative concentrations of 100% and 50% of HEX-labeled probe. In the 2D plot, 16 possible clusters are generated: clusters that contain only one target, clusters that simultaneously contain two targets and possibly clusters that contain three targets.
In the non-invasive prenatal testing field, multiplexing ddPCR using universal locked nucleic acid probes correctly identified several fetal aneuploidies [17], while in oncology this technique detected 4 different PIK3CA mutations on “liquid biopsy”, with a clinical impact in the management of metastatic breast cancer [18]. Moreover, an Italian group set a multiplexing ddPCR in patients affected by chronic myeloid leukemia (CML) with unusual BCR-ABL1 atypical transcripts, not quantifiable by standardized QT-PCR, with an optimal detection limit level (0.001%). The output of this technique is relevant, because it allows physicians to offer discontinuation of therapy with tyrosine kinase inhibitors even in cases with persistent deep molecular response whose atypical transcripts are difficult to be measured [19].
At the 2022 national meeting of the Italian Society of Experimental Hematology, our group presented a new kind of “higher order multiplexing ddPCR” able to simultaneously measure the expression of BMI1, EZH2, USP22 and GAPDH genes in 56 patients affected by aggressive B-cell lymphoma (DLBCL). This assay allowed us to analyze very small RNA quantities (the samples were paraffin-embedded and already employed for the cell of origin definition by the Nanostring technology) [20,21].
In conclusion, ddPCR, in its different versions, represents a new, widely applicable, specific, sensitive, and accurate quantitative technique. In the following manuscript, detailed uses of ddPCR in different hematological fields are described.
3. ddPCR Applications in Hematology
3.1. ddPCR in Acute Myeloid Leukemia and Myelodysplasias
Acute myeloid leukemia (AML) represents the prototype of a disease where the “target-therapy” is fundamental for improving patients’ survival [22]. In the recent years, the availability of the anti-CD33 monoclonal antibody gemtuzumab ozogamicin [23], of the FLT3 inhibitors (midostaurin and gilteritinib) [24,25] and of IDH1 and IDH2 inhibitors (ivosidenib and enasidenib) [26,27] significantly changed the therapeutic scenario. The increased probability of therapeutic success and the more defined disease genetic features prompted physicians to revise the WHO classification in 2016 [28] and to better define different prognostic classes (at low-, intermediate-, and poor-risk), with consequent different risk-adapted treatment strategies (chemotherapy only for low-risk patients, transplantation for high-risk cases and for those at intermediate-risk but still MRD-positive after the consolidation phase) [29]. Obviously, also the role of the MRD monitoring became more relevant and better standardized [30].
The acute promyelocytic leukemia (APL) was the first AML subtype where ddPCR played a relevant role: indeed, the positive clinical impact of restarting therapy at the re-appearance of the molecular transcript instead of at the hematological relapse is well known [31]. Consequently, the monitoring of PML-RAR transcript by a very sensitive technique represents a real clinical need. Recently, two Chinese groups set two new ddPCR assays: the first one measured in the same reaction both PML-RARα and ABL1, with a higher sensitivity in respect of QT-PCR [32], and the second one published another ddPCR method able to identify at the same time two types of transcripts: with a Limit of Detection (LOD) of 1 × 10−5, ddPCR recovered the PML-RARα fusion gene in 4% of patients already defined as negative by QT-PCR (whose LOD reached 1 × 10−4) [33].
Another ddPCR has been set for detection of PML-A216V mutation, already known to be responsible for the resistance to arsenic trioxide. Using ddPCR, 5/13 cases were recognized as mutated versus only 3 by Sanger sequencing; in addition, ddPCR anticipated the mutation appearance by 24, 3 and 4 months compared to Sanger sequencing [34].
In other AML types, ddPCR has been used at diagnosis for distinguishing between two “not otherwise specified” (NOS) forms: the expression levels of the ANXA3 and S100A9 genes were increased, whereas those of WT1 were decreased in the AML-M2 (according to the previous FAB classification) in respect of AML-M1. Moreover, STMN1 and ABL1 were upregulated in AMLs with FLT3 mutations, while CAT was over-expressed in the FLT3-wild-type cases [35].
In another work, ddPCR for RUNX1-RUNX1T1 rearrangement was used for MRD assessment in children affected by AML with t(8;21) and compared with flow cytometry and QT-PCR: the flow cytometry lost MRD in 21% of samples (positive by QT-PCR), and ddPCR resulted superior to QT-PCR in one quarter of cases. Finally, in 8 patients with disease progression, the fusion gene was detected by ddPCR but not by QT-PCR before hematological relapse [36].
Another ddPCR assay has been set for detecting and quantitating the NPM1 mutations, including the rarer ones. Indeed, according to the WHO classification [28], NPM1 mutations characterize a specific form of AML, and represent the most informative marker of MRD [29,37,38]. Approximately 95% of NPM1 mutations are represented by nucleotide insertions in exon 12, the most common being type A 75% of cases, and types B and D 15% of the mutated patients. The remaining 10% of patients have rare mutations not covered by the commercially available PCR kits; in this subgroup, ddPCR was able to detect all NPM1 mutation types, with a sensitivity of 1 × 10−4/5 × 10−5 [39], a positive predictive power of 100% and a negative predictive power of 94.5% [40]. ddPCR for NMP1 mutations was also applied to the allogeneic transplantation (alloSCT) setting: with a sensitivity of 0.01% (one log higher than QT-PCR), this technique identified as MRD-positive 33% of patients just before receiving graft. The PCR positivity was the only prognostic factor significantly associated with higher probability of relapse and death [41].
In the complex genomic scenario characterizing AML, ddPCR was also used to track DNMT3A, IDH1 and IDH2 mutations: indeed, one third of patients resulted positive for at least one mutation (DNMT3A > IDH2 > IDH1), even in hematological complete remission (CR). Moreover, among relapsing patients, 78% resulted ddPCR positive 60 days before, while 75% of patients who remained disease-free were persistently unmutated [42].
IDH1 and IDH2 have been reported to be frequently mutated in AML, Myelodysplastic Syndromes (MDS) and chronic Myeloproliferative Neoplasms (MPNs), the most frequent mutations being single-nucleotide variants involving the exon 4 at the arginine hotspot R140 or R172. Many efforts have been done to develop oral IDH1 and IDH2 inhibitors, with conflicting results as a monotherapy [43,44], and more promising results in combination with demethylating agents or intensive chemotherapy [45,46]. Being molecular targets for oral therapies, IDH1/2 have been investigated as markers of MRD, with consistent results when a high-sensibility technique such as ddPCR was used [47], also in post-transplantation setting [42]. A simple and time-sparing method called “drop-off ddPCR” (a multiplexing ddPCR) has been recently described and validated by our group for the simultaneous detection of the IDH2 most common mutations in codon 140 (Figure 3). With this technique, 60 AML patients at diagnosis have been screened for IDH2 mutations by the Sanger sequencing, amplification refractory mutation system (ARMS) PCR or ddPCR. With one log more of sensitivity, ddPCR and ARMS PCR identified IDH2 mutations in 21.6% of cases vs 13.5% of Sanger. Interestingly, ddPCR allowed to identify one of the 4 possible mutations in a single reaction versus the 4 needed reactions for the ARMS PCR. When IDH2 mutations have been monitored during follow-up, they predicted the hematological relapse in two third of cases, so making IDH2 attracting even as MRD marker [48]. c-KIT activating point mutations have been described in solid tumors, but they seem to have a relevant role both in AML, particularly in CFB-AML [49], and in the systemic mastocytosis [50]. Thus, ddPCR has been applied for detection of c-KIT mutations in AML, being associated with a higher relapse rate and poorer outcome [51,52].
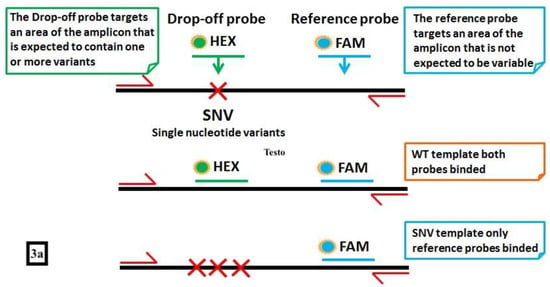
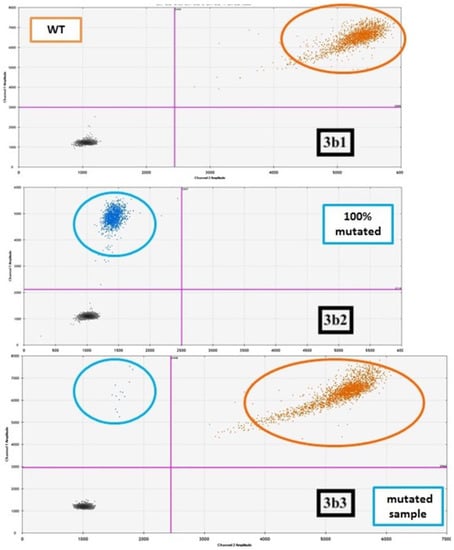
Figure 3.
An example of “drop-off” ddPCR (FAM/HEX Assay for IDH2 mutation). (a) This technique requires a single pair of probes to detect and quantify different mutations in a single reaction: the FAM-labeled probe binds a reference sequence distant from the target but within the same amplicon, while the HEX probe binds the wild-type sequence in the target site. Thus, wild-type samples present signals from both FAM and HEX probes, while the mutated ones display only the FAM signal. (b) In the 2D plot, samples with different IDH2 genotypes are represented, with channel 1 fluorescence (reference probe) plotted against channel 2 fluorescence (wild-type probe). The droplets are arranged according to the fluorescence levels. In (b1), a wild-type (WT) sample represented by a “double positive” population (in orange; reference and wild-type probe in the same droplet) (ref + wt). In (b2), a 100% IDH2-mutated case, where only the reference probe (blue = ref) matched with the IDH2 sequence. In (b3), a sample carrying the mutation in heterozygosity. This panel represents two droplets’ populations, the (with few events) mutated one (blue) (ref) and the (with a higher number of events) double positive (orange) (ref + wt) one.
If the importance of ddPCR as a diagnostic tool is well-recognized, in the recent years it has been used also for MRD assessment: Petterson et al. monitored MRD in 14 AML patients and were able to produce information about clonal/sub-clonal evolution during treatment and different disease phases [53]. Among the possible MRD markers, ddPCR has been applied for the detection of the Wilm’s Tumor 1 (WT1) gene [54]. In a series of 49 AML patients already in deep molecular response by QT-PCR, ddPCR was able to distinguish a subgroup with the best prognosis [55].
A further field of ddPCR application in myeloid malignancies includes the possibility of measuring methylated DNA [56], also analyzing the Alu repeats whose methylation levels are useful for evaluating the global DNA methylation. In the work performed by the Dr. Albano’s group, bone marrow samples from patients receiving azacytidine for intermediate-2/high-risk myelodysplasias were tested by ddPCR before and during treatment: as expected, a significant decrease of Alu sequences methylation after therapy compared to diagnosis was observed, so making ddPCR as an appealing instrument for DNA methylation assessment [57].
