Image Quantification for TSPO PET with a Novel Image-Derived Input Function Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Standard Protocol Approvals, Registrations, and Patient Consents
2.2. Participants
2.3. Data Acquisition for the Cohort with Arterial Blood Sampling Procedures
2.4. Data Acquisition for the Cohort without Arterial Blood Sampling
2.5. Image Post-Processing
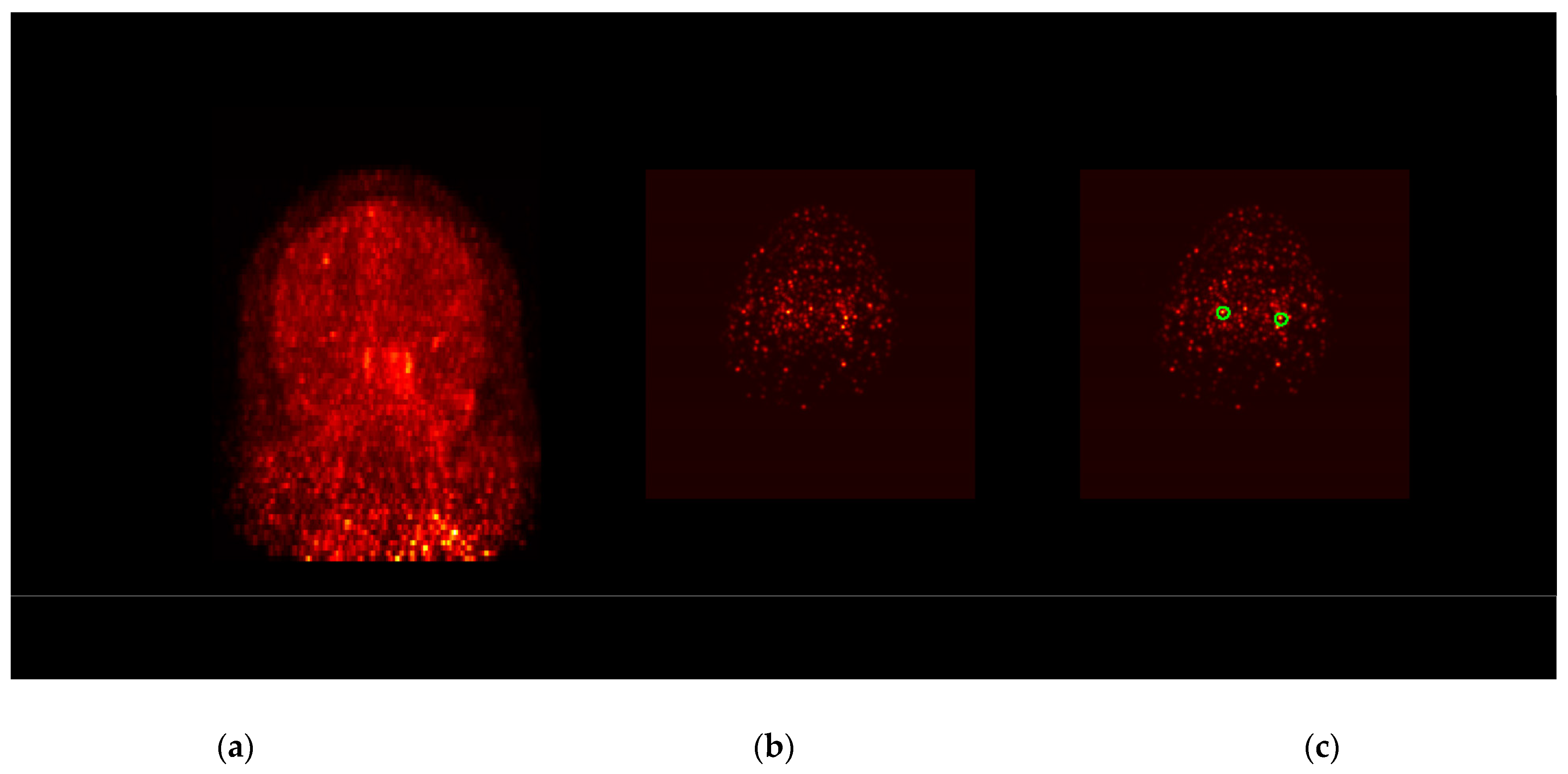
2.6. IDIF Extraction Procedure
2.7. Metabolite Correction
2.8. 18F-DPA-714 Quantification
2.9. Statistical Analysis
3. Results
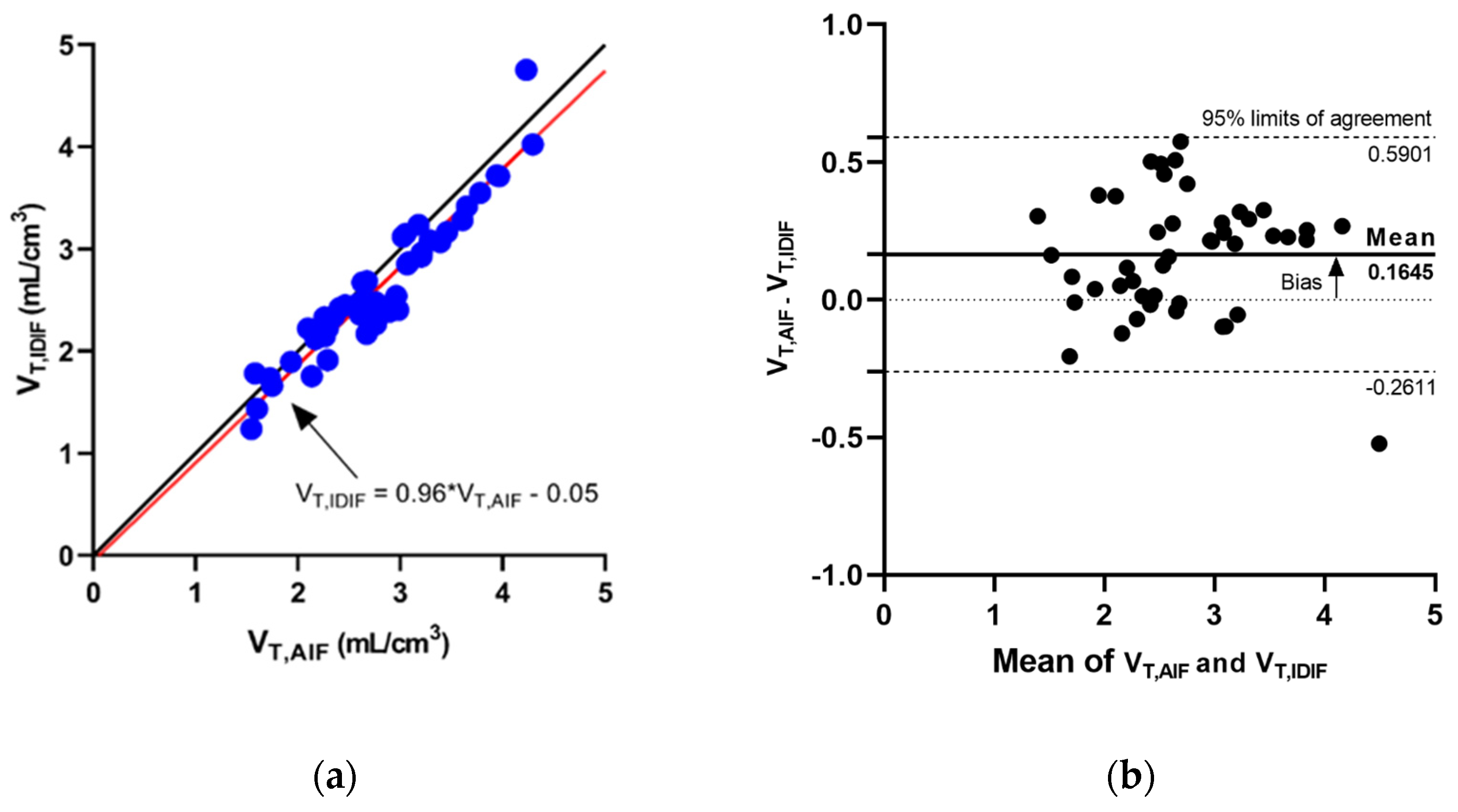
3.1. IDIF Predicts AIF
3.2. IDIF Method Distinguishes High-Affinity Binders from Mixed-Affinity Binders
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ranasoho, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef]
- Latta, C.H.; Brothers, H.M.; Wilcock, D.M. Neuroinflammation in Alzheimer’s disease; A source of heterogeneity and target for personalized therapy. Neuroscience 2015, 302, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, E.C.; Hunot, S. Neuroinflammation in Parkinson’s disease: A target for neuroprotection? Lancet Neurol. 2009, 8, 382–397. [Google Scholar] [CrossRef]
- Hirsch, E.C.; Standaert, D.G. Ten Unsolved Questions about Neuroinflammation in Parkinson’s Disease. Mov. Disord. 2021, 36, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Chaney, A.M.; Carlson, M.L.; Jackson, I.M.; Rao, A.; James, M.L. Neuroinflammation PET Imaging: Current Opinion and Future Directions. J. Nucl. Med. 2020, 61, 1107–1112. [Google Scholar] [CrossRef]
- Werry, E.L.; Bright, F.M.; Piguet, O.; Ittner, L.M.; Halliday, G.M.; Hodges, J.R.; Kiernan, M.C.; Loy, C.T.; Kril, J.J.; Kassiou, M. Recent Developments in TSPO PET Imaging as a Biomarker of Neuroinflammation in Neurodegenerative Disorders. Int. J. Mol. Sci. 2019, 20, 3161. [Google Scholar] [CrossRef] [PubMed]
- De Picker, L.J.; Haarman, B.C.M. Applicability, potential and limitations of TSPO PET imaging as a clinical immunopsychiatry biomarker. Eur. J. Nucl. Med. Mol. Imaging 2021, 49, 164–173. [Google Scholar] [CrossRef]
- Arlicot, N.; Vercouillie, J.; Ribeiro, M.-J.; Tauber, C.; Venel, Y.; Baulieu, J.-L.; Maia, S.; Corcia, P.; Stabin, M.G.; Reynolds, A.; et al. Initial evaluation in healthy humans of [18F]DPA-714, a potential PET biomarker for neuroinflammation. Nucl. Med. Biol. 2012, 39, 570–578. [Google Scholar] [CrossRef]
