Diagnostic Accuracy of Point-of-Care HCV Viral Load Assays for HCV Diagnosis: A Systematic Review and Meta-Analysis
Abstract: Background
1. Introduction
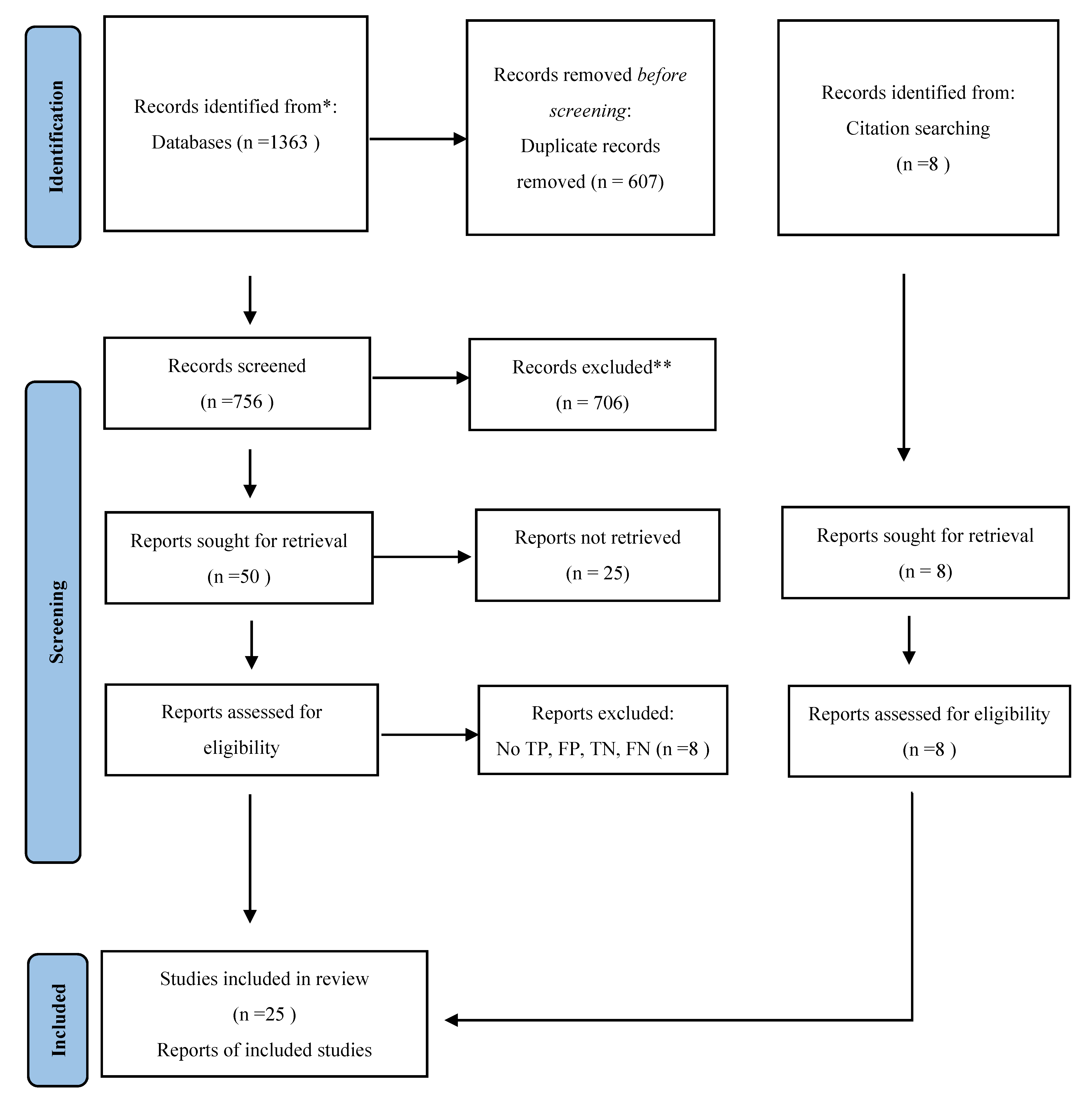
2. Methods
2.1. Research Question
2.2. Search Strategy
2.3. Selection Criteria
- (1)
- The study evaluated the diagnostic accuracy of any PoC HCV RNA assays against an acceptable quality-assured laboratory reference for HCV RNA determination, including Abbott RealTime HCV (Abbott Molecular, Des Plaines, USA), Generic HCV Charge Virale (Biocentric, Bandol, France), COBAS AmpliPrep/COBAS TaqMan HCV Qualitative or Quantitative Tests, Version 2.0 (Roche Molecular Diagnostics, Santa Clara, CA, USA), APTIMA HCV Quant Dx Assay (Hologic Inc., Marlborough, MA, USA), Artus HCV RG RT-PCR or QS-RGQ (Qiagen, Hilden, Germany), and HCV Real-TM Quant Dx (Sacace Biotechnologies, Como, Italy). These laboratory-based HCV virological tests are all commercially available products and have obtained CE mark certificates.
- (2)
- The study used a commercial PoC HCV viral load nucleic acid assay for point-of-care or near-patient nucleic acid testing. Assays include the Xpert HCV Viral load and the Xpert HCV VL Fingerstick assays (Cepheid, Sunnyvale, CA, USA), the Genedrive HCV ID Kit (Genedrive, Manchester, UK), the Truenat HCV RNA assay (Molbio Diagnostics, Goa, India), and the SAMBA II HCV Qualitative Whole Blood Test (Diagnostics for the New World, Birmingham, UK). These PoC HCV virological tests are all currently available in the market and have obtained CE mark certificates.
- (3)
- The study evaluated adults (>18 years of age), including key populations at high risk of HCV infection.
- (4)
- The study reported diagnostic accuracy or provided data to allow the calculation of diagnostic accuracy (True Positive [TP], False Positive [FP], True Negative [TN], and False Negative [FN]).