3.2. Digital PCR in Acute Lymphoblastic Leukemia
Acute Lymphoblastic Leukemia (ALL) is the most frequent childhood neoplasia with a dismal outcome when diagnosed in adults; in the last few years a better knowledge of B and T-ALL genetic landscape and advanced tools for MRD monitoring allowed to refine indications for alloSCT and ameliorate prognosis [58,59].
MRD monitoring in chromosome Philadelphia-positive (Ph’-positive) B-ALL is based on quantification of BCR-ABL1 transcript using QT-PCR; a recent Italian study applied ddPCR to patients enrolled into the GIMEMA LAL2116 trial, showing optimal sensitivity (1 × 10−5–5 × 10−6) and specificity (near to 100%). In follow-up samples, ddPCR was able to reduce the proportion of positive-not-quantifiable (PNQ) cases, which represent a grey zone in the clinical practice, significantly increasing the proportion of quantifiable samples. Therefore, of the 5 cases that were negative by QT-PCR and positive by ddPCR during follow-up, 4/5 experienced a relapse, confirming the clinical relevance of a deeper MRD monitoring [60]. Similar results were found in 2018 by Dr. Coccaro and colleagues [61] and by Dr. Guan and coworkers who applied ddPCR MRD monitoring in 10 relapsed/refractory Ph’-positive ALL patients treated with anti-CD19/CD22 CAR-T-cell cocktail therapy [62]. Finally, Dr. Martinez and colleagues proposed and validated a one-step ddPCR assay for the p190 BCR-ABL1 transcript that showed several advantages over QT-PCR: deeper sensitivity, no need for a standard curve, no need for standardization material to be shared between different laboratories for result comparison, and less propensity to the reaction inhibition [63].
In Ph’-positive neoplasms, the emergence of point mutations in Tyrosine Kinase Inhibitors (TKIs)-ligand domain of ABL1 may represent a major barrier for success of TKIs, with T315I mutations rendering cells sensible only to the 3rd generation TKI, ponatinib. If the role of ABL1 point mutations is well-recognized in Chronic Myeloid Leukemia (CML), in ALL the prognostic relevance of detecting small clones of T315I-mutated cells at diagnosis remains to be fully elucidated [64]. Nevertheless, in 2020 Dr. Akahoshi et al. reported that the detection of even a small amount of T315I mutation by ddPCR at the time of molecular relapse after autologous transplantation may provide appropriate information for identifying patients who are likely to develop hematological relapse [65].
About other rarer BCR-partner fusion genes, such as t(8;22) BCR/FGFR1, which confer to ALL a particularly aggressive disease course and dismal outcome, a standardized methods for MRD detection is lacking and response monitoring is usually performed with cytogenetic techniques or qualitative PCR [66]. In 2018, Dr. Coccaro et al. reported the absolute quantitation of the BCR/FGFR1 fusion gene by a new ddPCR assay with a LOD of 0.01% [67].
In Ph’-negative ALLs (B or T), which represent most ALLs in childhood, MRD detection is more complicated and based on the Immunoglobulin IgH, Ig, Ig and TCRs rearrangement analysis; nevertheless, a consistent fraction of samples with very-low MRD levels cannot be properly quantified and must be scored as positive not quantifiable (PNQ), that represent a clinical dilemma (are they MRD positive or negative? Is it worth to proceed with transplantation or not?) [68]. Trying to address this issue, Dr. Della Starza and coworkers proposed ddPCR as an alternative method for MRD monitoring [11,69] in samples from patients enrolled in the GIMEMA LAL1308 and in the AIEOP-BFM ALL 2000 trials, finding a concordance rate of 70% between QT-PCR and ddPCR. The greater accuracy of ddPCR allowed to quantitate samples defined as PNQ by QT-PCR in a quarter of cases. To allow a better standardization, the group also proposed a fixed-threshold of positive-droplet number to define a sample as negative, PNQ or positive [69].
Another gene mutated in about 15–20% of pediatric B-ALL and in 50% of adult ALL is the IKZF1 [70]. The presence of IKZF1 or BCR-ABL1 mutations has been reported to be an independent risk factor of poor prognosis [71]. In 2019, Dr. Hashiguchi and Dr. Onozawa described the application of ddPCR for detection and quantification of IKZF1 genomic aberrations, suggesting a possible its role even as MRD marker [72].
3.3. Digital PCR in Lymphoproliferative Disorders (Lymphomas and Multiple Myeloma)
The BRAF V600E mutation interests 70–100% of hairy cell leukemias (HCL) patients [73], but also up to 50% of cases of Langerhans cell histiocytosis, especially those with skin or central nervous system involvement [74]. The clinical translation of this finding is the possibility of administering vemurafenib, already employed in melanoma [75] and in non-small lung cell cancer [76], to relapsed/refractory hematological patients carrying the B-RAF mutation. In a basket study, vemurafenib was administered to 26 patients with Erdheim-Chester disease or Langerhans histiocytosis: the overall response rate (ORR) was 61.5%, with long-term survival [77]. In a series of 30 relapsed/refractory HCL patients, combination of vemurafenib with rituximab offered complete response (CR) to 87% of cases, with 65% of them achieving MRD-negativity [78]. Consequently, the assessment of the BRAFV600E mutation is relevant both from the diagnostic and the therapeutic point of view. In 2016, our group applied an innovative ddPCR assay for detection of BRAF mutation to a series of 47 HCL patients: the new approach was more sensitive than QT-PCR (LOD, 5 × 10−5 vs. 2.5 × 10−4) and when ddPCR was applied as MRD marker, it was able to detect as still MRD-positive 22% of cases otherwise defined as MRD-negative by IgH rearrangement and 5% of cases MRD-negative by QT-PCR [79]. In 2020, a Chinese group organized an inter-laboratory quality control for the use of ddPCR for the BRAF mutation assessment: with a LOD of 0.02%, ddPCR was more sensitive than NGS, whose LOD was 0.3%. About the reproducibility, the 8 participants laboratories demonstrated an appropriate technical competency to perform accurate ddPCR-based measurements, with droplet volume being an important factor influencing the reaction efficiency [80].
In chronic lymphocytic leukemia (CLL), identification of TP53 mutations, as well as of chromosome 17 deletions, that occur in about 5–10% of patients at diagnosis and more frequently at relapse, has a relevant clinical impact, making the chemo-immunotherapy not advisable for this kind of patients [81]. An Italian group proposed an interesting diagnostic workflow algorithm where a ddPCR with 6 probes for TP53 exons 5–7 was used as first screening step. This innovative approach, with a sensitivity of 1 × 10−3, resulted time- and cost-effective in comparison to NGS [82]. Analogously, the chemo-immunotherapy is not the best approach for cases with NOTCH1 mutations (that occur in approximately 10% of CLL patients at diagnosis, 20% at relapse and in over 30% of cases after Richter transformation) [83]. NOTCH1 mutations were detected by ddPCR in 53.4% of patients, and a significant reduction of mutation load was observed after successful treatment (from a median of 11.67% to 0.09%) [84]. Subsequently, another group used ddPCR for assessing NOTCH1 mutations in a larger series of CLL patients: with a LOD of 5 × 10−4, NOTCH1 mutations were detected in 25% of the whole series and in the 55% of patients with trisomy of chromosome 12, with a significant poor prognostic impact [85].
Another field where ddPCR resulted useful for identification of patients who might benefit of Bruton Kinase inhibitors [86] is that of lymphoproliferative disorders characterized by the presence of the MYD88L265P mutation. This genetic abnormality triggers the anti-apoptotic NF-kB pathway, activates the JAK-STAT3 and BTK signals, leading to the uncontrolled B cells proliferation. This mutation characterizes about 30% of Activated Diffuse Large B Cell Lymphomas (ABC-DLBCL), 52% of IgM monoclonal gammopathies (IgM-MGUS), 54% of the cutaneous DLBCL, 70% of primary DLBCL involving the central nervous system, and 90% of MW, while is absent in IgM multiple myeloma (MM) [87]. In 2018, our group contributed to set ddPCR for the identification of the MYD88 mutation; this technique detected mutation in 96% of MW and 87% of IgM-MGUS cases (vs 81% and 58% by QT-PCR, respectively); the concordance rate with QT-PCR amounted to 78% on bone marrow and 68% on peripheral blood samples; in the remaining cases, ddPCR confirmed its advantage. The most interesting finding of this work was the possible application of this molecular tool to the circulating tumor DNA (ctDNA) harvested from plasma [88] or in the cerebral spinal fluid [89]. In 2019, another group employed ddPCR for detecting MYD88L265P mutation in a cohort of 39 patients; with a sensitivity of 1 × 10−3, the authors identified the mutation in 90% of MW cases, in 44% of patients affected by LPL, in 5% of IgM MM, and no in CLL or mantle cell lymphoma (MCL) cases [90]. Another group proposed a new ddPCR assay able to detect and quantify the hot spots mutations of EZH2, STAT6, MYD88, and CCND3 that characterize about 20% of B-cell lymphomas, especially the germinal center DLBCL (GB-DLBCL) and follicular lymphoma (FL) that seem to be associated with resistance to treatment. ddPCR, with a sensitivity of 1 × 10−4, was accurate either on paraffin-embedded samples or on ctDNA (the “liquid biopsy”) [91].
Because BCL2/JH rearrangement can be found only in 60% of FL and the assessment of IgH rearrangement in this lymphoma is often difficult due to its hypermutated status [92], the possibility of assessing different molecular markers is intriguing. Among them, mutations of EZH2 are becoming relevant, even from the clinical point of view, after the recent introduction in the therapeutic armamentarium of the oral EZH2 inhibitor. In patients with relapsed/refractory FL, tazemetostat offered to EZH2-mutated patients 69% of OR and 13% of CR vs. 35% of ORR and 4% of CR to the wild-type subgroup [93]. A ddPCR for detecting EZH2 mutations was set; interestingly, in a patient carrying two different mutations in different tumor sites, the analysis of ctDNA revealed both EZH2 genomic aberrations, so demonstrating the optimal representativeness of liquid biopsy [94]. Even in early-stage FL, ddPCR for BCL2/IgH rearrangement was compared to classical QT-PCR: the concordance between the two techniques amounted to 92%, and the fusion gene was recovered by ddPCR in 18% of cases otherwise negative by QT-PCR [95].
In 2020, the European cooperative group for ddPCR published an interesting manuscript about the employ of ddPCR in 416 samples from 166 patients affected by MCL. Firstly, the authors observed a 90% of concordance rate among the 9 involved laboratories; then, they proposed some rules for performing and analyzing ddPCR reactions, such as starting from 500 ng of DNA, preferring 3 replicates, and considering as “positive” a sample showing at least 3 merged events, as “negative” that without events or with only one merged event, reserving the concept of “grey zone” (PNQ) to samples with two merged events. When ddPCR for IgH clonality and/or BCL1/IgH rearrangement was compared with QT-PCR, GeneScan PCR or flow cytometry, the respective sensitivities reached 1 × 10−5, 1 × 10−4, 5 × 10−2, and 1 × 10−4, showing once again the advantage of ddPCR in term of sensitivity [96,97].
