A Powerful Paradigm for Cardiovascular Risk Stratification Using Multiclass, Multi-Label, and Ensemble-Based Machine Learning Paradigms: A Narrative Review
Abstract
:1. Introduction
2. Search Strategy and Statistical Distributions
2.1. PRISMA Model
2.2. Statistical Distribution
3. Biological Link between Atherosclerosis and Cardiovascular Disease
4. Three Paradigms for Cardiovascular Risk Stratification
4.1. Multiclass-Based Cardiovascular Disease Risk Stratification System
4.1.1. CVD-Based Multiclass Risk Assessment System
4.1.2. Comparison between CVD Application and Non-CVD Application
4.1.3. Multiclass CVD Architecture for Office-Based CVD Risk Stratification
4.1.4. Multiclass CVD Architecture for Cardiac Stress Laboratories
4.2. Multi-Label-Based Cardiovascular Disease Classification
4.2.1. Covariates and Risk Factors for Multi-Label-Based CVD Classification
4.2.2. Multi-Label-Based Architectures for CVD Risk Stratification
4.3. Ensemble-Based Cardiovascular Disease Classification
4.3.1. Different Classifier Combination for Ensemble-Based CVD Risk Stratification
4.3.2. Comparison between the Three Types of CVD Risk Assessment Systems
4.4. Performance Evaluation Metrics for Multiclass, Multi-Label, and Ensemble Techniques
5. Bias Distribution in the ML System for Multiclass, Multi-Label, and Ensemble
6. CVD Risk Assessment through Mobile, E-Health, and Cloud Techniques
7. Critical Discussion
7.1. Principal Findings
7.2. Benchmarking Table
7.3. A Special Note on Non-Linear CVD Risk Stratification
7.4. A Special Note on Time-to-Event for Cardiovascular Risk Prediction
7.5. A Special Note on the Advantages of Machine Learning-Based Cardiovascular Risk Stratification
7.6. A Special Note on Deep Learning-Based Cardiovascular Risk Stratification
7.7. The Future of Cardiovascular Disease Risk Stratification
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Pseudo-Code for Multiclass Classification
Appendix A.1. Typical Online System for CVD Risk Stratification for Multiclass

Appendix A.2. Pseudo-Code for Multiclass

Appendix B. Pseudo-Code for Multi-Label Classification
Appendix B.1. Problem Transformation Methods for Multi-Label Prediction
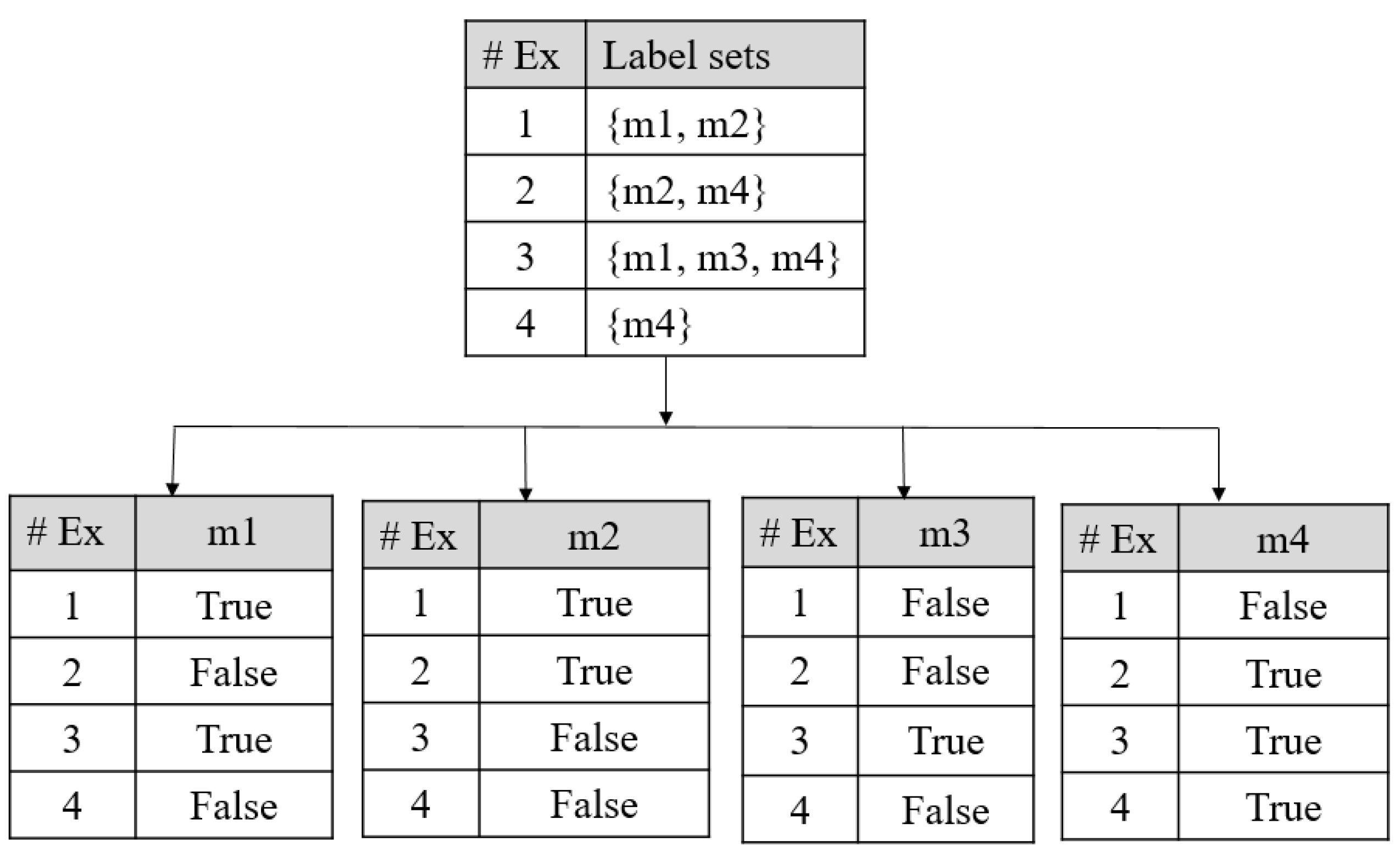
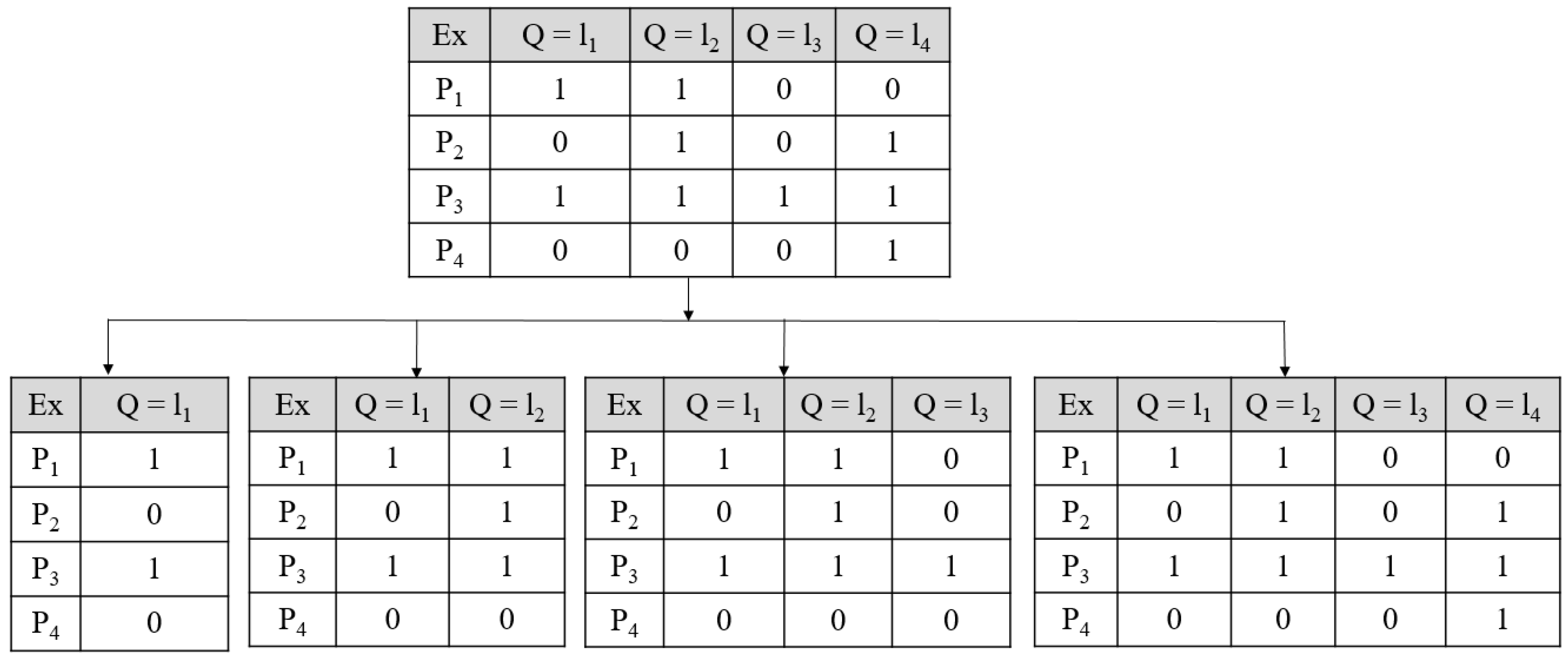
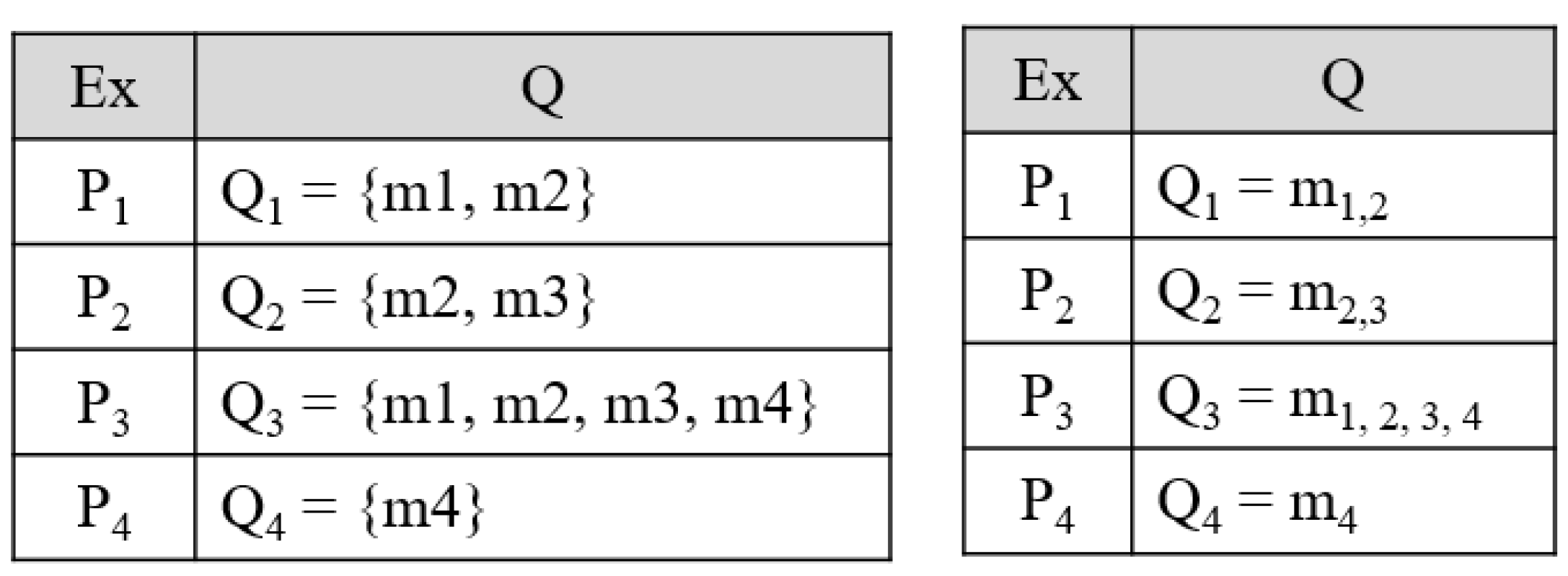
Appendix B.2. Algorithm Adaptation Methods for Multi-Label Prediction
Appendix B.3. Pseudo-Code for Multi-Label Classification Technique

Appendix C. Pseudo-Code for Ensemble Classification
Pseudo-Code for Ensemble-Based Technique

Appendix D. Comparison between 3 Paradigms