2.4. Data Extraction
2.5. Outcome and Analysis
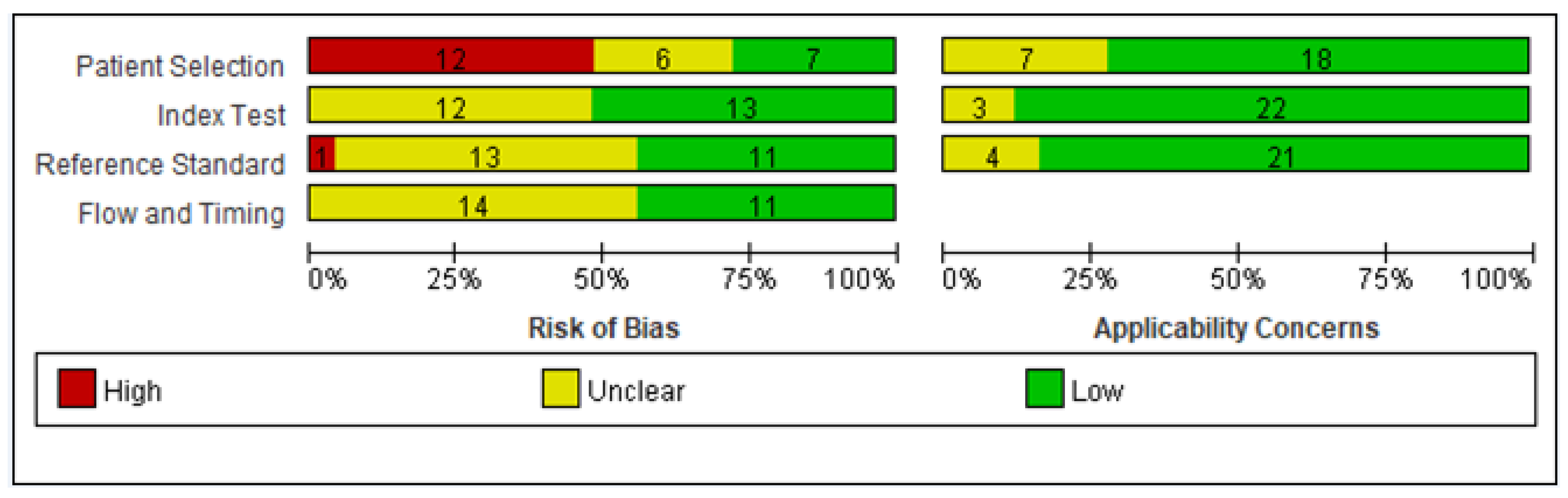
2.6. Risk of Bias Assessments
2.7. Grading Quality of Evidence
3. Results
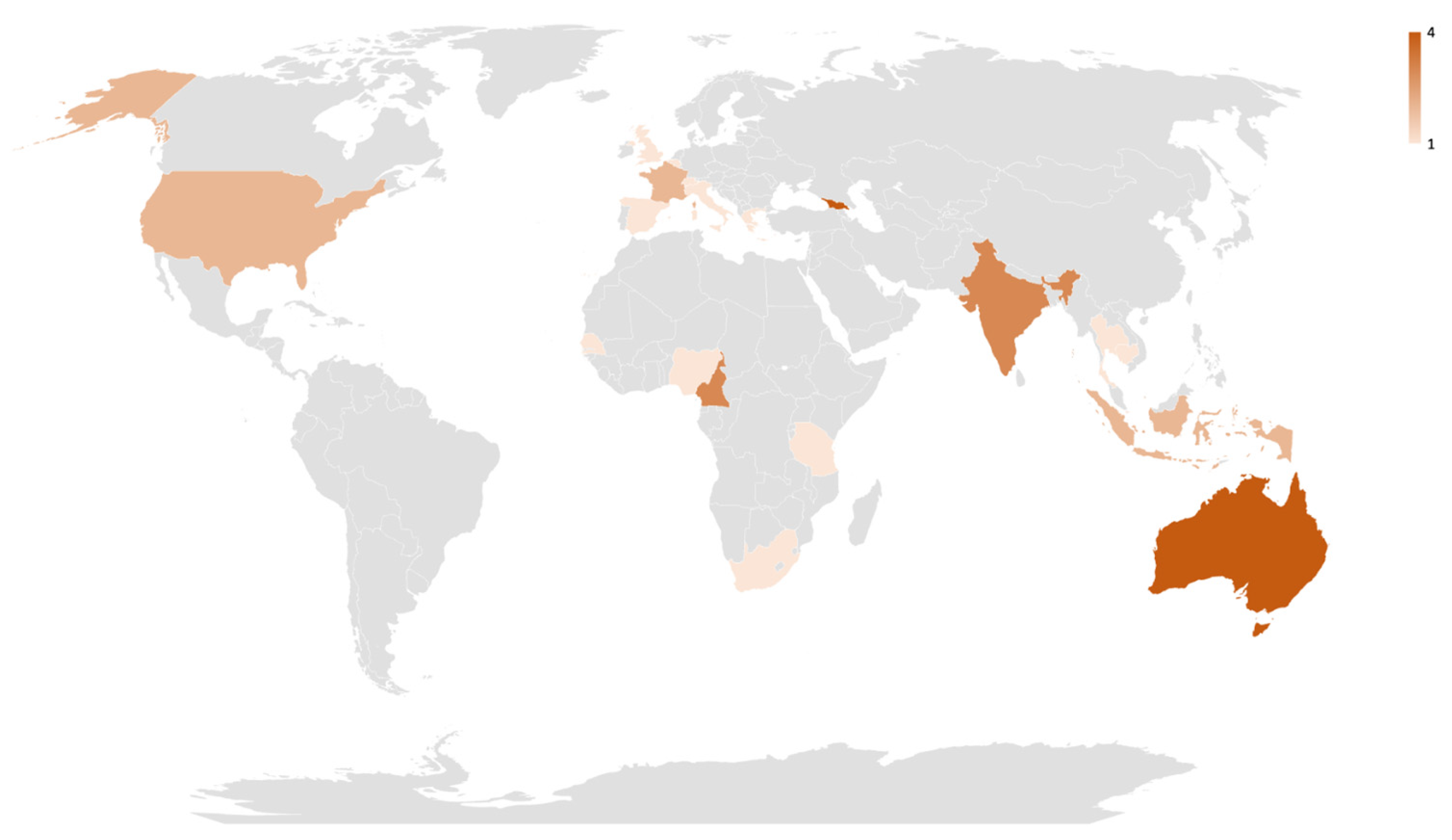
3.1. Study Characteristics
3.2. Quality Assessment
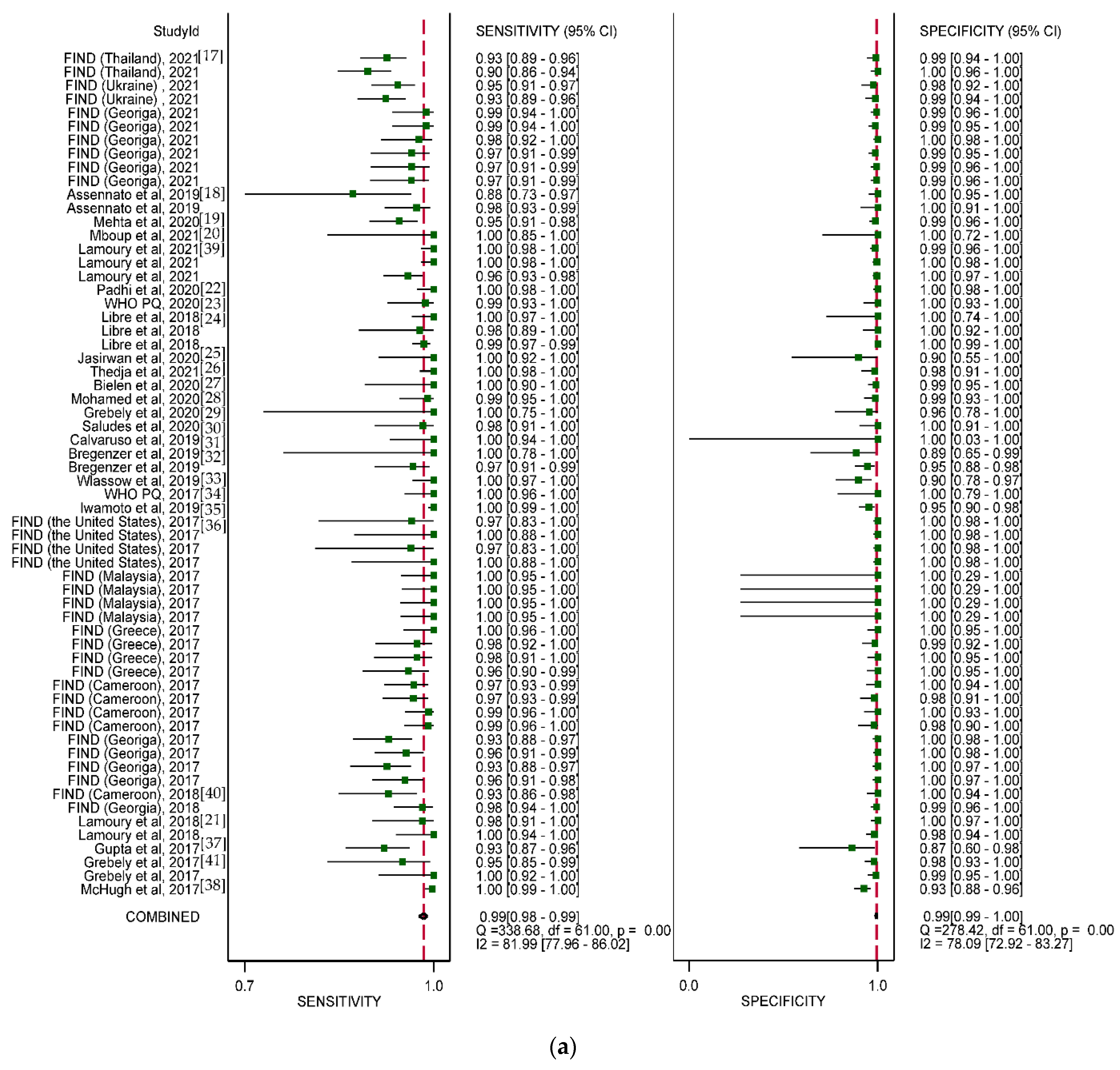
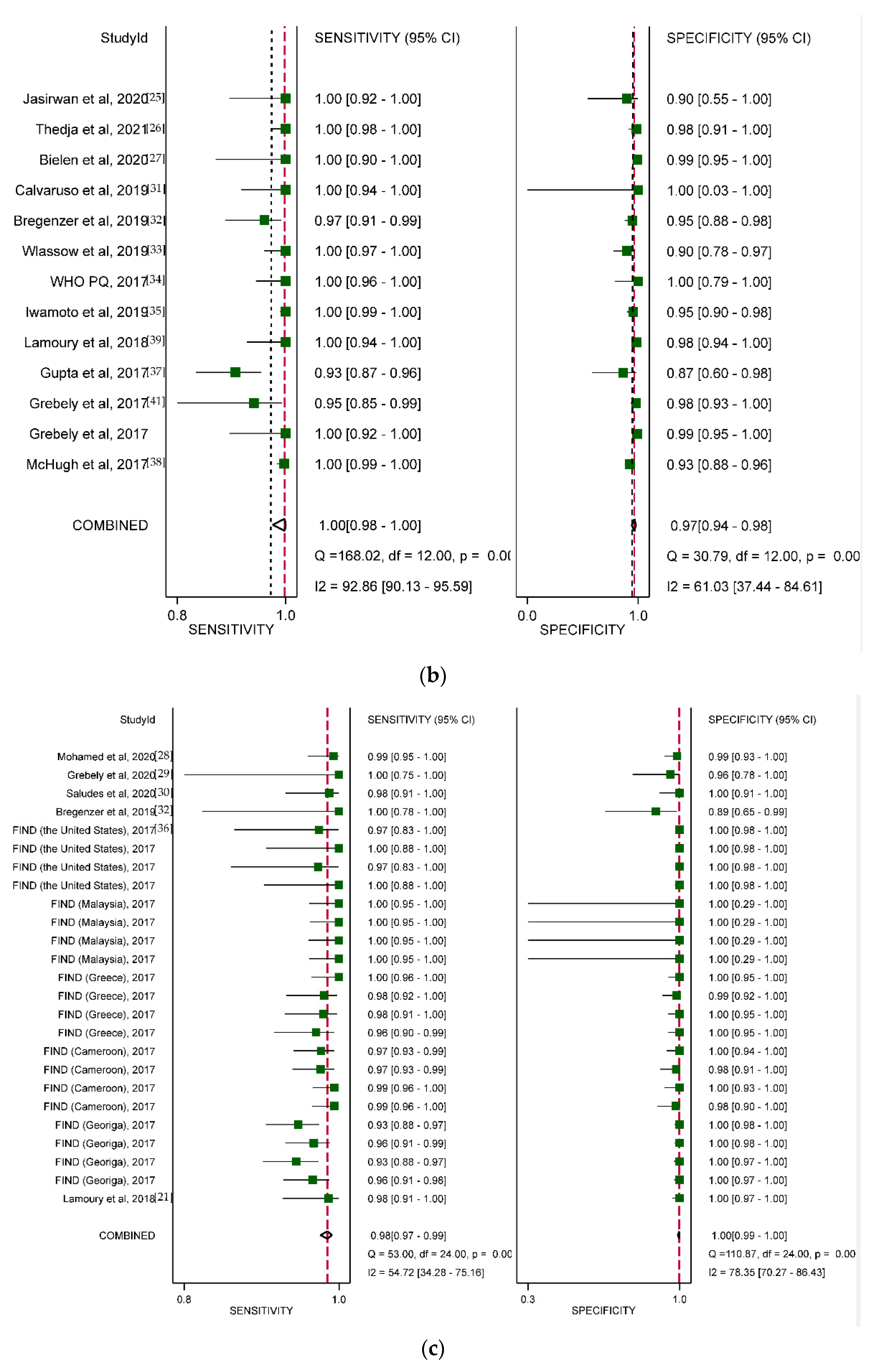
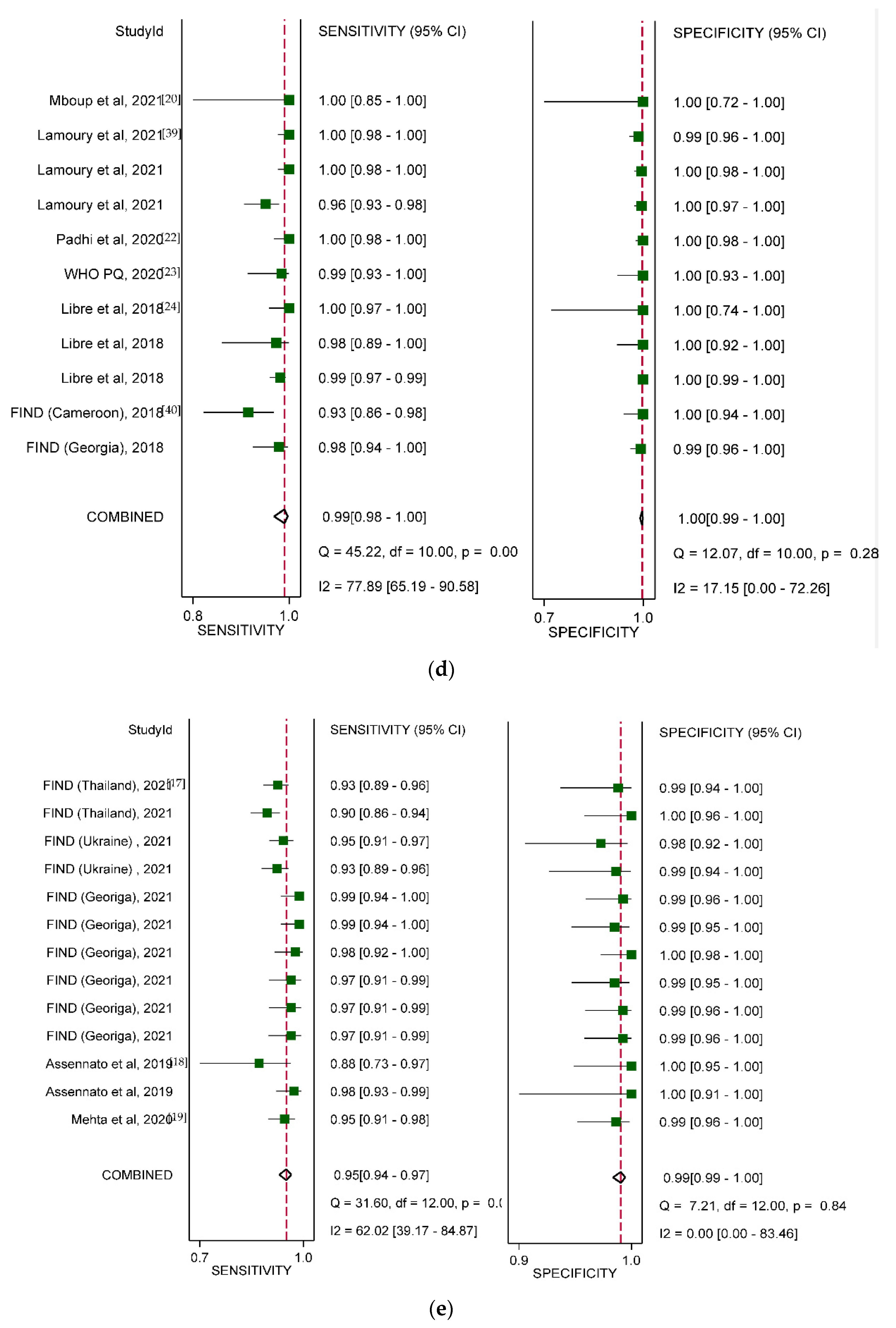
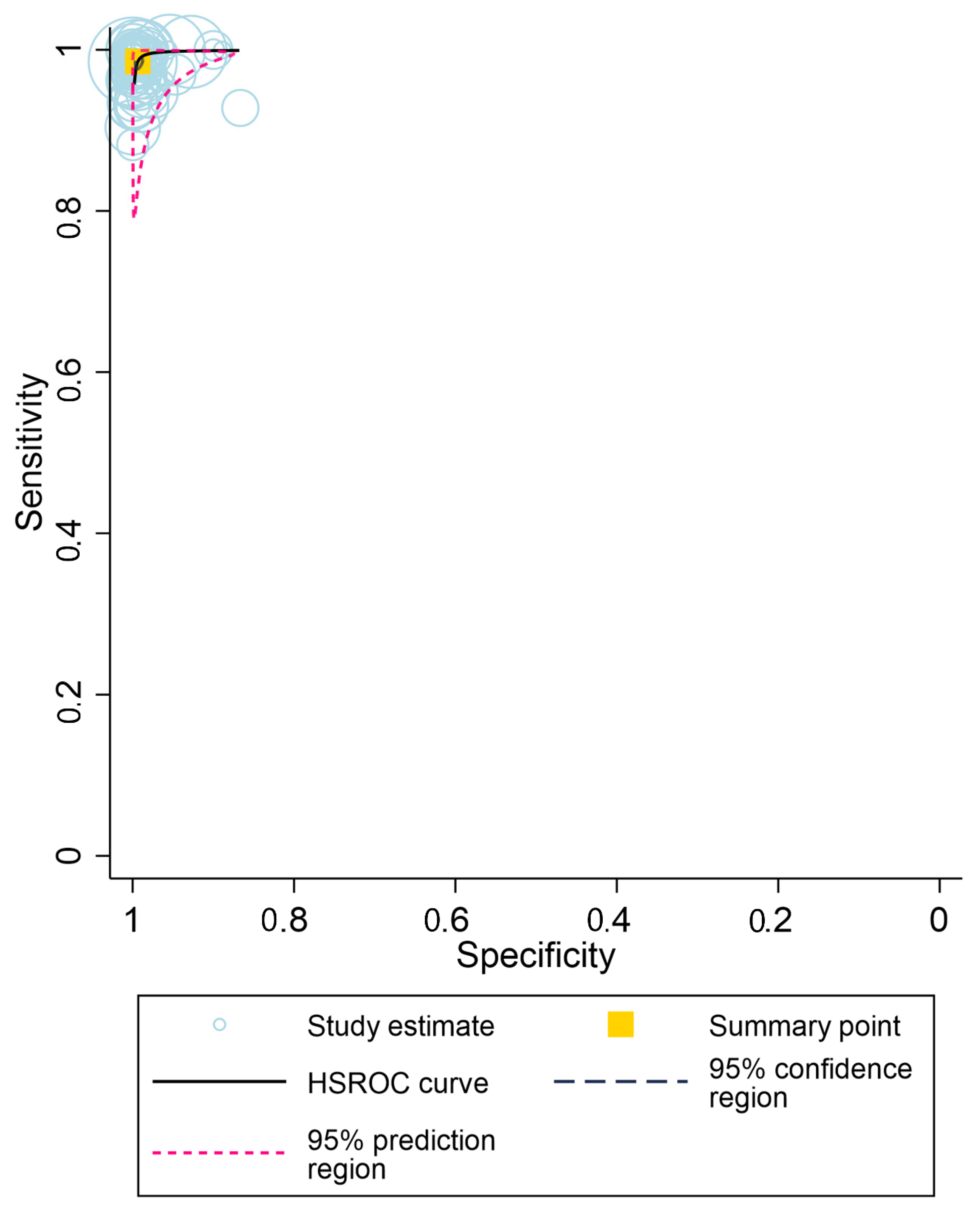
3.3. Overall Diagnostic Accuracy
3.4. Diagnostic Accuracy by Type of HCV VL PoC Assay
3.5. Subgroup Analysis by Study Setting, Population, and Specimen Type
3.6. Grading Quality of Evidence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Global Progress Report on Hiv, Viral Hepatitis And Sexually Transmitted Infections, 2021; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- WHO. Global Health Sector Strategu on Viral Hepatitis 2016–2021; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- Hutin, Y.J.F.; Bulterys, M.; Hirnschall, G.O. How far are we from viral hepatitis elimination service coverage targets? J. Int. AIDS Soc. 2018, 21, e25050. [Google Scholar] [CrossRef] [Green Version]
- WHO. WHO Global Hepatitis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Oru, E.; Trickey, A.; Shirali, R.; Kanters, S.; Easterbrook, P. Decentralisation, integration, and task-shifting in hepatitis C virus infection testing and treatment: A global systematic review and meta-analysis. Lancet Glob. Health 2021, 9, e431–e445. [Google Scholar] [CrossRef]
- Easterbrook, P.J.; Roberts, T.; Sands, A.; Peeling, R. Diagnosis of viral hepatitis. Curr. Opin. HIV AIDS 2017, 12, 302. [Google Scholar] [CrossRef]
- Trikey, A.; Fajardo, E.; Alemu, D.; Artenie, A.A.; Easterbrook, P. Impact of hepatitis C virus (HCV) point-of-care viral load testing compared to laboratory-based testing on uptake of testing and treatment, and turnaround times: A systematic review and meta-analysis. Int. J. Drug Policy 2021, 57, 11–17. [Google Scholar]
- ISO. Point-of-Care Testing (POCT)—Requirements for Quality and Competence; ISO: Geneva, Switzerland, 2006. [Google Scholar]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. BMJ 2009, 339, b2700. [Google Scholar] [CrossRef] [Green Version]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kim, K.W.; Choi, S.H.; Huh, J.; Park, S.H. Systematic review and meta-analysis of studies evaluating diagnostic test accuracy: A practical review for clinical researchers-part II. Statistical methods of meta-analysis. Korean J. Radiol. 2015, 16, 1188–1196. [Google Scholar] [CrossRef] [Green Version]
- Hoy, D.; Brooks, P.; Woolf, A.; Blyth, F.; March, L.; Bain, C.; Baker, P.; Smith, E.; Buchbinder, R. Assessing risk of bias in prevalence studies: Modification of an existing tool and evidence of interrater agreement. J. Clin. Epidemiol. 2012, 65, 934–939. [Google Scholar] [CrossRef]
- Sterne, J.A.; Hernan, M.A.; Reeves, B.C.; Savovic, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef] [Green Version]
- Siemieniuk, R.; Guyatt, G. BMJ Best Practice: What Is GRADE? Available online: https://bestpractice.bmj.com/info/toolkit/learn-ebm/what-is-grade/ (accessed on 9 September 2021).