- Corcia, P.; Tauber, C.; Vercouillie, J.; Arlicot, N.; Prunier, C.; Praline, J.; Nicolas, G.; Venel, Y.; Hommet, C.; Baulieu, J.-L.; et al. Molecular imaging of microglial activation in amyotrophic lateral sclerosis. PLoS ONE 2012, 7, e52941. [Google Scholar] [CrossRef]
- Kropholler, M.A.; Boellaard, R.; Schuitemaker, A.; Folkersma, H.; van Berckel, B.N.; Lammertsma, A.A. Evaluation of reference tissue models for the analysis of [11C](R)-PK11195 studies. J. Cereb. Blood Flow Metab. 2006, 26, 1431–1441. [Google Scholar] [CrossRef]
- Lyoo, C.H.; Ikawa, M.; Liow, J.-S.; Zoghbi, S.S.; Morse, C.L.; Pike, V.W.; Fujita, M.; Innis, R.B.; Kreisl, W.C. Cerebellum Can Serve As a Pseudo-Reference Region in Alzheimer Disease to Detect Neuroinflammation Measured with PET Radioligand Binding to Translocator Protein. J. Nucl. Med. 2015, 56, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Lavisse, S.; García-Lorenzo, D.; Peyronneau, M.-A.; Bodini, B.; Thiriez, C.; Kuhnast, B.; Comtat, C.; Remy, P.; Stankoff, B.; Bottlaender, M. Optimized Quantification of Translocator Protein Radioligand (1)(8)F-DPA-714 Uptake in the Brain of Genotyped Healthy Volunteers. J. Nucl. Med. 2015, 56, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.; Guo, Q.; Kalk, N.; Colasanti, A.; Kalogiannopoulou, D.; Dimber, R.; Lewis, Y.L.; Libri, V.; Barletta, J.; Ramada-Magalhaes, J.; et al. Determination of [(11)C]PBR28 binding potential in vivo: A first human TSPO blocking study. J. Cereb. Blood Flow Metab. 2014, 34, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Salinas, C.A.; Searle, G.E.; Gunn, R.N. The simplified reference tissue model: Model assumption violations and their impact on binding potential. J. Cereb. Blood Flow Metab. 2015, 35, 304–311. [Google Scholar] [CrossRef]
- Gerhard, A.; Trender-Gerhard, I.; Turkheimer, F.; Quinn, N.P.; Bhatia, K.P.; Brooks, D.J. In Vivo imaging of microglial activation with [11C](R)-PK11195 PET in progressive supranuclear palsy. Mov. Disord. 2006, 21, 89–93. [Google Scholar] [CrossRef]
- Edison, P.; Archer, H.A.; Gerhard, A.; Hinz, R.; Pavese, N.; Turkheimer, F.E.; Hammers, A.; Tai, Y.F.; Fox, N.; Kennedy, A.; et al. Microglia, amyloid, and cognition in Alzheimer’s disease: An [11C](R)PK11195-PET and [11C]PIB-PET study. Neurobiol. Dis. 2008, 32, 412–419. [Google Scholar] [CrossRef]
- Varrone, A.; Oikonen, V.; Forsberg, A.; Joutsa, J.; Takano, A.; Solin, O.; Haaparanta-Solin, M.; Nag, S.; Nakao, R.; Al-Tawil, N.; et al. Positron emission tomography imaging of the 18-kDa translocator protein (TSPO) with [18F]FEMPA in Alzheimer’s disease patients and control subjects. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 438–446. [Google Scholar] [CrossRef]