In the T angioimmunoblastic lymphoma, the ddPCR has been proposed for the G17V mutant RHOA (that hyperactivates the TCR signal so prompting the abnormal T cell proliferation); with a LOD of 1 × 10−4, ddPCR was able to recognize mutation in 4 cases that NGS defined as unmutated [98].
In Hodgkin’s lymphoma, ddPCR was used as confirmatory tool of STAT6 mutations on frozen biopsy tissue and ctDNA. NGS showed STAT6 mutations in about 30% of patients, being the most frequent recurrent mutations with those of XPO1 and B2M. With a sensitivity of 0.14%, ddPCR was able to recognize mutations in all cases already tested by NGS and in all cases ddPCR was able to detect mutations also on ctDNA [99].
Dr. Drandi and coworkers compared ddPCR to QT-PCR in a series of 69 patients with FL, MCL and MM: the concordance was good, and both techniques reached the LOD of 1 × 10−5; nevertheless, ddPCR was more accurate, because it was successful in 100% of cases, whereas QT-PCR failed in 4% of cases. This pivotal work clearly sustained the possibility of replacing ASO-PCR for IgH clonality with ddPCR [9].
Focusing on MM, it is incontrovertible that the prognostic value of MRD assessment is becoming a new target of treatment, thanks to the availability of drugs able to induce MRD eradication in up to 70% of patients [100]. A Japanese group recently revised the issue of MRD in autografts from 43 MM patients who underwent autologous stem cell transplantation comparing NGS (with a sensitivity of 1 × 10−7) to ASO-PCR (sensitivity 1 × 10−4/1 × 10−5) and to ddPCR (sensitivity 1 × 10−5). Correlation between ddPCR and ASO-PCR was satisfying (91%), with an advantage for ddPCR, while NGS resulted less performant [101] (Table 1).

Table 1.
ddPCR and lymphoproliferative neoplasms.
3.4. Digital PCR in Chronic Myeloid Leukemia
CML is a chronic myeloproliferative neoplasm characterized by the presence of Philadelphia chromosome (Ph’) and of BCR/ABL1 fusion gene originating from the t(9;22). TKIs (imatinib as first generation, dasatinib, nilotinib and bosutinib as second generation, ponatinib as third generation and asciminib, a new STAMP-inhibitor) are orally available drugs able to inhibit the chimeric protein function so leading to a long-term remission in more than 90% of patients [102]. Nevertheless, about one third of them must change TKI for scarce tolerability or treatment failure. In about 10% of cases, resistance to TKIs is due to point mutations in the kinase domain; among them, T315I confers resistance to all TKIs except for ponatinib and asciminib [103].
The correct management of CML patients is currently based on the serial quantitative molecular assessment of BCR-ABL1/ABL1 ratio, which results fundamental for continuing the same TKI (in patients with optimal response), changing drug (for failing cases) or for more strictly following cases with doubt or not stable response [104]. Nevertheless, in the last 10 years a great opportunity is opened for patients with deep and stable molecular response: the attempt of TKI discontinuation (treatment-free remission or TFR), that has success in about half of cases [105]. Many efforts have been made to correctly identify patients with high probability of TFR to reduce the failure occurrence [104,106]. Among them, it is necessary to correctly identify cases in real deep molecular response (because it is known that patients in less deep response are destined to rapidly fail TFR) [107,108]. In this context, ddPCR demonstrated e good correlation with QT-PCR (99.6%), but even a superiority in terms of LOD and level of quantification (LOQ) [106,109,110,111,112]. In addition to QT-PCR, the reproducibility of results was tested on the two different commercially available platforms: the QX200 Droplet Digital PCR System and the QuantStudio 3D Digital PCR System: the concordance raised to 98.7%, with consistent results [113].
In the ISAV trial, patients with undetectable BCR-ABL1 by ddPCR at time of TKI discontinuation more likely achieved a successful TFR. In that series, ddPCR, with a sensitivity up to 1 × 10−7, showed a significant negative predictive value; when ddPCR levels were combined with age, relapse rates were significantly different (100% for cases <45 years and ddPCR-positive vs 36% for patients >45 years and ddPCR-negative) [114].
Other two groups tempted to find a BCR-ABL1/ABL1 cut off that might predict the TFR success: using ddPCR, an Italian group proposed a cut-off of 0.468 copies/mL [115], while a French group proposed 0.0023% [116]. Notwithstanding the absence of a decisive and reproducible cut off, the North American multicentric prospective “LAST” study clearly confirmed the predictive power of ddPCR: indeed, the molecular recurrence was 10% when ddPCR combined with QT-PCR confirmed the deep molecular response instead of 50% when the deep response was assessed by QT-PCR only [117]. This finding was confirmed by other authors that used ddPCR for accurately identifying patients with deep response or undetectable fusion gene at the time of TKI discontinuation [118].
In addition to the better quantitation of BCR-ABL1 transcript, another promising use of ddPCR seems to be its use for screening BCR-ABL1 mutations. Indeed, it has been recently established that NGS seems to be the best technique for these mutations’ identification: in the “Next in CML” study, the percentage of mutated patients increased from 25% of Sanger to 47% of NGS. Interestingly, in 69 cases NGS allowed to identify the most appropriate TKI; in 10 patients, who resulted unmutated by Sanger, NGS detected the T315I mutation, with the immediate start of ponatinib [119].
In the context of the Italian “Campus CML” working group, 44 samples were screened for T315I by Sanger, NGS and ddPCR: in our hands, the minimum mutational burden detected was 0.02%; with this sensitivity, 25 samples were concordant between ddPCR and Sanger, while 5 cases resulted mutated by ddPCR but not by Sanger. In respect of NGS, 19 samples were concordant; 2 cases, mutated by NGS, resulted wild-type by ddPCR; on the other hand, other 2 cases wild-type by NGS was mutated by ddPCR. The VAF of these cases was 0.43% and 0.39%, values under the sensitivity limit of NGS. One of the 2 failing cases in ddPCR resulted mutated on genomic DNA but not on cDNA. These data, even if preliminary, sustain the possibility of using ddPCR for a rapid screening of T315I, with the immediate therapeutic change [120].
The possibility of employing ddPCR on genomic DNA to identify quiescent leukemia stem cells is another feature distinguishing ddPCR from QT-PCR, as well shown by Dr. Albano and his group [121] and might be worth of further investigation (Table 2).
3.5. Digital PCR in Chronic Myeloproliferative Neoplasms
The chronic myeloproliferative neoplasms (MPNs), including essential thrombocythemia (ET), polycythemia vera (PV) and myelofibrosis (MF), are frequently characterized by the JAK2 mutations [122,123]. Because the presence of these mutations (or, in unmutated cases, of mutations of Calreticulin (CALR) or MPL) is one of the diagnostic criteria [124], it is obvious that ddPCR was firstly set for the screening of JAK2 V617F mutation (that is more common than mutations at exon 12). In 2015, our group published an innovative ddPCR method for identifying and quantitating in a single reaction the JAK2 V617F mutation. In the 99 samples analyzed by both techniques, there was an optimal correlation between QT-PCR and ddPCR, with the latest technique showing half a log higher sensitivity than the former one (5 × 10−4 vs. 1 × 10−3). PV and MF presented a similar median mutation burden (40.45%), higher than that observed in ET (21.35%) [125], differences that we also confirmed by different grades of the spleen stiffness observed by ultrasonography [126,127,128]. Finally, a Korean group compared a ddPCR assay for JAK2 V617F mutation with the results from pyrosequencing, once again showing the superiority of ddPCR [129].
About CALR, it has been reported a ddPCR assay with a sensitivity of 0.01% able to quantitate the type 1 mutation; even in this case, ddPCR was predictive of the clinical outcome [130].

Table 2.
ddPCR and myeloid disorders.
Table 2.
ddPCR and myeloid disorders.
| Disease | Target | Reference |
|---|---|---|
| AML | NPM1 IDH1/IDH2 WT1 PML-RARa PML-A216V C-kit | [32,33,34,39,41,42,48,51,52,55] |
| CML | BCR-ABL1 T315I | [108,109,110,111,112,113,119] |
| MDS | Alu methylation | [57] |
| MPNs | JAK2 CALR | [123,124,127,129] |
3.6. Digital PCR in Transplant and Immunoterapies
Allogeneic hematopoietic stem cell transplantation (AlloSCT) is a potentially curative therapeutic option for several high-risk hematological malignancies (AML, ALL, MDS, lymphomas), especially if performed in CR. After AlloSCT the follow-up is principally based on chimerism and, when possible, on disease specific MRD markers or persistence of mutations: all these strategies allow to promptly detect and treat graft rejection or disease relapse [131]. Nevertheless, the correct timing, samples source—PB or BM—and techniques for chimerism evaluation as well as the exact threshold to distinguish complete donor chimerism from mixed chimerism are still matters of debate [132,133,134].
Currently, the standard methods to measure chimerism are QT-PCR-based analysis of Short Tandem Repeats (STR), with a sensitivity between 5% and 1%, according to the diversity of donor/recipient fingerprint [135]. During the last few years, several studies tried to apply ddPCR to the chimerism assessment, even for levels <1% [136,137]. One of the proposed strategies for children who underwent transplantation for primary immunodeficiency diseases included ddPCR for SRY and RPP30 genes that allowed detect the male/female chimerism. This method revealed accurate and was able to analyze very small amount of genomic material (less than 10 ng) [137]. With a sensitivity of 8 × 10−5, the correlation between STR and ddPCR was higher than 99%, thus supporting the use of ddPCR also for the chimerism assessment [138].
In AlloSCT for malignancies when a suitable MRD marker is available, the better clinical management could be obtained by integrating chimerism analysis with MRD monitoring; Dr. Waterhouse et al. reported a combined use of ddPCR for chimerism and MRD in a series of 70 patients who underwent transplantation, mainly for myeloid malignancies. The authors reported a high concordance between mixed chimerism detection and MRD values, when NPM1, DNMT3A, MLL-PTD, IDH1 and KRAS were monitored [139].
In line with these results, a Japanese group assessed by ddPCR (sensitivity 1 × 10−5) the presence of T315I mutation in 25 patients with Ph’-positive ALL who underwent alloSCT. The hematological relapse was predicted by the persistence/reapperance of the mutation after alloSCT, even at sub-clonal levels (median ratio T315I/ABL1 = 0.91%) [65].
Finally, a ddPCR assay was set for evaluating the immune reconstitution in MM patients after autologous transplantation. Indeed, during TCR rearrangement, excised DNA fragments create the TCR excision circles (TRECs) that have no clear functions but can be used for determining the thymus activity and output [140]. Our group used a new ddPCR for measuring TRECs in 9 patients with MM who underwent autologous transplantation and received high-dose zinc supplementation versus other 9 that did not receive zinc. Interestingly, zinc supplementation supported the immune reconstitution: indeed, TRECs significantly increased from day +30 until day +100 only in the zinc group (6.1-fold vs 1.8 in the control group) [141].