- Schünemann, H.J.; Oxman, A.D.; Brozek, J.; Glasziou, P.; Jaeschke, R.; Vist, G.E.; Williams, J.W.; Kunz, R.; Craig, J.; Montori, V.M. Grading quality of evidence and strength of recommendations for diagnostic tests and strategies. BMJ 2008, 336, 1106–1110. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Labra, A.; Brignardello-Petersen, R.; Santesso, N.; Neumann, I.; Mustafa, R.A.; Mbuagbaw, L.; Ikobaltzeta, I.E.; De Stio, C.; McCullagh, L.J.; Alonso-Coello, P. Improving GRADE evidence tables part 1: A randomized trial shows improved understanding of content in summary of findings tables with a new format. J. Clin. Epidemiol. 2016, 74, 7–18. [Google Scholar] [CrossRef]
- FIND. Multicentre Clinical Performance Evaluation of the Molbio Diagnostics Truenat™ HCV RNA Assay; HIV-NAT: Pathumwan, Bangkok, 2019. [Google Scholar]
- Assennato, S.; Ritvhie, A.; Sembongi, H.; Ivanova, E.; Stepchenkova, T.; Lee, H. SAMBA I I HCV: A New Point of Care Molecular Assy for HCV Diagnostic and DDA Monitoring. 2019. [Google Scholar]
- Mehta, V.; Ivanova, E.; Vetter, B.; Sarin, S.; Ramachandran, B.; Gupta, E. Clinical performance evaluation of the Molbio Diagnostics TruenatTM HCV RNA assay: A near point-of-care test to strengthen Hepatitis C virus diagnostics. In Proceedings of the 29th Annual Conference of Asian Pacific Association for the Study of the Liver, Bali, Indonesia, 4–8 March 2020. [Google Scholar]
- Mboup. Evaluation Report of the Clinical Performance of Genedrive Point-of-Care Test for Qualitative Detection of Hepatitis C Virus. 2021. [Google Scholar]
- Lamoury, F.M.J.; Bajis, S.; Hajarizadeh, B.; Marshall, A.D.; Martinello, M.; Ivanova, E.; Catlett, B.; Mowat, Y.; Marks, P.; Amin, J.; et al. Evaluation of the Xpert HCV Viral Load Finger-Stick Point-of-Care Assay. J. Infect. Dis. 2018, 217, 1889–1896. [Google Scholar] [CrossRef]
- Padhi, A.; Gupta, E.; Singh, G.; Agarwal, R.; Sharma, M.K.; Sarin, S.K. Evaluation of the Point of Care Molecular Diagnostic Genedrive HCV ID Kit for the detection of HCV RNA in clinical samples. Epidemiol. Infect. 2020, 18, 1–23. [Google Scholar] [CrossRef]
- WHO. WHO Prequalification of In Vitro Diagnostics; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Llibre, A.; Shimakawa, Y.; Mottez, E.; Ainsworth, S.; Buivan, T.P.; Firth, R.; Harrison, E.; Rosenberg, A.R.; Meritet, J.F.; Fontanet, A.; et al. Development and clinical validation of the Genedrive point-of-care test for qualitative detection of hepatitis C virus. Gut 2018, 67, 2017–2024. [Google Scholar] [CrossRef] [Green Version]
- Jasirwan, C.O.M.; Hasan, I.; Sulaiman, A.S.; Gani, R.A. Evaluation of GeneXpert for Quantification Viral Load Hepatitis C Virus. Indones. J. Gastroenterol. Hepatol. Dig. Endosc. 2020, 21, 182–187. [Google Scholar] [CrossRef]
- Thedja, M.D.; Wibowo, D.P.; El-Khobar, K.E.; Le, S.I.; Turyadi; Setiawan, L.; Murti, I.S.; Muljono, D.H. Improving Linkage to Care of Hepatitis C: Clinical Validation of GeneXpert(R) HCV Viral Load Point-of-Care Assay in Indonesia. Am. J. Trop. Med. Hyg. 2021, 105, 117–124. [Google Scholar] [CrossRef]
- Bielen, R.; Koc, O.M.; Busschots, D.; Verrando, R.; Nevens, F.; Robaeys, G. Validation of hepatitis C virus RNA detection using capillary blood by finger prick (GenXpert system)-Hepatitis C fingerprick study. J. Viral. Hepat. 2020, 27, 709–714. [Google Scholar] [CrossRef]