Comparison of ML-Based Multiclass, Multi-Label, and Ensemble CVD Classification
| SN | Attributes | Multiclass | Multi-Label | Ensemble | |||
| - | - | Characteristics | Characteristics | Characteristics | |||
| Total Studies | 14 | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | 8 | [83,84,85,86,87,88,89,90] | 32 [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | ||
| 1 | Data Size | 212–66,363 | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | 300–46,520 | [83,84,85,86,87,88,89,90] | 459–823,627 [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 2 | Risk Factors | Low | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | Large | [83,84,85,86,87,88,89,90] | Moderate [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 3 | Family History | Frequent Considered | [69,71,76,77,80,82] | Seldom Considered | [83,84,90] | Considered Intermittently [80,91,96,97,99,100,102,105,106,110,111,112,114,115,116,117,118,119,120] | |
| 4 | BMI | Less considered | [72,74,75,76,80] | Considered Moderately | [84,85,86] | Highly considered [46,47,48,49,50,51,52,80,91,93,94,95,96,97,99,100,102,106,107,112] | |
| 5 | Ethnicity | Less Considered | [72,74,75,76,80] | Considered Moderately | [84,85,86] | Highly Considered | |
| 6 | Type of data | OBBM and LBBM | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | OBBM, LBBM and Image | [83,84,85,86,87,88,89,90] | OBBM and LBBM [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 7 | Hypertension | Low Usage | [72,74,75,76,80] | High Usage | [83,84,85,86,87,88,89,90] | Moderate Usage [46,47,48,49,50,51,52,80,91,93,94,95,96,97,99,100,102,106,107,112] | |
| 8 | Smoking | Low Usage | [72,74,75,76,80] | High Usage | [83,84,85,86,87,88,89,90] | Moderate Usage [80,91,96,97,99,100,102,105,106,110,111,112,114,115,116,117,118,119,120] | |
| 9 | Multicenter | Low Usage | [72,74,75,76,80] | High Usage | [83,84,85,86,87,88,89,90] | Moderate Usage [80,91,96,97,99,100,102,105,106,110,111,112,114,115,116,117,118,119,120] | |
| 10 | MRI | Considered Moderately | [71,80] | Considered Moderately | [83,89] | Less Considered [80] | |
| 11 | ECG | Partial Considered | [72,74,75,78,79,81,82] | Strongly Considered | [83,86,87,89] | Not Considered | |
| 12 | CUSIP | Moderate Usage | Moderate Usage | Low Usage | |||
| 13 | # GT | Only 1 | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | Very high (10-4) | [83,84,85,86,87,88,89,90] | Average (1,2) | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] |
| 14 | # Algorithm | 🗶 | 🗸 | [83,84,85,86,87,88,89,90] | 🗶 | ||
| 15 | Type of Algorithm | 🗶 | - | 🗶 | |||