Another innovative field of ddPCR application is represented by the immunotherapy, and in particular by the administration of CAR-T cells to patients affected by CD19+ relapsed/refractory DLBCL, MCL or ALL [142]. Indeed, the shorter or longer persistence of CAR-T seems to be predictive of success [143], whereas it is not still clear the impact of CAR-T persistence on adverse events, such as the cytokines release syndrome (CRS) or immune effector cell-associated neurotoxicity syndrome (ICANS) [144]. In 2020, two different German groups developed new ddPCR assays for monitoring patients receiving CAR-T. In the first work, published by Dr. Fehse and coworkers, starting from 120 ng of DNA the authors reached a sensitivity of 0.01%. Interestingly, the CAR-T expansion above the median peak level of 11.2/mL was correlated with better clinical responses, whereas treatment was less effective in patients for whom CAR-T peaks were below the median [145].
In the paper by Dr. Mika et al. detection and quantification of CAR-T were feasible in all patients, once again with a sensitivity of 1 × 10−4. As expected, significant differences in CAR-T expansion were observed: in 4 patients the initial CAR-T expansion was followed by decreasing numbers of copies; in the other 3, CAR-T were still detectable after 9 months from infusion and the CAR-T persistence and expansion were associated with better clinical responses; in this series, higher levels of CAR-T correlated also with ICANS but not with CRS occurrence [146].
A third group added to the ddPCR for quantitating CAR-T a ddPCR assay for measuring IL-6 gene expression. Differently from that expected, IL-6 gene levels were not predictive for the development of CRS but might be useful for triggering tocilizumab treatment at the first clinical signs of CRS. From the CAR-T expansion and clearance point of view, 4 different patterns have been described by these authors: that of rapid increase and rapid decrease with complete disappearance of CAR-T, that rapid increase and slow decrease with higher persistence, that of rapid increase and rapid decrease with lower persistence, and that of slow increase but rapid decrease with almost disappearance. Interestingly, patients assigned to the category “rapid increase and slow decrease with higher persistence” seemed to have the best response rate, but also a higher risk of CRS, independently from the IL-6 gene expression [147] (Table 3).

Table 3.
ddPCR and immunotherapies.
4. Conclusions
Born about 20 years ago, ddPCR is a new version of QT-PCR, more sensitive, specific, and accurate. With a LOD ranging from 10−4 and 10−5 according to different assays, ddPCR allows to quantitate about one quarter of samples already defined as PNQ by QT-PCR, so making more easily the patients’ management and follow-up. The versatility of this technique makes it available for measuring gene expression (without the need of a standard curve or plasmids), but also for detecting single or multiple point mutations, either on cDNA but also on genomic DNA, both on bone marrow, peripheral blood or liquid biopsy.
As above reported, many are the hematological contexts where ddPCR has been used and implemented: acute leukemias, where it is able to quantitate NPM1 mutations but also WT1 expression; Ph’-positive leukemias, where it is used for measuring more accurately the BCR-ABL1/ABL1 ratio to also identify the patients best candidate to TFR but also for BCR-ABL1 mutations detection; the MPNs, where JAK2 and CALR mutations have a clear diagnostic role, and the lymphoma/myeloma setting, where IgH and TCR clonality can be combined with BCL2/JH and BCL1/JH fusion genes for assessing MRD. Finally, ddPCR can be used for chimerism determination and for monitoring immune reconstitution and CAR-T persistence in patients who receive transplantation or the new immunotherapies (Table 4).

Table 4.
Comparison among different quantitative molecular techniques.
In respect of NGS, ddPCR is more sensitive (at least 2 logs), but also more easily optimizable and fast in producing results. Indeed, we must consider that for reducing NGS costs more than one sample must be loaded into the cartridge, with increased time for results availability. On the other hand, we must keep in mind that ddPCR is not able to recognize all possible nucleotide changes, and that is appliable essentially to few known hot spot mutations. Nevertheless, as in the case of T315I, a rapid identification of a mutation that might induce a rapid therapeutic change might be worth of consideration.
Probably, the best diagnostic algorithm must put together all available sensitive and specific molecular techniques (QT-PCR, ddPCR, NGS) to help physician to do a “reasoned” therapeutic or follow-up approach that would lead to reach the modern goal of hematologists: the “patient-tailored” therapy. In this scenario, we must consider ddPCR as a good allied.
Author Contributions
S.G., S.B. and R.C. wrote the manuscript; M.D.R. and F.G. revised the manuscript, did tables and figures. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are not publicly available due to privacy.
Acknowledgments
Authors thank AIL for support to patients and research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jerič Kokelj, B.; Štalekar, M.; Vencken, S.; Dobnik, D.; Kogovšek, P.; Stanonik, M.; Arnol, M.; Ravnikar, M. Feasibility of Droplet Digital PCR Analysis of Plasma Cell-Free DNA from Kidney Transplant Patients. Front. Med. 2021, 8, 748668. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Tao, Y.; Shao, Z.; Zheng, Y.; Zhang, R.; Yang, X.; Liu, J.; Li, X.; Sun, R. A Comparison of Blood Pathogen Detection Among Droplet Digital PCR, Metagenomic Next-Generation Sequencing, and Blood Culture in Critically Ill Patients with Suspected Bloodstream Infections. Front. Microbiol. 2021, 12, 641202. [Google Scholar] [CrossRef]
- Yan, I.K.; Lohray, R.; Patel, T. Droplet Digital PCR for Quantitation of Extracellular RNA. Methods Mol. Biol. 2018, 1740, 155–162. [Google Scholar] [CrossRef]
- Cirillo, P.D.R.; Margiotti, K.; Mesoraca, A.; Giorlandino, C. Quantification of circulating microRNAs by droplet digital PCR for cancer detection. BMC Res. Notes 2020, 13, 351. [Google Scholar] [CrossRef]
- Liu, C.; Shi, Q.; Peng, M.; Lu, R.; Li, H.; Cai, Y.; Chen, J.; Xu, J.; Shen, B. Evaluation of droplet digital PCR for quantification of SARS-CoV-2 Virus in discharged COVID-19 patients. Aging 2020, 12, 20997–21003. [Google Scholar] [CrossRef]
- Vasudevan, H.N.; Xu, P.; Servellita, V.; Miller, S.; Liu, L.; Gopez, A.; Chiu, C.Y.; Abate, A.R. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci. Rep. 2021, 11, 780. [Google Scholar] [CrossRef]
- Drandi, D.; Kubiczkova-Besse, L.; Ferrero, S.; Dani, N.; Passera, R.; Mantoan, B.; Gambella, M.; Monitillo, L.; Saraci, E.; Ghione, P.; et al. Minimal Residual Disease Detection by Droplet Digital PCR in Multiple Myeloma, Mantle Cell Lymphoma, and Follicular Lymphoma: A Comparison with Real-Time PCR. J. Mol. Diagn. 2015, 17, 652–660. [Google Scholar] [CrossRef]
- Drandi, D.; Genuardi, E.; Dogliotti, I.; Ferrante, M.; Jiménez, C.; Guerrini, F.; Schirico, M.L.; Mantoan, B.; Muccio, V.; Lia, G.; et al. Highly sensitive MYD88L265P mutation detection by droplet digital polymerase chain reaction in Waldenström macroglobulinemia. Haematologica 2018, 103, 1029–1037. [Google Scholar] [CrossRef]
- Della Starza, I.; Nunes, V.; Cavalli, M.; De Novi, L.A.; Ilari, C.; Apicella, V.; Vitale, A.; Testi, A.M.; Del Giudice, I.; Chiaretti, S.; et al. Comparative analysis between RQ-PCR and digital-droplet-PCR of immunoglobulin/T-cell receptor gene rearrangements to monitor minimal residual disease in acute lymphoblastic leukaemia. Br. J. Haematol. 2016, 174, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, H. Comparison of Minimal Residual Disease Detection by Multiparameter Flow Cytometry, ASO-qPCR, Droplet Digital PCR, and Deep Sequencing in Patients with Multiple Myeloma Who Underwent Autologous Stem Cell Transplantation. J. Clin. Med. 2017, 6, 91, Erratum in J. Clin. Med. 2017, 6, 106. [Google Scholar] [CrossRef] [PubMed]
- dMIQE Group; Huggett, J.F. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029, Erratum in Clin. Chem. 2020, 66, 1464. [Google Scholar] [CrossRef] [PubMed]
- Taly, V.; Pekin, D.; Benhaim, L.; Kotsopoulos, S.K.; Le Corre, D.; Li, X.; Atochin, I.; Link, D.R.; Griffiths, A.D.; Pallier, K.; et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 2013, 59, 1722–1731. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.N.; Hughes, S.M.; Roychoudhury, P.; Amstuz, C.; Zhu, H.; Huang, M.L.; Lehman, D.A.; Jerome, K.R.; Hladik, F. HIV reservoir quantification by five-target multiplex droplet digital PCR. STAR Protoc. 2021, 2, 100885. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- de Kock, R.; Baselmans, M.; Scharnhorst, V.; Deiman, B. Sensitive detection and quantification of SARS-CoV-2 by multiplex droplet digital RT-PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 807–813. [Google Scholar] [CrossRef]