- Mohamed, Z.; Mbwambo, J.; Rwegasha, J.; Mgina, N.; Doulla, B.; Mwakale, P.; Tuaillon, E.; Chevaliez, S.; Shimakawa, Y.; Taylor-Robinson, S.D.; et al. In-field evaluation of Xpert(R) HCV viral load Fingerstick assay in people who inject drugs in Tanzania. Liver Int. 2020, 40, 514–521. [Google Scholar] [CrossRef] [Green Version]
- Grebely, J.; Read, P.; Cunningham, E.B.; Weltman, M.; Matthews, G.V.; Dunlop, A.; Montebello, M.; Martinello, M.; Gilliver, R.; Marks, P.; et al. Elbasvir and grazoprevir for hepatitis C virus genotype 1 infection in people with recent injecting drug use (DARLO-C): An open-label, single-arm, phase 4, multicentre trial. Health Sci. Rep. 2020, 3, e151. [Google Scholar] [CrossRef]
- Saludes, V.; Antuori, A.; Lazarus, J.V.; Folch, C.; Gonzalez-Gomez, S.; Gonzalez, N.; Ibanez, N.; Colom, J.; Matas, L.; Casabona, J.; et al. Evaluation of the Xpert HCV VL Fingerstick point-of-care assay and dried blood spot HCV-RNA testing as simplified diagnostic strategies among people who inject drugs in Catalonia, Spain. Int. J. Drug Policy 2020, 80, 102734. [Google Scholar] [CrossRef]
- Calvaruso, V.; Bronte, F.; Ferraro, D.; Reina, G.; Conte, E.; Rini, F.; Magro, B.; Petta, S.; Di Marco, V.; Craxi, A. Point-of-care HCV RNA testing in the setting of DAA therapy: HCV-FiS (HEpatitis C Virus Fingerstick Study). Liver Int. 2019, 39, 2240–2243. [Google Scholar] [CrossRef]
- Bregenzer, A.; Warmann, N.; Ottiger, C.; Fux, C.A. Rapid point-of-care HCV RNA quantification in capillary whole blood for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection. Swiss. Med. Wkly. 2019, 149, w20137. [Google Scholar] [CrossRef]
- Wlassow, M.; Poiteau, L.; Roudot-Thoraval, F.; Rosa, I.; Soulier, A.; Hezode, C.; Ortonne, V.; Pawlotsky, J.M.; Chevaliez, S. The new Xpert HCV viral load real-time PCR assay accurately quantifies hepatitis C virus RNA in serum and whole-blood specimens. J. Clin. Virol. 2019, 117, 80–84. [Google Scholar] [CrossRef]
- WHO. WHO Prequalification of In Vitro Diagnostics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Iwamoto, M.; Calzia, A.; Dublineau, A.; Rouet, F.; Nouhin, J.; Yann, S.; Pin, S.; Sun, C.; Sann, K.; Dimanche, C.; et al. Field evaluation of GeneXpert((R)) (Cepheid) HCV performance for RNA quantification in a genotype 1 and 6 predominant patient population in Cambodia. J. Viral Hepat. 2019, 26, 38–47. [Google Scholar] [CrossRef] [Green Version]
- FIND. Evaluation of the Diagnostic Performance of the Xpert® Fingerstick HCV Viral Load (VL) Assay; HIV-NAT: Bangkok, Thailand, 2017. [Google Scholar]
- Gupta, E.; Agarwala, P.; Kumar, G.; Maiwall, R.; Sarin, S.K. Point -of -care testing (POCT) in molecular diagnostics: Performance evaluation of GeneXpert HCV RNA test in diagnosing and monitoring of HCV infection. J. Clin. Virol. 2017, 88, 46–51. [Google Scholar] [CrossRef]
- McHugh, M.P.; Wu, A.H.B.; Chevaliez, S.; Pawlotsky, J.M.; Hallin, M.; Templeton, K.E. Multicenter Evaluation of the Cepheid Xpert Hepatitis C Virus Viral Load Assay. J. Clin. Microbiol. 2017, 55, 1550–1556. [Google Scholar] [CrossRef] [Green Version]