| 16 | # Classifiers | Ranging from 1–4 | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | Ranging from 1–9 | [83,84,85,86,87,88,89,90] | Ranging from 1–10 [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| SN | Attributes | Multiclass | Multi-label | Ensemble | |||
| - | - | Characteristics | Characteristics | Characteristics | |||
| 17 | Classifier Type | SVM, RF, CNN DT, k-NN Agatston classifier, Elastic Net, NN, NB, XGBoost SVM, ELM, OAO, OAA, DDAG, ECOC [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | RF, SVM, DT, KNN, LDA, LR, XGBoost, AdaBoost, GBA, Basic RNN, GRU RNN CNN, AAM [83,84,85,86,87,88,89,90] | kNN, GaussNB, LDA, QDA, RF, MLP, CNN, LSTM, GRU, BiLSTM, BiGRU Bagging, XGBoost, Adaboost, DNN, NB, NN, RS, GAMs, Elastic Net, GBMs, DT, CART, MARS, Logistic, EB, SMO, Boosting, MLDS, AVEn, MVEn, WAVEn, HTSA [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |||
| 18 | # Classes | 🗸 | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | 🗶 | 🗶 | ||
| 19 | Hyperparameters Used | 🗸 | [79] | 🗸 | [83,84,90] | 🗸 | [92,98,99,100] |
| 20 | Protocol | K-10 | [64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82] | K-10, K, K-5 | [83,84,85,86,87,88,89,90] | K-10, k, K-5 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] |
| 21 | # PE parameters | Ranging from 1–5 | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | Ranging from 1–8 | [83,84,85,86,87,88,89,90] | Ranging from 1–8 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] |
| 22 | Precision | 🗸 | [72,73,77,81,82] | 🗶 | 🗸 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 23 | PPV | 🗶 | 🗸 | [84,86] | 🗸 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 24 | NPV | 🗶 | 🗸 | [84,86] | 🗸 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 25 | FPR | 🗶 | 🗸 | [84,90] | 🗸 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 26 | FNR | 🗶 | 🗸 | [84] | 🗸 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 27 | Hamming Loss | 🗶 | 🗸 | [87] | 🗶 | ||
| 28 | C-index | 🗶 | 🗸 | [83] | 🗶 | ||
| 29 | Statistical Analysis | 🗶 | 🗸 | [83,84,85,86,87,88,89,90] | 🗸 | [80,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |
| 30 | Power Analysis | 🗶 | 🗸 | [83,84] | 🗶 | ||
| 31 | Hazard Analysis | 🗶 | 🗸 | [83] | 🗶 | ||
| 32 | Survival Test | 🗶 | 🗸 | [83] | 🗶 | ||
Appendix E. Performance Evaluation Metrics
Performance Evaluation Metrics Descriptions
| SN | Label-Based Performance Metrics | Mathematical Expression |
|---|---|---|
| 1 | Sensitivity (Sen), % | |
| 2 | Specificity (Spec), % | |
| 3 | Positive Predictive Rate (PPR), % | |
| 4 | Negative Predictive Rate (NPR), % | |
| 5 | False Predictive Value (FPV), % | |
| 6 | False Negative Value (FNV), % | |