- Hillmer, A.T.; Sandiego, C.M.; Hannestad, J.; Angarita, G.; Kumar, A.; McGovern, E.M.; Huang, Y.; O’Connor, K.C.; Carson, R.; O’Malley, S.S.; et al. In Vivo imaging of translocator protein, a marker of activated microglia, in alcohol dependence. Mol. Psychiatry 2017, 22, 1759–1766. [Google Scholar] [CrossRef]
- Mabrouk, R.; Rusjan, P.; Mizrahi, R.; Jacobs, M.F.; Koshimori, Y.; Houle, S.; Ko, J.H.; Strafella, A.P. Image derived input function for [18F]-FEPPA: Application to quantify translocator protein (18 kDa) in the human brain. PLoS ONE 2014, 9, e115768. [Google Scholar] [CrossRef]
- Zanotti-Fregonara, P.; Liow, J.-S.; Fujita, M.; Dusch, E.; Zoghbi, S.S.; Luong, E.; Boellaard, R.; Pike, V.W.; Comtat, C.; Innis, R.B. Image-derived input function for human brain using high resolution PET imaging with [C](R)-rolipram and [C]PBR28. PLoS ONE 2011, 6, e17056. [Google Scholar] [CrossRef]
- Macaskill, M.G.; Walton, T.; Williams, L.; Morgan, T.E.F.; Alcaide-Corral, C.J.; Dweck, M.R.; Gray, G.A.; Newby, D.E.; Lucatelli, C.; Sutherland, A.; et al. Kinetic modelling and quantification bias in small animal PET studies with [18F]AB5186, a novel 18 kDa translocator protein radiotracer. PLoS ONE 2019, 14, e0217515. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.H.; Muzic, R.F., Jr. Spillover and partial-volume correction for image-derived input functions for small-animal 18F-FDG PET studies. J. Nucl. Med. 2008, 49, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Wimberley, C.; Nguyen, D.L.; Truillet, C.; Peyronneau, M.-A.; Gulhan, Z.; Tonietto, M.; Boumezbeur, F.; Boisgard, R.; Chalon, S.; Bouilleret, V.; et al. Longitudinal mouse-PET imaging: A reliable method for estimating binding parameters without a reference region or blood sampling. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2589–2601. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.R.; Yeo, A.J.; Gunn, R.N.; Song, K.; Wadsworth, G.; Lewis, A.; Rhodes, C.; Pulford, D.J.; Bennacef, I.; Parker, C.A.; et al. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J. Cereb. Blood Flow Metab. 2012, 32, 1–5. [Google Scholar] [CrossRef]
- Lavisse, S.; Goutal, S.; Wimberley, C.; Tonietto, M.; Bottlaender, M.; Gervais, P.; Kuhnast, B.; Peyronneau, M.-A.; Barret, O.; Lagarde, J.; et al. Increased microglial activation in patients with Parkinson disease using [(18)F]-DPA714 TSPO PET imaging. Parkinsonism Relat. Disord. 2021, 82, 29–36. [Google Scholar] [CrossRef]