- Lamoury, F.M.J.; Njouom, R.; Amougou-Atsama, M.; Yiagnigni Mfopou, E.; Berishvili, N.; Sologashvili, M.; Fajardo, E.; Malobela, A.; Mace, A.; Chirehwa, M.; et al. Diagnostic Performance and Usability of the Genedrive((R)) HCV ID Kit in Two Decentralized Settings in Cameroon and Georgia. Diagnostics 2021, 11, 746. [Google Scholar] [CrossRef]
- FIND. Prospective Evaluation of the Genedrive® HCV ID Kit in Georgia; HIV-NAT: Bangkok, Thailand, 2018. [Google Scholar]
- Bajis, S.; Dore, G.J.; Hajarizadeh, B.; Cunningham, E.B.; Maher, L.; Grebely, J. Interventions to enhance testing, linkage to care and treatment uptake for hepatitis C virus infection among people who inject drugs: A systematic review. Int. J. Drug Policy 2017, 47, 34–46. [Google Scholar] [CrossRef]
- Chevaliez, S.; Wlassow, M.; Volant, J.; Roudot-Thoraval, F.; Bachelard, A.; Poiteau, L.; Trabut, J.-B.; Hézode, C.; Bourdel, A.; Dominguez, S. Assessing molecular point-of-care testing and dried blood spot for hepatitis C virus screening in people who inject drugs. Open Forum Infect. Dis. 2020, 7, 196. [Google Scholar] [CrossRef]
- Bristow, C.C.; Kojima, N.; Lee, S.-J.; Leon, S.R.; Ramos, L.B.; Konda, K.A.; Brown, B.; Caceres, C.F.; Klausner, J.D. HIV and syphilis testing preferences among men who have sex with men and among transgender women in Lima, Peru. PLoS ONE 2018, 13, e0206204. [Google Scholar] [CrossRef]
- Trickey, A.; Fajardo, E.; Alemu, D.; Artenie, A.A.; Easterbrook, P. Impact of hepatitis C virus (HCV) point-of-care viral load testing compared to laboratory-based testing on uptake of testing and treatment, and turnaround times: A systematic review and meta-analysis. In Proceedings of the WHO HCV Guideline Development Meeting, Geneva, Switzerland, 18 October 2021. [Google Scholar]
- Kronfli, N.; Linthwaite, B.; Kouyoumdjian, F.; Klein, M.B.; Lebouché, B.; Sebastiani, G.; Cox, J. Interventions to increase testing, linkage to care and treatment of hepatitis C virus (HCV) infection among people in prisons: A systematic review. Int. J. Drug Policy 2018, 57, 95–103. [Google Scholar] [CrossRef]
- Harris, M.; Rhodes, T.; Martin, A. Taming systems to create enabling environments for HCV treatment: Negotiating trust in the drug and alcohol setting. Soc. Sci. Med. 2013, 83, 19–26. [Google Scholar] [CrossRef]
- Operskalski, E.A.; Kovacs, A. HIV/HCV co-infection: Pathogenesis, clinical complications, treatment, and new therapeutic technologies. Curr. HIV/AIDS Rep. 2011, 8, 12–22. [Google Scholar] [CrossRef] [Green Version]
- Pappalardo, B.L. Influence of maternal human immunodeficiency virus (HIV) co-infection on vertical transmission of hepatitis C virus (HCV): A meta-analysis. Int. J. Epidemiol. 2003, 32, 727–734. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Consolidated Guidelines on HIV Testing Services; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- World Health Organization. Xpert MTB/RIF Assay for the Diagnosis of Pulmonary and Extrapulmonary TB in Adults and Children; WHO: Geneva, Switzerland, 2013. [Google Scholar]
| Xpert HCV Viral Load (n = 16) No. Studies (% of Studies, Total Sample) | Genedrive HCV ID Kit (n = 6) No. Studies (% of Studies, Total Sample) | Truenat and SAMBA (n = 3) No. Studies (% of Studies, Total Sample) | |
|---|---|---|---|
| Publication year (range) | 2017–2021 | 2018–2021 | 2019–2021 |