| 7 | False Discovery Value, % | |
| 8 | F1-Score, % | |
| 9 | Accuracy (ACC), % |
| SN | Sample-Based Performance Metrics | Mathematical Expression |
|---|---|---|
| 1 | Hamming Loss, HL | |
| 2 | Jaccard Score, JS | |
| 3 | Precision, Pe | |
| 4 | Recall, Re | |
| 5 | F1-score, F1 | |
| 6 |
Appendix F. Power Analysis
Power Analysis for Multi-Label and Ensemble-Based CVD Risk Stratification
Appendix G. CVD Risk Assessment through Mobile, E-Health, and Cloud Techniques
Characteristic of Mobile and Cloud-Based CVD Systems
| C0 | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | ||||
| SN | Authors/Citations | ST | Year | Journal | DS | Diseases | FDA | SV | Comparator | |||
| 1 | Buss et al. [197] | SR | 2020 | JMIR | 7 ED | CVD, DIA | 🗶 | 🗶 | No (i.e., standard care), await list control, intervention | |||
| 2 | Villarreal et al. [198] | SR | 2020 | AIF | 44 | CVD | 🗶 | 🗶 | CVD, No CVD | |||
| 3 | Xiao et al. [199] | R | 2017 | TM | 151 | CVD | 🗶 | 🗶 | CVD, No CVD | |||
| 4 | Saba et al. [200] | R | 2018 | IHJ | 100 | CVD | 🗶 | 🗸 | CVD, No CVD | |||
| 5 | Lillo-Castellano et al. [208] | R | 2015 | JBHI | 6848 | CVD | 🗸 | 🗸 | CVD, No CVD | |||
| 6 | Huda et al. [201] | R | 2020 | TENSYMP | BIHAD | CVD | 🗶 | 🗸 | Normal ECG, Abnormal ECG | |||
| 7 | Sakellarios et al. [209] | R | 2018 | EMBC | 236 | CAD | 🗶 | 🗸 | No CAD, OCAD, Non-OCAD | |||
| 8 | Singh et al. [202] | R | 2019 | IEEEc | 2 | CVDa | 🗶 | 🗸 | Arrhythmia, CVD | |||
| 9 | Spanakis et al. [203] | R | 2020 | EMBC | 🗶 | CHF | 🗶 | 🗸 | CHF, No CHF | |||
| 10 | Paredes et al. [204] | R | 2018 | BIBM | 1600 | MI, CVD | 🗶 | 🗸 | Acute MI, No MI | |||
| 11 | Freyer et al. [205] | R | 2021 | AJH | 🗶 | AF | 🗶 | 🗸 | AF, No AF | |||
| 12 | Giansanti et al. [206] | S | 2021 | mHealth | 🗶 | CVD | 🗶 | 🗶 | Use of AI, non-use of AI | |||
| 13 | Park et al. [207] | R | 2014 | IEEEa | 🗶 | Arrhythmia | 🗶 | 🗶 | Arrhythmia, CVD | |||
| SN | Authors/Citations | Non ML/ML | Cloud | Mob | Sea | DE | Analysis | # O | OT | # C | Classifier | |
| 1 | Buss et al. [197] | Non-ML | 🗶 | 🗸 | 🗸 | 🗸 | 🗸 | 2 | Dia, CVD | 3 | 🗶 | |
| 2 | Villarreal et al. [198] | Non-ML | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | CVD | 2 | 🗶 | |
| 3 | Xiao et al. [199] | Non-ML | 🗶 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | CVD | 2 | 🗶 | |
| 4 | Saba et al. [200] | Non-ML | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | CVD | 2 | 🗶 | |
| 5 | Lillo-Castellano et al. [208] | ML | 🗸 | 🗶 | 🗸 | 🗸 | 🗸 | 1 | CVD | 2 | k-NN | |
| 6 | Huda et al. [201] | ML, DL | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | Arrhythmia | 2 | SVM, CNN | |