- Tanner, C.; Gilley, D.; Goetz, C. A brief screening questionnaire for parkinsonism. Ann. Neurol. 1990, 28, 267–268. [Google Scholar]
- Sekine, T.; ter Voert, E.E.; Warnock, G.; Buck, A.; Huellner, M.W.; Veit-Haibach, P.; Delso, G. Clinical Evaluation of Zero-Echo-Time Attenuation Correction for Brain 18F-FDG PET/MRI: Comparison with Atlas Attenuation Correction. J. Nucl. Med. 2016, 57, 1927–1932. [Google Scholar] [CrossRef]
- Rousset, O.G.; Ma, Y.; Evans, A.C. Correction for partial volume effects in PET: Principle and validation. J. Nucl. Med. 1998, 39, 904–911. [Google Scholar]
- Thomas, B.; Cuplov, V.; Bousse, A.; Mendes, A.; Thielemans, K.; Hutton, B.F.; Erlandsson, K. PETPVC: A toolbox for performing partial volume correction techniques in positron emission tomography. Phys. Med. Biol. 2016, 61, 7975–7993. [Google Scholar] [CrossRef]
- Fischl, B. FreeSurfer. Neuroimage 2012, 62, 774–781. [Google Scholar] [CrossRef]
- Krejza, J.; Arkuszewski, M.; Kasner, S.E.; Weigele, J.; Ustymowicz, A.; Hurst, R.W.; Cucchiara, B.L.; Messe, S.R. Carotid artery diameter in men and women and the relation to body and neck size. Stroke 2006, 37, 1103–1105. [Google Scholar] [CrossRef] [PubMed]
- Vendrow, J.; Haddock, J.; Rebrova, E.; Needell, D. On a guided nonnegative matrix factorization. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021. [Google Scholar]
- Zhou, Y.; Li, Z.; Yang, X.; Zhang, L.; Srivastava, S.; Jin, R.; Chan, C. Using Knowledge Driven Matrix Factorization to Reconstruct Modular Gene Regulatory Network. In Proceedings of the Twenty-Third AAAI Conference on Artificial Intelligence, AAAI 2008, Chicago, IL, USA, 13–17 July 2008. [Google Scholar]
- Feng, D.; Wong, K.-P.; Wu, C.-M.; Siu, W.-C. A technique for extracting physiological parameters and the required input function simultaneously from PET image measurements: Theory and simulation study. IEEE Trans. Inf. Technol. Biomed. 1997, 1, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-C.; Phelps, M.E.; Hoffman, E.J.; Sideris, K.; Selin, C.J.; Kuhl, D.E. Noninvasive determination of local cerebral metabolic rate of glucose in man. Am. J. Physiol.-Endocrinol. Metab. 1980, 238, E69–E82. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lin, S.F.; Gallezot, J.D.; Chan, C.; Prasad, R.; Thorn, S.L.; Stacy, M.R.; Huang, Y.; Zonouz, T.H.; Liu, Y.