- Tan, C.; Chen, X.; Wang, F.; Wang, D.; Cao, Z.; Zhu, X.; Lu, C.; Yang, W.; Gao, N.; Gao, H.; et al. A multiplex droplet digital PCR assay for non-invasive prenatal testing of fetal aneuploidies. Analyst 2019, 144, 2239–2247. [Google Scholar] [CrossRef] [PubMed]
- Corné, J.; Le Du, F.; Quillien, V.; Godey, F.; Robert, L.; Bourien, H.; Brunot, A.; Crouzet, L.; Perrin, C.; Lefeuvre-Plesse, C.; et al. Development of multiplex digital PCR assays for the detection of PIK3CA mutations in the plasma of metastatic breast cancer patients. Sci. Rep. 2021, 11, 17316. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Petiti, J.; Lo Iacono, M.; Dragani, M.; Pironi, L.; Fantino, C.; Rapanotti, M.C.; Quarantelli, F.; Izzo, B.; Divona, M.; Rege-Cambrin, G.; et al. Novel Multiplex Droplet Digital PCR Assays to Monitor Minimal Residual Disease in Chronic Myeloid Leukemia Patients Showing Atypical BCR-ABL1 Transcripts. J. Clin. Med. 2020, 9, 1457. [Google Scholar] [CrossRef]
- Bettelli, S.; Marcheselli, R.; Pozzi, S.; Marcheselli, L.; Papotti, R.; Forti, E.; Cox, M.C.C.; Di Napoli, A.; Tadmor, T.; Mansueto, G.R.; et al. Cell of origin (COO), BCL2/MYC status and IPI define a group of patients with Diffuse Large B-cell Lymphoma (DLBCL) with poor prognosis in a real-world clinical setting. Leuk. Res. 2021, 104, 106552. [Google Scholar] [CrossRef]
- Galimberti, S.; Guerrini, F.; Volpe, G.; Grassi, S.; Ciabatti, E.; Forti, E.; Rapotti, R.; Bettelli, S.; Sacchi, S. A new digital PCR method for measuring the expression value of Polycomb genes in DLBCL. Haematologica 2022, 107, 35. [Google Scholar]
- DiNardo, C.D.; Wei, A.H. How I treat acute myeloid leukemia in the era of new drugs. Blood 2020, 135, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.; Pautas, C.; Terré, C.; Raffoux, E.; Turlure, P.; Caillot, D.; Legrand, O.; Thomas, X.; Gardin, C.; Gogat-Marchant, K.; et al. Gemtuzumab ozogamicin for de novo acute myeloid leukemia: Final efficacy and safety updates from the open-label, phase III ALFA-0701 trial. Haematologica 2019, 104, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.E.; Martinelli, G.; Cortes, J.E.; Neubauer, A.; Berman, E.; Paolini, S.; Montesinos, P.; Baer, M.R.; Larson, R.A.; Ustun, C.; et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3-Mutated AML. N. Engl. J. Med. 2019, 381, 1728–1740. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Stein, A.S.; Stein, E.M.; Fathi, A.T.; Frankfurt, O.; Schuh, A.C.; Döhner, H.; Martinelli, G.; Patel, P.A.; Raffoux, E.; et al. Mutant Isocitrate Dehydrogenase 1 Inhibitor Ivosidenib in Combination with Azacitidine for Newly Diagnosed Acute Myeloid Leukemia. J. Clin. Oncol. 2021, 39, 57–65, Erratum in J. Clin. Oncol. 2021, 39, 341. [Google Scholar] [CrossRef]
- Stein, E.M.; DiNardo, C.D.; Pollyea, D.A.; Fathi, A.T.; Roboz, G.J.; Altman, J.K.; Stone, R.M.; DeAngelo, D.J.; Levine, R.L.; Flinn, I.W.; et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017, 130, 722–731. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Schuurhuis, G.J.; Heuser, M.; Freeman, S.; Béné, M.C.; Buccisano, F.; Cloos, J.; Grimwade, D.; Haferlach, T.; Hills, R.K.; Hourigan, C.S.; et al. Minimal/measurable residual disease in AML: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2018, 131, 1275–1291. [Google Scholar] [CrossRef]
- Sanz, M.A.; Fenaux, P.; Tallman, M.S.; Estey, E.H.; Löwenberg, B.; Naoe, T.; Lengfelder, E.; Döhner, H.; Burnett, A.K.; Chen, S.J.; et al. Management of acute promyelocytic leukemia: Updated recommendations from an expert panel of the European LeukemiaNet. Blood 2019, 133, 1630–1643. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Cui, M.; Yu, S.; Wang, H.; Jing, R. Droplet digital PCR for quantification of PML-RARα in acute promyelocytic leukemia: A comprehensive comparison with real-time PCR. Anal. Bioanal. Chem. 2019, 411, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.W.; Chen, S.Z.; Zhu, X.Y.; Xu, X.X.; Liu, Y. Development and validation of a droplet digital PCR assay for the evaluation of PML-RARα fusion transcripts in acute promyelocytic leukemia. Mol. Cell Probes. 2020, 53, 101617. [Google Scholar] [CrossRef]
- Alfonso, V.; Iaccarino, L.; Ottone, T.; Cicconi, L.; Lavorgna, S.; Divona, M.; Cairoli, R.; Cristiano, A.; Ciardi, C.; Travaglini, S.; et al. Early and sensitive detection of PML-A216V mutation by droplet digital PCR in ATO-resistant acute promyelocytic leukemia. Leukemia 2019, 33, 1527–1530. [Google Scholar] [CrossRef]
- Handschuh, L.; Kaźmierczak, M.; Milewski, M.C.; Góralski, M.; Łuczak, M.; Wojtaszewska, M.; Uszczyńska-Ratajczak, B.; Lewandowski, K.; Komarnicki, M.; Figlerowicz, M. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int. J. Oncol. 2018, 52, 656–678. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zong, S.; Yi, M.; Liu, C.; Wang, B.; Duan, Y.; Cheng, X.; Ruan, M.; Zhang, L.; Zou, Y.; et al. Minimal residual disease monitoring via AML1-ETO breakpoint tracing in childhood acute myeloid leukemia. Transl. Oncol. 2021, 14, 101119. [Google Scholar] [CrossRef] [PubMed]
- Forghieri, F.; Comoli, P.; Marasca, R.; Potenza, L.; Luppi, M. Minimal/Measurable Residual Disease Monitoring in NPM1-Mutated Acute Myeloid Leukemia: A Clinical Viewpoint and Perspectives. Int. J. Mol. Sci. 2018, 19, 3492. [Google Scholar] [CrossRef]
- Gorello, P.; Cazzaniga, G.; Alberti, F.; Dell’Oro, M.G.; Gottardi, E.; Specchia, G.; Roti, G.; Rosati, R.; Martelli, M.F.; Diverio, D.; et al. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia 2006, 20, 1103–1108. [Google Scholar] [CrossRef]
- Mencia-Trinchant, N.; Hu, Y.; Alas, M.A.; Ali, F.; Wouters, B.J.; Lee, S.; Ritchie, E.K.; Desai, P.; Guzman, M.L.; Roboz, G.J.; et al. Minimal Residual Disease Monitoring of Acute Myeloid Leukemia by Massively Multiplex Digital PCR in Patients with NPM1 Mutations. J. Mol. Diagn. 2017, 19, 537–548. [Google Scholar] [CrossRef]
- Pettersson, L.; Johansson Alm, S.; Almstedt, A.; Chen, Y.; Orrsjö, G.; Shah-Barkhordar, G.; Zhou, L.; Kotarsky, H.; Vidovic, K.; Asp, J.; et al. Comparison of RNA- and DNA-based methods for measurable residual disease analysis in NPM1-mutated acute myeloid leukemia. Int. J. Lab. Hematol. 2021, 43, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Bill, M.; Grimm, J.; Jentzsch, M.; Kloss, L.; Goldmann, K.; Schulz, J.; Beinicke, S.; Häntschel, J.; Cross, M.; Vucinic, V.; et al. Digital droplet PCR-based absolute quantification of pre-transplant NPM1 mutation burden predicts relapse in acute myeloid leukemia patients. Ann. Hematol. 2018, 97, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Brambati, C.; Galbiati, S.; Xue, E.; Toffalori, C.; Crucitti, L.; Greco, R.; Sala, E.; Crippa, A.; Chiesa, L.; Soriani, N.; et al. Droplet digital polymerase chain reaction for DNMT3A and IDH1/2 mutations to improve early detection of acute myeloid leukemia relapse after allogeneic hematopoietic stem cell transplantation. Haematologica 2016, 101, e157–e161. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gong, Y. Isocitrate dehydrogenase inhibitors in acute myeloid leukemia. Biomark. Res. 2019, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- McMurry, H.; Fletcher, L.; Traer, E. IDH Inhibitors in AML-Promise and Pitfalls. Curr. Hematol. Malig. Rep. 2021, 16, 207–217. [Google Scholar] [CrossRef]
- Venugopal, S.; Takahashi, K.; Daver, N.; Maiti, A.; Borthakur, G.; Loghavi, S.; Short, N.J.; Ohanian, M.; Masarova, L.; Issa, G.; et al. Efficacy and safety of enasidenib and azacitidine combination in patients with IDH2 mutated acute myeloid leukemia and not eligible for intensive chemotherapy. Blood Cancer J. 2022, 12, 10. [Google Scholar] [CrossRef]
- Stein, E.M.; DiNardo, C.D.; Fathi, A.T.; Mims, A.S.; Pratz, K.W.; Savona, M.R.; Stein, A.S.; Stone, R.M.; Winer, E.S.; Seet, C.S.; et al. Ivosidenib or enasidenib combined with intensive chemotherapy in patients with newly diagnosed AML: A phase 1 study. Blood 2021, 137, 1792–1803. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Petrova, L.; Vrbacky, F.; Lanska, M.; Zavrelova, A.; Zak, P.; Hrochova, K. IDH1 and IDH2 mutations in patients with acute myeloid leukemia: Suitable targets for minimal residual disease monitoring? Clin. Biochem. 2018, 61, 34–39, Erratum in Clin Biochem. 2019, 63, 161. [Google Scholar] [CrossRef] [PubMed]
- Grassi, S.; Guerrini, F.; Ciabatti, E.; Puccetti, R.; Salehzadeh, S.; Metelli, M.R.; Di Vita, A.; Domenichini, C.; Caracciolo, F.; Orciuolo, E.; et al. Digital Droplet PCR is a Specific and Sensitive Tool for Detecting IDH2 Mutations in Acute Myeloid LeuKemia Patients. Cancers 2020, 12, 1738. [Google Scholar] [CrossRef]