| 7 | Sakellarios et al. [209] | ML | 🗸 | 🗶 | 🗸 | 🗸 | 🗸 | 1 | CVD | 3 | SVM | |
| 8 | Singh et al. [202] | DL | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | CVDa | 2 | CNN | |
| 9 | Spanakis et al. [203] | IoT | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | CHF | 2 | 🗶 | |
| 10 | Paredes et al. [204] | CI | 🗶 | 🗸 | 🗸 | 🗸 | 🗸 | 2 | CVD, MI | 2 | Bayesian | |
| 11 | Freyer et al. [205] | Non-ML | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | AF | 2 | 🗶 | |
| 12 | Giansanti et al. [206] | AI | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | CVD | 2 | 🗶 | |
| 13 | Park et al. [207] | ML | 🗶 | 🗸 | 🗸 | 🗸 | 🗸 | 1 | Arrhythmia | 2 | DT, RF | |
| C0 | C19 | C20 | C21 | C22 | C23 | C24 | C25 | C26 | C27 | C28 | C29 | |
| SN | Authors/Citations | CV | Protocol | # PE | SEN | SPEC | Acc | Pre | F1 S | PV | SS | ROC |
| 1 | Buss et al. [197] | 🗶 | 🗶 | 0 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 2 | Villarreal et al. [198] | 🗶 | 🗶 | 0 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 3 | Xiao et al. [199] | 🗶 | 🗶 | 1 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 2.87 | 🗶 |
| 4 | Saba et al. [200] | 🗸 | 🗶 | 1 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 1 |
| 5 | Lillo-Castellano et al. [208] | 🗸 | K | 1 | 🗶 | 🗶 | 90 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 6 | Huda et al. [201] | 🗸 | 🗶 | 1 | 🗶 | 🗶 | 96 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 7 | Sakellarios et al. [209] | 🗸 | 🗶 | 3 | 44 | 98.7 | 85.1 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 8 | Singh et al. [202] | 🗶 | 🗶 | 1 | 🗶 | 🗶 | 97 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 9 | Spanakis et al. [203] | 🗶 | 🗶 | 1 | 🗶 | 🗶 | 1 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 10 | Paredes et al. [204] | 🗸 | 🗶 | 0 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 11 | Freyer et al. [205] | 🗸 | 🗶 | 1 | 🗶 | 🗶 | 1 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 12 | Giansanti et al. [206] | 🗸 | 🗶 | 0 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
| 13 | Park et al. [207] | 🗸 | 🗶 | 3 | 1 | 1 | 1 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 |
Appendix H. Miscellaneous Figures
Appendix H.1. Anatomical Link between the Carotid Artery and Aortic Arch and Typical Neural Network

| SN | Abb * | Definition | SN | Abb * | Definition |
|---|---|---|---|---|---|
| 1 | ACC | American college of cardiology | 42 | IPN | Intraplaque neovascularization |
| 2 | AD | Alzheimer’s | 43 | KNN | K-nearest neighbor |
| 3 | AHA | American heart association | 44 | LBBM | Laboratory-based biomarker |