-H.; et al. Quantitative Analysis of Dynamic 123I-mIBG SPECT Imaging Data in Healthy Humans with a Population-Based Metabolite Correction Method. J. Nucl. Med. 2016, 57, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Buchert, R.; Dirks, M.; Schütze, C.; Wilke, F.; Mamach, M.; Wirries, A.-K.; Pflugrad, H.; Hamann, L.; Langer, L.B.; Wetzel, C.; et al. Reliable quantification of (18)F-GE-180 PET neuroinflammation studies using an individually scaled population-based input function or late tissue-to-blood ratio. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2887–2900. [Google Scholar] [CrossRef]
- Feeney, C.; Scott, G.; Raffel, J.; Roberts, S.; Coello, C.; Jolly, A.; Searle, G.; Goldstone, A.; Brooks, D.; Nicholas, R.S.; et al. Kinetic analysis of the translocator protein positron emission tomography ligand [18F] GE-180 in the human brain. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 2201–2210. [Google Scholar] [CrossRef]
- Logan, J.; Fowler, J.S.; Volkow, N.D.; Wolf, A.P.; Dewey, S.L.; Schlyer, D.J.; MacGregor, R.R.; Hitzemann, R.; Bendriem, B.; Gatley, S.J.; et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(-)-cocaine PET studies in human subjects. J. Cereb. Blood Flow Metab. 1990, 10, 740–747. [Google Scholar] [CrossRef]
- Logan, J. Graphical analysis of PET data applied to reversible and irreversible tracers. Nucl. Med. Biol. 2000, 27, 661–670. [Google Scholar] [CrossRef]
- Golla, S.S.; Boellaard, R.; Oikonen, V.; Hoffmann, A.; van Berckel, B.N.; Windhorst, A.D.; Virta, J.; Haaparanta-Solin, M.; Luoto, P.; Savisto, N.; et al. Quantification of [18F]DPA-714 binding in the human brain: Initial studies in healthy controls and Alzheimer’s disease patients. J. Cereb. Blood Flow Metab. 2015, 35, 766–772. [Google Scholar] [CrossRef]
- Wimberley, C.; Lavisse, S.; Hillmer, A.; Hinz, R.; Turkheimer, F.; Zanotti-Fregonara, P. Kinetic modeling and parameter estimation of TSPO PET imaging in the human brain. Eur. J. Nucl. Med. Mol. Imaging 2021, 49, 246–256. [Google Scholar] [CrossRef]
- Vera, J.H.; Guo, Q.; Cole, J.H.; Boasso, A.; Greathead, L.; Kelleher, P.; Rabiner, E.A.; Kalk, N.; Bishop, C.; Gunn, R.N.; et al. Neuroinflammation in treated HIV-positive individuals: A TSPO PET study. Neurology 2016, 86, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Dimber, R.; Guo, Q.; Bishop, C.; Adonis, A.; Buckley, A.; Kocsis, A.; Owen, D.; Kalk, N.; Newbould, R.; Gunn, R.N.; et al. Evidence of Brain Inflammation in Patients with Human T-Lymphotropic Virus Type 1-Associated Myelopathy (HAM): A Pilot, Multimodal Imaging Study Using 11C-PBR28 PET, MR T1-Weighted, and Diffusion-Weighted Imaging. J. Nucl. Med. 2016, 57, 1905–1912. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Terada, T.; Yokokura, M.; Yoshikawa, E.; Futatsubashi, M.; Kono, S.; Konishi, T.; Miyajima, H.; Hashizume, T.; Ouchi, Y. Extrastriatal spreading of microglial activation in Parkinson’s disease: A positron emission tomography study. Ann. Nucl. Med. 2016, 30, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Matheson, G.J.; Plaven-Sigray, P.; Forsberg, A.; Varrone, A.; Farde, L.; Cervenka, S. Assessment of simplified ratio-based approaches for quantification of PET [(11)C]PBR28 data. EJNMMI Res. 2017, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- García-Lorenzo, D.; Lavisse, S.; Leroy, C.; Wimberley, C.; Bodini, B.; Remy, P.; Veronese, M.; Turkheimer, F.; Stankoff, B.; Bottlaender, M. Validation of an automatic reference region extraction for the quantification of [(18)F]DPA-714 in dynamic brain PET studies. J. Cereb. Blood Flow Metab. 2018, 38, 333–346. [Google Scholar] [CrossRef]
- Schubert, J.; Tonietto, M.; Turkheimer, F.; Zanotti-Fregonara, P.; Veronese, M. Supervised clustering for TSPO PET imaging. Eur. J. Nucl. Med. Mol. Imaging 2021, 49, 257–268. [Google Scholar] [CrossRef]
- Zanotti-Fregonara, P.; Kreisl, W.C.; Innis, R.B.; Lyoo, C.H. Automatic Extraction of a Reference Region for the Noninvasive Quantification of Translocator Protein in Brain Using (11)C-PBR28. J. Nucl. Med. 2019, 60, 978–984. [Google Scholar] [CrossRef]
- Plaven-Sigray, P.; Matheson, G.J.; Cselenyi, Z.; Jucaite, A.; Farde, L.; Cervenka, S. Test-retest reliability and convergent validity of (R)-[(11)C]PK11195 outcome measures without arterial input function. EJNMMI Res. 2018, 8, 102. [Google Scholar] [CrossRef]
- Su, Y.; Blazey, T.M.; Snyder, A.Z.; Raichle, M.E.; Hornbeck, R.C.; Aldea, P.; Morris, J.C.; Benzinger, T.L.S. Quantitative amyloid imaging using image-derived arterial input function. PLoS ONE 2015, 10, e0122920. [Google Scholar] [CrossRef]
- Kuttner, S.; Wickstrøm, K.K.; Lubberink, M.; Tolf, A.; Burman, J.; Sundset, R.; Jenssen, R.; Appel, L.; Axelsson, J. Cerebral blood flow measurements with (15)O-water PET using a non-invasive machine-learning-derived arterial input function. J. Cereb. Blood Flow Metab. 2021, 41, 2229–2241. [Google Scholar] [CrossRef]