- Ayatollahi, H.; Shajiei, A.; Sadeghian, M.H.; Sheikhi, M.; Yazdandoust, E.; Ghazanfarpour, M.; Shams, S.F.; Shakeri, S. Prognostic Importance of C-KIT Mutations in Core Binding Factor Acute Myeloid Leukemia: A Systematic Review. Hematol. Oncol. Stem Cell Ther. 2017, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pardanani, A. Systemic mastocytosis in adults: 2021 Update on diagnosis, risk stratification and management. Am. J. Hematol. 2021, 96, 508–525. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, Z.; Wang, W.; Zhu, G.; Guo, J.; Chen, X.; Zheng, C.; Xu, Z.; Chang, J.; Ren, F.; et al. Monitoring of clonal evolution of double C-KIT exon 17 mutations by Droplet Digital PCR in patients with core-binding factor acute myeloid leukemia. Leuk. Res. 2018, 69, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Tsujimoto, S.; Miyake, M.; Uchiyama, Y.; Ikeda, J.; Yoshitomi, M.; Shimosato, Y.; Tokumasu, M.; Matsuo, H.; Yoshida, K.; et al. Droplet digital polymerase chain reaction assay for the detection of the minor clone of KIT D816V in paediatric acute myeloid leukaemia especially showing RUNX1-RUNX1T1 transcripts. Br. J. Haematol. 2021, 194, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, L.; Chen, Y.; George, A.M.; Rigo, R.; Lazarevic, V.; Juliusson, G.; Saal, L.H.; Ehinger, M. Subclonal patterns in follow-up of acute myeloid leukemia combining whole exome sequencing and ultrasensitive IBSAFE digital droplet analysis. Leuk. Lymphoma. 2020, 61, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Bussaglia, E.; Pratcorona, M.; Carricondo, M.; Sansegundo, L.; Rubio, M.A.; Monter, A.; Brell, A.; Badell, I.; Esteve, J.; Arnan, M.; et al. Application of a digital PCR method for WT1 to myeloid neoplasms in CR and deep ELN WT1 molecular response (<10 copies). Ann. Hematol. 2020, 99, 765–772. [Google Scholar] [CrossRef]
- Koizumi, Y.; Furuya, D.; Endo, T.; Asanuma, K.; Yanagihara, N.; Takahashi, S. Quantification of Wilms’ tumor 1 mRNA by digital polymerase chain reaction. Int. J. Hematol. 2018, 107, 230–234. [Google Scholar] [CrossRef]
- Yu, M.; Heinzerling, T.J.; Grady, W.M. DNA Methylation Analysis Using Droplet Digital PCR. Methods Mol. Biol. 2018, 1768, 363–383. [Google Scholar] [CrossRef]
- Orsini, P.; Impera, L.; Parciante, E.; Cumbo, C.; Minervini, C.F.; Minervini, A.; Zagaria, A.; Anelli, L.; Coccaro, N.; Casieri, P.; et al. Droplet digital PCR for the quantification of Alu methylation status in hematological malignancies. Diagn. Pathol. 2018, 13, 98. [Google Scholar] [CrossRef]
- Marks, D.I.; Rowntree, C. Management of adults with T-cell lymphoblastic leukemia. Blood 2017, 129, 1134–1142, Erratum in Blood 2017, 129, 2204. [Google Scholar] [CrossRef]
- Hein, K.; Short, N.; Jabbour, E.; Yilmaz, M. Clinical Value of Measurable Residual Disease in Acute Lymphoblastic Leukemia. Blood Lymphat. Cancer 2022, 12, 7–16. [Google Scholar] [CrossRef]
- Ansuinelli, M.; Della Starza, I.; Lauretti, A.; Elia, L.; Siravo, V.; Messina, M.; De Novi, L.A.; Taherinasab, A.; Canichella, M.; Guarini, A.; et al. Applicability of droplet digital polymerase chain reaction for minimal residual disease monitoring in Philadelphia-positive acute lymphoblastic leukaemia. Hematol. Oncol. 2021, 39, 680–686. [Google Scholar] [CrossRef]
- Coccaro, N.; Anelli, L.; Zagaria, A.; Casieri, P.; Tota, G.; Orsini, P.; Impera, L.; Minervini, A.; Minervini, C.F.; Cumbo, C.; et al. Droplet Digital PCR Is a Robust Tool for Monitoring Minimal Residual Disease in Adult Philadelphia-Positive Acute Lymphoblastic Leukemia. J. Mol. Diagn. 2018, 20, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zhang, M.; Zhang, W.; Wang, J.; Shen, K.; Zhang, K.; Yang, L.; Huang, L.; Wang, N.; Xiao, M.; et al. Clinical Utility of Droplet Digital PCR to Monitor BCR-ABL1 Transcripts of Patients with Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia Post-chimeric Antigen Receptor19/22 T-Cell Cocktail Therapy. Front. Oncol. 2021, 11, 646499. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.J.; Kang, Q.; Nennig, D.; Bailey, N.G.; Brown, N.A.; Betz, B.L.; Tewari, M.; Thyagarajan, B.; Bachanova, V.; Mroz, P. One-Step Multiplexed Droplet Digital Polymerase Chain Reaction for Quantification of p190 BCR-ABL1 Fusion Transcript in B-Lymphoblastic Leukemia. Arch. Pathol. Lab. Med. 2022, 146, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Vitale, A.; Poerio, A.; Gnani, A.; Colarossi, S.; Iacobucci, I.; Cimino, G.; Elia, L.; Lonetti, A.; Vignetti, M.; et al. Philadelphia-positive acute lymphoblastic leukemia patients already harbor BCR-ABL kinase domain mutations at low levels at the time of diagnosis. Haematologica 2011, 96, 552–557. [Google Scholar] [CrossRef]
- Akahoshi, Y.; Nakasone, H.; Kawamura, K.; Kusuda, M.; Kawamura, S.; Takeshita, J.; Yoshino, N.; Misaki, Y.; Yoshimura, K.; Gomyo, A.; et al. Detection of T315I using digital polymerase chain reaction in allogeneic transplant recipients with Ph-positive acute lymphoblastic anemia in the dasatinib era. Exp. Hematol. 2020, 81, 60–67, Erratum in Exp. Hematol. 2020, 85, 71. [Google Scholar] [CrossRef]
- Wang, W.; Tang, G.; Kadia, T.; Lu, X.; Li, Y.; Huang, L.; Montenegro-Garreaud, X.; Miranda, R.N.; Wang, S.A. Cytogenetic Evolution Associated With Disease Progression in Hematopoietic Neoplasms With t(8;22)(p11;q11)/BCR-FGFR1 Rearrangement. J. Natl. Compr. Cancer Netw. 2016, 14, 708–711. [Google Scholar] [CrossRef]
- Coccaro, N.; Tota, G.; Zagaria, A.; Anelli, L.; Casieri, P.; Impera, L.; Minervini, A.; Minervini, C.F.; Orsini, P.; Cumbo, C.; et al. Monitoring minimal residual disease by ddPCR in acute lymphoblastic leukemia associated with the FGFR1 gene rearrangement. Int. J. Lab. Hematol. 2018, 40, e117–e120. [Google Scholar] [CrossRef]
- van der Velden, V.H.; Cazzaniga, G.; Schrauder, A.; Hancock, J.; Bader, P.; Panzer-Grumayer, E.R.; Flohr, T.; Sutton, R.; Cave, H.; Madsen, H.O.; et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: Guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007, 21, 604–611. [Google Scholar] [CrossRef]
- Della Starza, I.; Nunes, V.; Lovisa, F.; Silvestri, D.; Cavalli, M.; Garofalo, A.; Campeggio, M.; De Novi, L.A.; Soscia, R.; Oggioni, C.; et al. Droplet Digital PCR Improves IG-/TR-based MRD Risk Definition in Childhood B-cell Precursor Acute Lymphoblastic Leukemia. Hemasphere 2021, 5, e543. [Google Scholar] [CrossRef]
- Medeiros, B.C. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 2009, 360, 1787. [Google Scholar] [CrossRef]
- Martinelli, G.; Iacobucci, I.; Storlazzi, C.T.; Vignetti, M.; Paoloni, F.; Cilloni, D.; Soverini, S.; Vitale, A.; Chiaretti, S.; Cimino, G.; et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: A GIMEMA AL WP report. J. Clin. Oncol. 2009, 27, 5202–5207. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, J.; Onozawa, M.; Okada, K.; Amano, T.; Hatanaka, K.C.; Nishihara, H.; Sato, N.; Teshima, T. Quantitative detection of IKZF1 deletion by digital PCR in patients with acute lymphoblastic leukemia. Int. J. Lab. Hematol. 2019, 41, e38–e40. [Google Scholar] [CrossRef] [PubMed]
- Maitre, E.; Cornet, E.; Troussard, X. Hairy cell leukemia: 2020 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019, 94, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Galindo, C.; Allen, C.E. Langerhans cell histiocytosis. Blood 2020, 135, 1319–1331. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef]
- Mazieres, J.; Cropet, C.; Montané, L.; Barlesi, F.; Souquet, P.J.; Quantin, X.; Dubos-Arvis, C.; Otto, J.; Favier, L.; Avrillon, V.; et al. Vemurafenib in non-small-cell lung cancer patients with BRAFV600 and BRAFnonV600 mutations. Ann. Oncol. 2020, 31, 289–294. [Google Scholar] [CrossRef]
- Diamond, E.L.; Subbiah, V.; Lockhart, A.C.; Blay, J.Y.; Puzanov, I.; Chau, I.; Raje, N.S.; Wolf, J.; Erinjeri, J.P.; Torrisi, J.; et al. Vemurafenib for BRAF V600-Mutant Erdheim-Chester Disease and Langerhans Cell Histiocytosis: Analysis of Data from the Histology-Independent, Phase 2, Open-label VE-BASKET Study. JAMA Oncol. 2018, 4, 384–388, Erratum in JAMA Oncol. 2019, 5, 122. [Google Scholar] [CrossRef]
- Tiacci, E.; De Carolis, L.; Simonetti, E.; Capponi, M.; Ambrosetti, A.; Lucia, E.; Antolino, A.; Pulsoni, A.; Ferrari, S.; Zinzani, P.L.; et al. Vemurafenib plus Rituximab in Refractory or Relapsed Hairy-Cell Leukemia. N. Engl. J. Med. 2021, 384, 1810–1823. [Google Scholar] [CrossRef]
- Guerrini, F.; Paolicchi, M.; Ghio, F.; Ciabatti, E.; Grassi, S.; Salehzadeh, S.; Ercolano, G.; Metelli, M.R.; Del Re, M.; Iovino, L.; et al. The Droplet Digital PCR: A New Valid Molecular Approach for the Assessment of B-RAF V600E Mutation in Hairy Cell Leukemia. Front. Pharmacol. 2016, 7, 363. [Google Scholar] [CrossRef]
- Dong, L.; Wang, X.; Wang, S.; Du, M.; Niu, C.; Yang, J.; Li, L.; Zhang, G.; Fu, B.; Gao, Y.; et al. Interlaboratory assessment of droplet digital PCR for quantification of BRAF V600E mutation using a novel DNA reference material. Talanta 2020, 207, 120293. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef] [PubMed]
- Frazzi, R.; Bizzarri, V.; Albertazzi, L.; Cusenza, V.Y.; Coppolecchia, L.; Luminari, S.; Ilariucci, F. Droplet digital PCR is a sensitive tool for the detection of TP53 deletions and point mutations in chronic lymphocytic leukaemia. Br. J. Haematol. 2020, 189, e49–e52. [Google Scholar] [CrossRef] [PubMed]
- Stilgenbauer, S.; Schnaiter, A.; Paschka, P.; Zenz, T.; Rossi, M.; Döhner, K.; Bühler, A.; Böttcher, S.; Ritgen, M.; Kneba, M.; et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: Results from the CLL8 trial. Blood 2014, 123, 3247–3254. [Google Scholar] [CrossRef] [PubMed]
- Minervini, A.; Francesco Minervini, C.; Anelli, L.; Zagaria, A.; Casieri, P.; Coccaro, N.; Cumbo, C.; Tota, G.; Impera, L.; Orsini, P.; et al. Droplet digital PCR analysis of NOTCH1 gene mutations in chronic lymphocytic leukemia. Oncotarget 2016, 7, 86469–86479. [Google Scholar] [CrossRef][Green Version]