| Study setting according to WHO geographic regions | |||
| European region | 7 (35.0%, 1764) 1 | 3 (33.3%, 1707) 2 | 2 (40.0%, 672) 3 |
| Western Pacific region | 6 (30.0%, 1139) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| Americas region | 2 (10.0%, 816) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| South-East Asian region | 3 (15.0%, 451) | 1 (11.1%, 320) | 2 (40.0%, 696) |
| African region | 2 (10.0%, 385) | 5 (55.6%, 869) | 0 (0.0%, 0) |
| Eastern Mediterranean region | 0 (0.0%, 0) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| Not reported | 0 (0.0%, 0) | 0 (0.0%, 0) | 1 (20.0%, 160) |
| Study country income classification 4 | |||
| High income | 11 (61.1%, 2135) 5 | 1 (11.1%, 1011) 6 | 0 (0.0%, 0) |
| Upper-middle income | 1 (5.6%, 380) | 3 (33.3%, 822) | 1 (20.0%, 594) 7 |
| Lower-middle income | 5 (27.8%, 1238) | 5 (55.6%, 1063) | 3 (60.0%, 774) |
| Low income | 1 (5.6%, 188) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| Not reported | 0 (0.0%, 0) | 0 (0.0%, 0) | 1 (20.0%, 160) |
| Year of specimen collection | |||
| 2012–2014 | 2 (12.5%, 783) | 0 (0.0%, 0,) | 0 (0.0%, 0) |
| 2015–2016 | 4 (25.0%, 543) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| 2017–2019 | 9 (56.3%, 2514) | 4 (66.7%, 908) | 1 (33.3%, 907) |
| Not reported | 1 (6.3%, 101) | 2 (33.3%, 1562) | 2 (66.7%, 621) |
| Study setting for recruitment | |||
| HCV clinics | 9 (52.9%, 2835) 8 | 2 (28.6%, 449) | 1 (33.3%, 907) |
| Drug treatment sites | 6 (35.3%, 803) | 2 (28.6%, 696)9 | 0 (0.0%, 0) |
| Not reported | 2 (11.8%, 303) | 3 (42.9%, 1325) | 2 (66.7%, 621) |
| Study design | |||
| Case-control | 1 (6.3%, 101) | 3 (50.0%, 1299) | 0 (0.0%, 0) |
| Cross-sectional | 7 (43.8%, 2699) | 3 (50.0%, 2023) | 3 (100.0%, 1528) |
| Cohort study | 8 (50.0%, 1141) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| HCV antibody status and population group | |||
| Known HCV-positive | 6 (37.5%, 1712) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| Unknown HCV status | 10 (62.5%, 2229) | 6 (100.0%, 2470) | 3 (100.0%, 1528) |
| Other high-risk | 2 (20.0%, 1156) | 1 (16.7%, 425) | 1 (25.0%, 313) 10 |
| PWID | 6 (60.0%, 803) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| General population | 2 (20.0%, 270) | 5 (83.3%, 2045) | 3 (75.0%, 1215) |
| Specimen type | |||
| Fingerstick capillary whole blood 11 | 9 (47.4%, 1960) 12 | 0 (0.0%, 0) | 2 (28.6%, 1018) 13 |
| Serum or plasma | 8 (42.1%, 2088) | 6 (100.0%, 2470) | 3 (42.9%, 1412) |
| Venous whole blood | 2 (10.5%, 1098) | 0 (0.0%, 0) | 2 (28.6%, 359) |
| Frozen or fresh samples | |||
| Fresh samples only | 11 (68.8%, 2849) | 3 (42.9%, 947) 15 | 2 (50.0%, 1067) 16 |
| Frozen samples only | 3 (18.8%, 424) | 4 (57.1%, 1523) | 2 (50.0%, 461) |
| Fresh or frozen samples14 | 1 (6.3%, 614) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| Not reported | 1 (6.3%, 54) | 0 (0.0%, 0) | 0 (0.0%, 0) |
| Xpert HCV VL Fingerprick assay | |||
| Yes | 6 (33.3%, 1453) 17 | N/A | N/A |
| No | 12 (66.7%, 2655) | N/A | N/A |