| 4 | AI | Artificial intelligence | 45 | LP | Label Powerset |
| 5 | ANOVA | Analysis of variance | 46 | LSTM | Long short-term memory network |
| 6 | APG | Acceleration Plethysmogram | 47 | LVD | Large vessel disease |
| 7 | ASCVD | Atherosclerotic cardiovascular disease | 48 | MCI | Mild cognitive impairment |
| 8 | AUC | Area-under-the-curve | 49 | MedUSE | Medication use |
| 9 | BCVD | Binary CVD | 50 | MI | Myocardial Infarction |
| 10 | BMI | Body mass index | 51 | ML | Machine learning |
| 11 | BR | Binary recursive | 52 | MLARM | Multi-label adaptive resonance asso & map |
| 12 | CAC | Coronary artery calcification | 53 | MLkNN | Multi-label k nearest neighbor |
| 13 | RetiCAC | Deep learning Retinal CAC score | 54 | MPH | Maximum plaque height |
| 14 | CAD | Coronary artery disease | 55 | MRI | Magnetic resonance imaging |
| 15 | CAS | Coronary artery syndrome | 56 | NPV | Negative predictive value |
| 16 | CC | Classifier chain | 57 | Non-ML | Non-machine learning |
| 17 | CCVRC | Conventional cardiovascular risk cal # | 58 | OBBM | Office-based biomarker |
| 18 | CHD | Coronary Heart Disease | 59 | PCA | principal component analysis |
| 19 | CHD | Chronic Heart Conditions | 60 | PCE | Pooled cohort equation |
| 20 | cIMT | Carotid intima-media thickness | 61 | PE | Performance evaluation matrices |
| 21 | CKD | Chronic kidney disease | 62 | PMCI | Progressive MCI |
| 22 | CT | Computed tomography | 63 | PPV | Positive predictive value |
| 23 | CUSIP | Carotid ultrasound image phenotype | 64 | PTC | Plaque tissue characterization |
| 24 | CV | Cross-validation | 65 | QRISK3 | QResearch cardiovascular risk algorithm |
| 25 | CVD | Cardiovascular disease | 66 | RA | Rheumatoid arthritis |
| 26 | CVE | Cardiovascular events | 67 | RakEL | Random k-label set |
| 27 | DL | Deep learning | 68 | #RC | Risk classes |
| 28 | DM | Diabetes mellitus | 69 | RF | Random forest |
| 29 | DT | Decision tree | 70 | RoB | Risk-of-bias |
| 30 | ECG | Electrocardiogram | 71 | ROC | Receiver operating-characteristics |
| 31 | EEGS | Event-equivalent gold standard | 72 | RRS | Reynolds risk score |
| 32 | ESC | European society of cardiology | 73 | SCD | Sudden cardiac death |
| 33 | FH | Family history | 74 | SCG | Seismocardiography (SCG-Z) |
| 34 | FNR | False-negative rate | 75 | SCORE | Systematic coronary risk evaluation |
| 35 | FPR | False-positive rate | 76 | SCMI | Significant memory concern |
| 36 | FRS | Framingham risk score | 77 | SMOTE | Synthetic minority over-sampling tech. |
| 37 | GCG | Gyrocardiography | 78 | SVM | Support vector machine |