- Hoofd, C.; Huang, S.J.; Gusscott, S.; Lam, S.; Wong, R.; Johnston, A.; Ben-Neriah, S.; Steidl, C.; Scott, D.W.; Bruyere, H.; et al. Ultrasensitive Detection of NOTCH1 c.7544_7545delCT Mutations in Chronic Lymphocytic Leukemia by Droplet Digital PCR Reveals High Frequency of Subclonal Mutations and Predicts Clinical Outcome in Cases with Trisomy 12. J. Mol. Diagn. 2020, 22, 571–578. [Google Scholar] [CrossRef]
- Gertz, M.A. Waldenström macroglobulinemia: 2021 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2021, 96, 258–269. [Google Scholar] [CrossRef]
- Yu, X.; Li, W.; Deng, Q.; Li, L.; Hsi, E.D.; Young, K.H.; Zhang, M.; Li, Y. MYD88 L265P Mutation in Lymphoid Malignancies. Cancer Res. 2018, 78, 2457–2462. [Google Scholar] [CrossRef]
- Ferrante, M.; Furlan, D.; Zibellini, S.; Borriero, M.; Candido, C.; Sahnane, N.; Uccella, S.; Genuardi, E.; Alessandria, B.; Bianchi, B.; et al. MYD88L265P Detection in IgM Monoclonal Gammopathies: Methodological Considerations for Routine Implementation. Diagnostics 2021, 11, 779. [Google Scholar] [CrossRef]
- Chen, K.; Ma, Y.; Ding, T.; Zhang, X.; Chen, B.; Guan, M. Effectiveness of digital PCR for MYD88L265P detection in vitreous fluid for primary central nervous system lymphoma diagnosis. Exp. Ther. Med. 2020, 20, 301–308. [Google Scholar] [CrossRef]
- Willenbacher, E.; Willenbacher, W.; Wolf, D.G.; Zelger, B.; Peschel, I.; Manzl, C.; Haun, M.; Brunner, A. Digital PCR in bone marrow trephine biopsies is highly sensitive for MYD88L265P detection in lymphomas with plasmacytic/plasmacytoid differentiation. Br. J. Haematol. 2019, 186, 189–191. [Google Scholar] [CrossRef]
- Alcaide, M.; Yu, S.; Bushell, K.; Fornika, D.; Nielsen, J.S.; Nelson, B.H.; Mann, K.K.; Assouline, S.; Johnson, N.A.; Morin, R.D. Multiplex Droplet Digital PCR Quantification of Recurrent Somatic Mutations in Diffuse Large B-Cell and Follicular Lymphoma. Clin Chem. 2016, 62, 1238–1247. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, S.; Luminari, S.; Ciabatti, E.; Grassi, S.; Guerrini, F.; Dondi, A.; Marcheselli, L.; Ladetto, M.; Piccaluga, P.P.; Gazzola, A.; et al. Minimal residual disease after conventional treatment significantly impacts on progression-free survival of patients with follicular lymphoma: The FIL FOLL05 trial. Clin. Cancer Res. 2014, 20, 6398–6405. [Google Scholar] [CrossRef] [PubMed]
- von Keudell, G.; Salles, G. The role of tazemetostat in relapsed/refractory follicular lymphoma. Ther. Adv. Hematol. 2021, 12, 20406207211015882. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Bátai, B.; Balogh, A.; Illés, S.; Mikala, G.; Nagy, N.; Kiss, L.; Kotmayer, L.; Matolcsy, A.; Alpár, D.; et al. Quantitative Analysis and Monitoring of EZH2 Mutations Using Liquid Biopsy in Follicular Lymphoma. Genes 2020, 11, 785. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, M.; De Novi, L.A.; Della Starza, I.; Cappelli, L.V.; Nunes, V.; Pulsoni, A.; Del Giudice, I.; Guarini, A.; Foà, R. Comparative analysis between RQ-PCR and digital droplet PCR of BCL2/IGH gene rearrangement in the peripheral blood and bone marrow of early stage follicular lymphoma. Br. J. Haematol. 2017, 177, 588–596. [Google Scholar] [CrossRef]
- Novella, E.; Giaretta, I.; Felice, F.; Madeo, D.; Piccin, A.; Castaman, G.; Rodeghiero, F. Fluorescent polymerase chain reaction and capillary electrophoresis for IgH rearrangement and minimal residual disease evaluation in multiple myeloma. Haematologica 2002, 87, 1157–1164. [Google Scholar]
- Drandi, D.; Alcantara, M.; Benmaad, I.; Söhlbrandt, A.; Lhermitte, L.; Zaccaria, G.; Ferrante, M.; Genuardi, E.; Mantoan, B.; Villarese, P.; et al. Droplet Digital PCR Quantification of Mantle Cell Lymphoma Follow-up Samples from Four Prospective Trials of the European MCL Network. Hemasphere 2020, 4, e347. [Google Scholar] [CrossRef]
- Tanzima Nuhat, S.; Sakata-Yanagimoto, M.; Komori, D.; Hattori, K.; Suehara, Y.; Fukumoto, K.; Fujisawa, M.; Kusakabe, M.; Matsue, K.; Wakamatsu, H.; et al. Droplet digital polymerase chain reaction assay and peptide nucleic acid-locked nucleic acid clamp method for RHOA mutation detection in angioimmunoblastic T-cell lymphoma. Cancer Sci. 2018, 109, 1682–1689. [Google Scholar] [CrossRef]
- Bessi, L.; Viailly, P.J.; Bohers, E.; Ruminy, P.; Maingonnat, C.; Bertrand, P.; Vasseur, N.; Beaussire, L.; Cornic, M.; Etancelin, P.; et al. Somatic mutations of cell-free circulating DNA detected by targeted next-generation sequencing and digital droplet PCR in classical Hodgkin lymphoma. Leuk. Lymphoma. 2019, 60, 498–502. [Google Scholar] [CrossRef]
- Bertamini, L.; D’Agostino, M.; Gay, F. MRD Assessment in Multiple Myeloma: Progress and Challenges. Curr. Hematol. Malig. Rep. 2021, 16, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, H.; Wee, R.K.; Zaimoku, Y.; Murata, R.; Zheng, J.; Moorhead, M.; Carlton, V.E.H.; Kong, K.A.; Takezako, N.; Ito, S.; et al. A comparison of minimal residual disease detection in autografts among ASO-qPCR, droplet digital PCR, and next-generation sequencing in patients with multiple myeloma who underwent autologous stem cell transplantation. Br. J. Haematol. 2018, 183, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Apperley, J.; Milojkovic, D.; Cross, N.C.P.; Foroni, L.; Byrne, J.; Goringe, A.; Rao, A.; Khorashad, J.; de Lavallade, H.; et al. A British Society for Haematology Guideline on the diagnosis and management of chronic myeloid leukaemia. Br. J. Haematol. 2020, 191, 171–193. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef] [PubMed]
- Saussele, S.; Richter, J.; Guilhot, J.; Gruber, F.X.; Hjorth-Hansen, H.; Almeida, A.; Janssen, J.J.W.M.; Mayer, J.; Koskenvesa, P.; Panayiotidis, P.; et al. Discontinuation of tyrosine kinase inhibitor therapy in chronic myeloid leukaemia (EURO-SKI): A prespecified interim analysis of a prospective, multicentre, non-randomised, trial. Lancet Oncol. 2018, 19, 747–757. [Google Scholar] [CrossRef]
- Atallah, E.; Sweet, K. Treatment-Free Remission: The New Goal in CML Therapy. Curr. Hematol. Malig. Rep. 2021, 16, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Baccarani, M.; Abruzzese, E.; Accurso, V.; Albano, F.; Annunziata, M.; Barulli, S.; Beltrami, G.; Bergamaschi, M.; Binotto, G.; Bocchia, M.; et al. Managing chronic myeloid leukemia for treatment-free remission: A proposal from the GIMEMA CML WP. Blood Adv. 2019, 3, 4280–4290. [Google Scholar] [CrossRef]
- Dragani, M.; Rege Cambrin, G.; Berchialla, P.; Dogliotti, I.; Rosti, G.; Castagnetti, F.; Capodanno, I.; Martino, B.; Cerrano, M.; Ferrero, D.; et al. A Retrospective Analysis about Frequency of Monitoring in Italian Chronic Myeloid Leukemia Patients after Discontinuation. J. Clin. Med. 2020, 9, 3692. [Google Scholar] [CrossRef]
- Chung, H.J.; Hur, M.; Yoon, S.; Hwang, K.; Lim, H.S.; Kim, H.; Moon, H.W.; Yun, Y.M. Performance Evaluation of the QXDx BCR-ABL %IS Droplet Digital PCR Assay. Ann. Lab. Med. 2020, 40, 72–75. [Google Scholar] [CrossRef]
- Wang, W.J.; Zheng, C.F.; Liu, Z.; Tan, Y.H.; Chen, X.H.; Zhao, B.L.; Li, G.X.; Xu, Z.F.; Ren, F.G.; Zhang, Y.F.; et al. Droplet digital PCR for BCR/ABL(P210) detection of chronic myeloid leukemia: A high sensitive method of the minimal residual disease and disease progression. Eur. J. Haematol. 2018, 101, 291–296. [Google Scholar] [CrossRef]
- Franke, G.N.; Maier, J.; Wildenberger, K.; Cross, M.; Giles, F.J.; Müller, M.C.; Hochhaus, A.; Niederwieser, D.; Lange, T. Comparison of Real-Time Quantitative PCR and Digital Droplet PCR for BCR-ABL1 Monitoring in Patients with Chronic Myeloid Leukemia. J. Mol. Diagn. 2020, 22, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.A.; Olmedillas, S.; Serrano-López, J.; Lainez-González, D.; Castaño, T.; Iñiguez, R.; Lopez-Lorenzo, J.L.; García, A.; Atance, M.; Sánchez, R.N.S.; et al. Comparison of Droplet Digital PCR versus qPCR Measurements on the International Scale for the Molecular Monitoring of Chronic Myeloid Leukemia Patients. Mol. Diagn. Ther. 2020, 24, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Fava, C.; Bernardi, S.; Gottardi, E.M.; Lorenzatti, R.; Galeotti, L.; Ceccherini, F.; Cordoni, F.; Daraio, F.; Giugliano, E.; Jovanovski, A.; et al. Alignment of Qx100/Qx200 Droplet Digital (Bio-Rad) and QuantStudio 3D (Thermofisher) Digital PCR for Quantification of BCR-ABL1 in Ph+ Chronic Myeloid Leukemia. Diseases 2021, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Vagge, E.; le Coutre, P.; Abruzzese, E.; Martino, B.; Pungolino, E.; Elena, C.; Pierri, I.; Assouline, S.; D’Emilio, A.; et al. Age and dPCR can predict relapse in CML patients who discontinued imatinib: The ISAV study. Am. J. Hematol. 2015, 90, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Malagola, M.; Zanaglio, C.; Polverelli, N.; Dereli Eke, E.; D’Adda, M.; Farina, M.; Bucelli, C.; Scaffidi, L.; Toffoletti, E.; et al. Digital PCR improves the quantitation of DMR and the selection of CML candidates to TKIs discontinuation. Cancer Med. 2019, 8, 2041–2055. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, F.E.; Dulucq, S.; Boureau, L.; Cony-Makhoul, P.; Charbonnier, A.; Escoffre-Barbe, M.; Rigal-Huguet, F.; Coiteux, V.; Varet, B.; Dubruille, V.; et al. Evaluation of Residual Disease and TKI Duration Are Critical Predictive Factors for Molecular Recurrence after Stopping Imatinib First-line in Chronic Phase CML Patients. Clin. Cancer Res. 2019, 25, 6606–6613. [Google Scholar] [CrossRef]