| Xpert HCV Viral Load | Genedrive HCV ID Kit | Truenat and SAMBA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary Sensitivity % (95% CI) | p-Value | Summary Specificity % (95% CI) | p-Value | p for Joint Model | Summary Sensitivity % (95% CI) | p-Value | Summary Specificity % (95% CI) | p-Value | p for Joint Model | Summary Sensitivity % (95% CI) | p-Value | Summary Specificity % (95% CI) | p-Value | p for Joint Model | |
| Overall | 99 (98–99) | 99 (98–100) | 99 (98–100) | 100 (99–100) | 95 (94–97) | 99 (99–100) | |||||||||
| Study setting according to WHO geographic regions | |||||||||||||||
| European region | 98 (97–99) | Ref | 99 (97–100) | Ref | Ref | 99 (97–100) | Ref | 100 (99–100) | Ref | Ref | 96 (94–98) | Ref | 99 (98–100) | Ref | Ref |
| Western Pacific region | 100 (96–100) | <0.001 *** | 98 (96–99) | 0.42 | 0.02 * | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Americas region | 99 (97–100) | 0.04 * | 100 (73–100) | 0.11 | 0.16 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| South-East Asian region | 98 (97–99) | <0.001 *** | 99 (97–100) | 0.75 | 0.78 | 100 (96–100) | 0.64 | 99 (98–100) | 0.10 | 0.94 | 93 (90–96) | <0.001 *** | 99 (98–100) | <0.01 ** | 0.14 |
| African region | 100 (66–100) | 0.79 | 95 (81–99) | <0.01 ** | 0.28 | 100 (98–100) | <0.001 *** | 100 (98–100) | <0.001 *** | 0.13 | N/A | N/A | N/A | N/A | N/A |
| Study country income classification | |||||||||||||||
| High income | 99 (98–99) | Ref | 99 (98–100) | Ref | Ref | 99 (97–99) | Ref | 100 (99–100) | Ref | Ref | N/A | N/A | N/A | N/A | N/A |
| Upper-middle income | 96 (95–98) | 0.08 | 100 (99–100) | <0.001 *** | 0.09 | 100 (95–100) | 0.9 | 99 (98–100) | <0.001 *** | 0.07 | 96 (94–98) | Ref | 99 (99–100) | Ref | Ref |
| Lower-middle income | 99 (97–100) | 0.04 * | 97 (95–99) | 0.7 | 0.52 | 100 (96–100) | 0.46 | 100 (99–100) | <0.001 *** | 0.2 | 94 (92–95) | <0.001 *** | 99 (97–99) | 0.02 * | 0.23 |
| Low income | 99 (95–100) | 0.47 | 99 (93–100) | 0.02 * | 0.98 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Year of specimen collection | |||||||||||||||
| 2012–2014 | 100 (99–100) | Ref | 92 (88–95) | Ref | Ref | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 2015–2016 | 98 (95–100) | <0.001 *** | 99 (95–100) | <0.001 *** | <0.01 ** | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 2017–2019 | 99 (98–99) | <0.01 ** | 100 (99–100) | 0.03 | 0.05 * | 100 (96–100) | N/A | 99 (99–100) | N/A | N/A | 95 (94–97) | Ref | 99 (99–100) | Ref | Ref |
| Study setting for recruitment | |||||||||||||||
| HCV clinics | 99 (98–100) | Ref | 99 (97–100) | Ref | Ref | 100 (98–100) | Ref | 100 (98–100) | Ref | Ref | 95 (94–97) | Ref | 99 (99–100) | Ref | Ref |
| Drug treatment sites | 99 (97–100) | 0.08 | 99 (98–99) | 0.01 ** | 0.56 | 100 (95–100) | <0.01 ** | 99 (98–100) | <0.001 *** | 0.19 | N/A | N/A | N/A | N/A | N/A |
| Study design | |||||||||||||||
| Cross-sectional | 99 (98–99) | Ref | 100 (99–100) | Ref | Ref | 99 (95–100) | Ref | 99 (99–100) | Ref | Ref | 95 (94–97) | Ref | 99 (99–100) | Ref | Ref |
| Cohort study | 99 (96–99) | 0.02 * | 98 (96–99) | 0.65 | 0.05 * | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Casecontrol | 100 (100–100) | <0.001 *** | 100 (100–100) | <0.001 *** | 0.3 | 99 (99–99) | 0.84 | 100 (100–100) | <0.001 *** | 0.11 | N/A | N/A | N/A | N/A | N/A |
| HCV antibody status and Population group | |||||||||||||||