| 38 | GUI | Graphical user interface | 79 | TPA | Total plaque area |
| 39 | HTN | Hypertension | 80 | US | Ultrasound |
| 40 | IM | Image modalities | 81 | WHO | World health organization |
| 41 | IMTV | Intima-media thickness variability | - | - | - |
References
- Kaptoge, S.; Pennells, L.; de Bacquer, D.; Cooney, M.T.; Kavousi, M.; Stevens, G.; Riley, L.M.; Savin, S.; Khan, T.; Altay, S. World Health Organization cardiovascular disease risk charts: Revised models to estimate risk in 21 global regions. Lancet Glob. Health 2019, 7, e1332–e1345. [Google Scholar] [CrossRef]
- Dunbar, S.B.; Khavjou, O.A.; Bakas, T.; Hunt, G.; Kirch, R.A.; Leib, A.R.; Morrison, R.S.; Poehler, D.C.; Roger, V.L.; Whitsel, L.P. Projected costs of informal caregiving for cardiovascular disease: 2015 to 2035: A policy statement from the American Heart Association. Circulation 2018, 137, e558–e577. [Google Scholar] [CrossRef]
- Banchhor, S.K.; Londhe, N.D.; Araki, T.; Saba, L.; Radeva, P.; Khanna, N.N.; Suri, J.S. Calcium detection, its quantification, and grayscale morphology-based risk stratification using machine learning in multimodality big data coronary and carotid scans: A review. Comput. Biol. Med. 2018, 101, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, V.; Jamthikar, A.D.; Gupta, D.; Shanu, N.; Puvvula, A.; Khanna, N.N.; Saba, L.; Omerzum, T.; Viskovic, K.; Mavrogeni, S. Low-cost preventive screening using carotid ultrasound in patients with diabetes. Front. Biosci. 2020, 25, 1132–1171. [Google Scholar]
- Jamthikar, A.D.; Puvvula, A.; Gupta, D.; Johri, A.M.; Nambi, V.; Khanna, N.N.; Saba, L.; Mavrogeni, S.; Laird, J.R.; Pareek, G. Cardiovascular disease and stroke risk assessment in patients with chronic kidney disease using integration of estimated glomerular filtration rate, ultrasonic image phenotypes, and artificial intelligence: A narrative review. Int. Angiol. J. Int. Union Angiol. 2020, 40, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, V.; Jamthikar, A.D.; Gupta, D.; Puvvula, A.; Khanna, N.N.; Saba, L.; Viskovic, K.; Mavrogeni, S.; Turk, M.; Laird, J.R. Integration of estimated glomerular filtration rate biomarker in image-based cardiovascular disease/stroke risk calculator: A south Asian-Indian diabetes cohort with moderate chronic kidney disease. Int. Angiol. 2020, 39, 290–306. [Google Scholar] [CrossRef]
- Jamthikar, A.D.; Gupta, D.; Puvvula, A.; Johri, A.M.; Khanna, N.N.; Saba, L.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M. Cardiovascular risk assessment in patients with rheumatoid arthritis using carotid ultrasound B-mode imaging. Rheumatol. Int. 2020, 40, 1921–1939. [Google Scholar] [CrossRef]