| All (n = 5) | PD (n = 3) | HC (n = 2) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| VT,AIF | VT,IDIF | Error % | VT,AIF | VT,IDIF | Error % | VT,AIF | VT,IDIF | Error % | |
| Putamen | 2.44 ± 0.52 | 2.30 ± 0.52 | −5.7 ± 7.7 | 2.61 ± 0.47 | 2.43 ± 0.59 | −7.7 ± 9.7 | 2.19 ± 0.95 | 2.12 ± 0.79 | −2.7 ± 5.4 |
| Caudate | 1.91 ± 0.48 | 1.81 ± 0.50 | −5.4 ± 11.9 | 1.91 ± 0.60 | 1.83 ± 0.61 | −3.8 ± 16.4 | 1.93 ± 0.70 | 1.79 ± 0.73 | −7.6 ± 3.8 |
| Thalamus | 2.97 ± 0.63 | 2.77 ± 0.61 | −6.7 ± 7.0 | 3.12 ± 0.71 | 2.89 ± 0.75 | −7.6 ± 9.4 | 2.75 ± 1.17 | 2.59 ± 1.06 | −5.3 ± 1.8 |
| Hippocampus | 2.61 ± 0.58 | 2.42 ± 0.44 | −6.3 ± 9.3 | 2.79 ± 0.62 | 2.57 ± 0.45 | −6.6 ± 12.6 | 2.34 ± 0.96 | 2.19 ± 0.84 | −6.0 ± 3.0 |
| Frontal | 2.70 ± 0.49 | 2.55 ± 0.48 | −5.1 ± 7.5 | 2.79 ± 0.46 | 2.65 ± 0.54 | −5.2 ± 9.5 | 2.56 ± 0.88 | 2.41 ± 0.77 | −5.0 ± 3.2 |
| Temporal | 3.45 ± 0.86 | 3.34 ± 1.08 | −4.5 ± 11.5 | 3.73 ± 0.92 | 3.65 ± 1.33 | −4.2 ± 15.7 | 3.03 ± 1.29 | 2.86 ± 1.12 | −4.8 ± 2.8 |
| Occipital | 2.96 ± 0.55 | 2.78 ± 0.55 | −6.1 ± 7.3 | 3.15 ± 0.45 | 2.95 ± 0.57 | −6.5 ± 9.9 | 2.69 ± 1.05 | 2.52 ± 0.92 | −5.5 ± 4.8 |
| Parietal | 3.13 ± 0.52 | 2.97 ± 0.47 | −5.0 ± 7.0 | 3.26 ± 0.45 | 3.08 ± 0.51 | −5.7 ± 8.7 | 2.93 ± 0.97 | 2.79 ± 0.80 | −3.9 ± 4.1 |
| Cerebellum | 3.16 ± 0.60 | 2.95 ± 0.58 | −6.4 ± 7.1 | 3.35 ± 0.55 | 3.11 ± 0.67 | −7.4 ± 9.7 | 2.87 ± 1.14 | 2.70 ± 0.97 | −5.0 ± 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Y.-H.D.; McConathy, J.E.; Yacoubian, T.A.; Zhang, Y.; Kennedy, R.E.; Standaert, D.G. Image Quantification for TSPO PET with a Novel Image-Derived Input Function Method. Diagnostics 2022, 12, 1161. https://doi.org/10.3390/diagnostics12051161
Fang Y-HD, McConathy JE, Yacoubian TA, Zhang Y, Kennedy RE, Standaert DG. Image Quantification for TSPO PET with a Novel Image-Derived Input Function Method. Diagnostics. 2022; 12(5):1161. https://doi.org/10.3390/diagnostics12051161
Chicago/Turabian StyleFang, Yu-Hua Dean, Jonathan E. McConathy, Talene A. Yacoubian, Yue Zhang, Richard E. Kennedy, and David G. Standaert. 2022. "Image Quantification for TSPO PET with a Novel Image-Derived Input Function Method" Diagnostics 12, no. 5: 1161. https://doi.org/10.3390/diagnostics12051161
APA StyleFang, Y.-H. D., McConathy, J. E., Yacoubian, T. A., Zhang, Y., Kennedy, R. E., & Standaert, D. G. (2022). Image Quantification for TSPO PET with a Novel Image-Derived Input Function Method. Diagnostics, 12(5), 1161. https://doi.org/10.3390/diagnostics12051161