- Atallah, E.; Schiffer, C.A.; Radich, J.P.; Weinfurt, K.P.; Zhang, M.J.; Pinilla-Ibarz, J.; Kota, V.; Larson, R.A.; Moore, J.O.; Mauro, M.J.; et al. Assessment of Outcomes After Stopping Tyrosine Kinase Inhibitors Among Patients with Chronic Myeloid Leukemia: A Nonrandomized Clinical Trial. JAMA Oncol. 2021, 7, 42–50. [Google Scholar] [CrossRef]
- Colafigli, G.; Scalzulli, E.; Porrazzo, M.; Diverio, D.; Loglisci, M.G.; Latagliata, R.; Guarini, A.; Foà, R.; Breccia, M. Digital droplet PCR at the time of TKI discontinuation in chronic-phase chronic myeloid leukemia patients is predictive of treatment-free remission outcome. Hematol. Oncol. 2019, 37, 652–654. [Google Scholar] [CrossRef]
- Soverini, S.; Bavaro, L.; De Benedittis, C.; Martelli, M.; Iurlo, A.; Orofino, N.; Sica, S.; Sorà, F.; Lunghi, F.; Ciceri, F.; et al. Prospective assessment of NGS-detectable mutations in CML patients with nonoptimal response: The NEXT-in-CML study. Blood 2020, 135, 534–541, Erratum in Blood 2022, 139, 1601. [Google Scholar] [CrossRef]
- Galimberti, S.; Guerrini, F.; Grassi, S.; Bocchia, M.; Bavaro, L.; Ciabatti, E.; Dragani, M.; Gottardi, E.M.; Izzo, B.; Lunghi, F.; et al. DIGITAL Droplet PCR Is a Fast and Effective Tool for Detecting T315i Mutation in Chronic Myeloid Leukemia. EHA Libr. 2020, 294653, EP735. [Google Scholar]
- Cumbo, C.; Impera, L.; Minervini, C.F.; Orsini, P.; Anelli, L.; Zagaria, A.; Coccaro, N.; Tota, G.; Minervini, A.; Casieri, P.; et al. Genomic BCR-ABL1 breakpoint characterization by a multi-strategy approach for “personalized monitoring” of residual disease in chronic myeloid leukemia patients. Oncotarget 2018, 9, 10978–10986. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Barbui, T. Polycythemia vera and essential thrombocythemia: 2021 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2020, 95, 1599–1613. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A. Primary myelofibrosis: 2021 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2021, 96, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Barbui, T.; Thiele, J.; Gisslinger, H.; Kvasnicka, H.M.; Vannucchi, A.M.; Guglielmelli, P.; Orazi, A.; Tefferi, A. The 2016 WHO classification and diagnostic criteria for myeloproliferative neoplasms: Document summary and in-depth discussion. Blood Cancer J. 2018, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Fontanelli, G.; Baratè, C.; Ciabatti, E.; Guerrini, F.; Grassi, S.; Del Re, M.; Morganti, R.; Petrini, I.; Arici, R.; Barsotti, S.; et al. Real-Time PCR and Droplet Digital PCR: Two techniques for detection of the JAK2(V617F) mutation in Philadelphia-negative chronic myeloproliferative neoplasms. Int. J. Lab. Hematol. 2015, 37, 766–773. [Google Scholar] [CrossRef]
- Benedetti, E.; Tavarozzi, R.; Morganti, R.; Bruno, B.; Bramanti, E.; Baratè, C.; Balducci, S.; Iovino, L.; Ricci, F.; Ricchiuto, V.; et al. Organ Stiffness in the Work-Up of Myelofibrosis and Philadelphia-Negative Chronic Myeloproliferative Neoplasms. J. Clin. Med. 2020, 9, 2149. [Google Scholar] [CrossRef]
- Zheng, C.F.; Zhao, X.X.; Chen, X.H.; Liu, Z.; Wang, W.J.; Luo, M.; Ren, Y.; Wang, H.W. Quantification of JAK2V617F mutation load by droplet digital PCR can aid in diagnosis of myeloproliferative neoplasms. Int. J. Lab. Hematol. 2021, 43, 645–650. [Google Scholar] [CrossRef]
- La Rocca, F.; Grieco, V.; Ruggieri, V.; Zifarone, E.; Villani, O.; Zoppoli, P.; Russi, S.; Laurino, S.; Falco, G.; Calice, G.; et al. Superiority of Droplet Digital PCR Over Real-Time Quantitative PCR for JAK2V617F Allele Mutational Burden Assessment in Myeloproliferative Neoplasms: A Retrospective Study. Diagnostics 2020, 10, 143. [Google Scholar] [CrossRef]
- Wieseler, B.; Kerekes, M.F.; Vervoelgyi, V.; McGauran, N.; Kaiser, T. Impact of document type on reporting quality of clinical drug trials: A comparison of registry reports, clinical study reports, and journal publications. BMJ 2012, 344, d8141. [Google Scholar] [CrossRef]
- Anelli, L.; Zagaria, A.; Coccaro, N.; Tota, G.; Minervini, A.; Casieri, P.; Impera, L.; Minervini, C.F.; Brunetti, C.; Ricco, A.; et al. Droplet digital PCR assay for quantifying of CALR mutant allelic burden in myeloproliferative neoplasms. Ann. Hematol. 2016, 95, 1559–1560. [Google Scholar] [CrossRef]
- Baron, F.; Sandmaier, B.M. Chimerism and outcomes after allogeneic hematopoietic cell transplantation following nonmyeloablative conditioning. Leukemia 2006, 20, 1690–1700. [Google Scholar] [CrossRef] [PubMed]
- Bader, P.; Niethammer, D.; Willasch, A.; Kreyenberg, H.; Klingebiel, T. How and when should we monitor chimerism after allogeneic stem cell transplantation? Bone Marrow Transplant. 2005, 35, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Antin, J.H.; Childs, R.; Filipovich, A.H.; Giralt, S.; Mackinnon, S.; Spitzer, T.; Weisdorf, D. Establishment of complete and mixed donor chimerism after allogeneic lymphohematopoietic transplantation: Recommendations from a workshop at the 2001 Tandem Meetings of the International Bone Marrow Transplant Registry and the American Society of Blood and Marrow Transplantation. Biol. Blood Marrow Transplant. 2001, 7, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Mika, T.; Baraniskin, A.; Ladigan, S.; Wulf, G.; Dierks, S.; Haase, D.; Schork, K.; Turewicz, M.; Eisenacher, M.; Schmiegel, W.; et al. Digital droplet PCR-based chimerism analysis for monitoring of hematopoietic engraftment after allogeneic stem cell transplantation. Int. J. Lab. Hematol. 2019, 41, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, M.; Bernard, M.; Danic, B.; Dauriac, C.; Birebent, B.; Lapart, C.; Lamy, T.; Le Prisé, P.Y.; Beauplet, A.; Bories, D.; et al. Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction. Blood 2002, 99, 4618–4625. [Google Scholar] [CrossRef] [PubMed]
- Stahl, T.; Böhme, M.U.; Kröger, N.; Fehse, B. Digital PCR to assess hematopoietic chimerism after allogeneic stem cell transplantation. Exp. Hematol. 2015, 43, 462–468.e1. [Google Scholar] [CrossRef] [PubMed]
- Okano, T.; Tsujita, Y.; Kanegane, H.; Mitsui-Sekinaka, K.; Tanita, K.; Miyamoto, S.; Yeh, T.W.; Yamashita, M.; Terada, N.; Ogura, Y.; et al. Droplet Digital PCR-Based Chimerism Analysis for Primary Immunodeficiency Diseases. J. Clin. Immunol. 2018, 38, 300–306. [Google Scholar] [CrossRef]
- Kliman, D.; Castellano-Gonzalez, G.; Withers, B.; Street, J.; Tegg, E.; Mirochnik, O.; Lai, J.; Clancy, L.; Gottlieb, D.; Blyth, E. Ultra-Sensitive Droplet Digital PCR for the Assessment of Microchimerism in Cellular Therapies. Biol. Blood Marrow Transplant. 2018, 24, 1069–1078. [Google Scholar] [CrossRef]
- Waterhouse, M.; Pfeifer, D.; Duque-Afonso, J.; Follo, M.; Duyster, J.; Depner, M.; Bertz, H.; Finke, J. Droplet digital PCR for the simultaneous analysis of minimal residual disease and hematopoietic chimerism after allogeneic cell transplantation. Clin. Chem. Lab. Med. 2019, 57, 641–647. [Google Scholar] [CrossRef]
- Somech, R. T-cell receptor excision circles in primary immunodeficiencies and other T-cell immune disorders. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 517–524. [Google Scholar] [CrossRef]
- Iovino, L.; Mazziotta, F.; Carulli, G.; Guerrini, F.; Morganti, R.; Mazzotti, V.; Maggi, F.; Macera, L.; Orciuolo, E.; Buda, G.; et al. High-dose zinc oral supplementation after stem cell transplantation causes an increase of TRECs and CD4+ naïve lymphocytes and prevents TTV reactivation. Leuk. Res. 2018, 70, 20–24. [Google Scholar] [CrossRef]
- Holstein, S.A.; Lunning, M.A. CAR T-Cell Therapy in Hematologic Malignancies: A Voyage in Progress. Clin. Pharmacol. Ther. 2020, 107, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Kersten, M.J.; Spanjaart, A.M.; Thieblemont, C. CD19-directed CAR T-cell therapy in B-cell NHL. Curr. Opin. Oncol. 2020, 32, 408–417. [Google Scholar] [CrossRef]
- Adkins, S. CAR T-Cell Therapy: Adverse Events and Management. J. Adv. Pract. Oncol. 2019, 10 (Suppl. 3), 21–28. [Google Scholar] [CrossRef]
- Fehse, B.; Badbaran, A.; Berger, C.; Sonntag, T.; Riecken, K.; Geffken, M.; Kröger, N.; Ayuk, F.A. Digital PCR Assays for Precise Quantification of CD19-CAR-T Cells after Treatment with Axicabtagene Ciloleucel. Mol. Ther. Methods Clin. Dev. 2020, 16, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Mika, T.; Maghnouj, A.; Klein-Scory, S.; Ladigan-Badura, S.; Baraniskin, A.; Thomson, J.; Hasenkamp, J.; Hahn, S.A.; Wulf, G.; Schroers, R. Digital-Droplet PCR for Quantification of CD19-Directed CAR T-Cells. Front. Mol. Biosci. 2020, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Pabst, T.; Joncourt, R.; Shumilov, E.; Heini, A.; Wiedemann, G.; Legros, M.; Seipel, K.; Schild, C.; Jalowiec, K.; Mansouri Taleghani, B.; et al. Analysis of IL-6 serum levels and CAR T cell-specific digital PCR in the context of cytokine release syndrome. Exp. Hematol. 2020, 88, 7–14.e3. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).