| Known HCV-positive | 100 (96–100) | Ref | 94 (90–97) | Ref | Ref | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Unknown HCV status | 98 (97–99) | 0.95 | 99 (99–100) | <0.001 *** | 0.01 ** | 99 (98–100) | N/A | 100 (99–100) | N/A | N/A | 95 (94–97) | Ref | 99 (99–100) | Ref | Ref |
| Other high risk | 98 (97–99) | Ref | 100 (99–100) | Ref | Ref | 97 (92–100) | Ref | 100 (99–100) | Ref | Ref | 94 (91–96) | Ref | 98 (95–100) | Ref | Ref |
| PWID | 99 (97–100) | <0.001 *** | 99 (98–99) | 0.48 | 0.25 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| General population | 100 (100–100) | <0.001 *** | 98 (89–100) | <0.001 *** | 0.03 * | 100 (98–100) | 0.18 | 100 (99–100) | 0.31 | 0.20 | 96 (94–97) | <0.001 *** | 99 (99–100) | <0.01 ** | 0.26 |
| Specimen type | |||||||||||||||
| Fingerstick capillary whole blood | 98 (97–99) | Ref | 99 (98–100) | Ref | Ref | N/A | N/A | N/A | N/A | N/A | 92 (89–95) | Ref | 100 (98–100) | Ref | Ref |
| Serum or plasma | 100 (97–100) | 0.02 * | 96 (93–98) | 0.87 | 0.01 * | 99 (98–100) | N/A | 100 (99–100) | N/A | N/A | 100 (97–100) | <0.001 *** | 96 (93–98) | 0.55 | 0.07 |
| Venous whole blood | 98 (96–99) | <0.001 *** | 100 (98–100) | 0.78 | 0.87 | N/A | N/A | N/A | N/A | N/A | 96 (93–98) | <0.001 *** | 99 (97–100) | 0.53 | 0.34 |
| Frozen or Fresh samples | |||||||||||||||
| Fresh samples only | 99 (98–99) | Ref | 99 (99–100) | Ref | Ref | 99 (95–100) | Ref | 99 (98–100) | Ref | Ref | 96 (94–97) | Ref | 99 (99–100) | Ref | Ref |
| Frozen samples only | 100 (55–100) | 0.86 | 91 (81–96) | 0.02 * | 0.04 * | 99 (98–100) | 0.48 | 100 (100–100) | <0.001 *** | 0.02 * | 93 (88–99) | 0.03 * | 99 (98–100) | 0.16 | 0.6 |
| Fresh or frozen samples | 99 (95–100) | 0.98 | 99 (93–100) | <0.01 ** | 0.17 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Xpert HCV VL Fingerprick assay | |||||||||||||||
| Yes | 98 (97–99) | Ref | 100 (99–100) | Ref | Ref | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| No | 100 (98–100) | <0.001 *** | 97 (94–98) | 0.84 | <0.01 ** | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, W.; Tao, Y.; Fajardo, E.; Reipold, E.I.; Chou, R.; Tucker, J.D.; Easterbrook, P. Diagnostic Accuracy of Point-of-Care HCV Viral Load Assays for HCV Diagnosis: A Systematic Review and Meta-Analysis. Diagnostics 2022, 12, 1255. https://doi.org/10.3390/diagnostics12051255
Tang W, Tao Y, Fajardo E, Reipold EI, Chou R, Tucker JD, Easterbrook P. Diagnostic Accuracy of Point-of-Care HCV Viral Load Assays for HCV Diagnosis: A Systematic Review and Meta-Analysis. Diagnostics. 2022; 12(5):1255. https://doi.org/10.3390/diagnostics12051255
Chicago/Turabian StyleTang, Weiming, Yusha Tao, Emmanuel Fajardo, Elena Ivanova Reipold, Roger Chou, Joseph D. Tucker, and Philippa Easterbrook. 2022. "Diagnostic Accuracy of Point-of-Care HCV Viral Load Assays for HCV Diagnosis: A Systematic Review and Meta-Analysis" Diagnostics 12, no. 5: 1255. https://doi.org/10.3390/diagnostics12051255
APA StyleTang, W., Tao, Y., Fajardo, E., Reipold, E. I., Chou, R., Tucker, J. D., & Easterbrook, P. (2022). Diagnostic Accuracy of Point-of-Care HCV Viral Load Assays for HCV Diagnosis: A Systematic Review and Meta-Analysis. Diagnostics, 12(5), 1255. https://doi.org/10.3390/diagnostics12051255