- Konstantonis, G.; Singh, K.V.; Sfikakis, P.P.; Jamthikar, A.D.; Kitas, G.D.; Gupta, S.K.; Saba, L.; Verrou, K.; Khanna, N.N.; Ruzsa, Z. Cardiovascular disease detection using machine learning and carotid/femoral arterial imaging frameworks in rheumatoid arthritis patients. Rheumatol. Int. 2022, 42, 215–239. [Google Scholar] [CrossRef]
- Porcu, M.; Mannelli, L.; Melis, M.; Suri, J.S.; Gerosa, C.; Cerrone, G.; Defazio, G.; Faa, G.; Saba, L. Carotid plaque imaging profiling in subjects with risk factors (diabetes and hypertension). Cardiovasc. Diagn. Ther. 2020, 10, 1005–1018. [Google Scholar] [CrossRef]
- Saba, L.; Micheletti, G.; Brinjikji, W.; Garofalo, P.; Montisci, R.; Balestrieri, A.; Suri, J.; DeMarco, J.; Lanzino, G.; Sanfilippo, R. Carotid intraplaque-hemorrhage volume and its association with cerebrovascular events. Am. J. Neuroradiol. 2019, 40, 1731–1737. [Google Scholar] [CrossRef]
- Acharya, U.R.; Molinari, F.; Sree, S.V.; Chattopadhyay, S.; Ng, K.-H.; Suri, J.S. Automated diagnosis of epileptic EEG using entropies. Biomed. Signal Process. Control 2012, 7, 401–408. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Alvin, A.P.C.; Suri, J.S. Use of principal component analysis for automatic classification of epileptic EEG activities in wavelet framework. Expert Syst. Appl. 2012, 39, 9072–9078. [Google Scholar] [CrossRef]
- El-Hasnony, I.M.; Elzeki, O.M.; Alshehri, A.; Salem, H. Multi-Label Active Learning-Based Machine Learning Model for Heart Disease Prediction. Sensors 2022, 22, 1184. [Google Scholar] [CrossRef]
- Oresko, J.J.; Jin, Z.; Cheng, J.; Huang, S.; Sun, Y.; Duschl, H.; Cheng, A.C. A wearable smartphone-based platform for real-time cardiovascular disease detection via electrocardiogram processing. IEEE Trans. Inf. Technol. Biomed. 2010, 14, 734–740. [Google Scholar] [CrossRef]
- Panhuyzen-Goedkoop, N.M.; Wellens, H.J.; Verbeek, A.L.; Jørstad, H.T.; Smeets, J.R.; Peters, R.J.G. ECG criteria for the detection of high-risk cardiovascular conditions in master athletes. Eur. J. Prev. Cardiol. 2020, 7, 1529–1538. [Google Scholar] [CrossRef]
- Myers, P.D.; Scirica, B.M.; Stultz, C.M. Machine learning improves risk stratification after acute coronary syndrome. Sci. Rep. 2017, 7, 12692. [Google Scholar] [CrossRef]
- Cuadrado-Godia, E.; Jamthikar, A.D.; Gupta, D.; Khanna, N.N.; Araki, T.; Maniruzzaman, M.; Saba, L.; Nicolaides, A.; Sharma, A.; Omerzu, T. Ranking of stroke and cardiovascular risk factors for an optimal risk calculator design: Logistic regression approach. Comput. Biol. Med. 2019, 108, 182–195. [Google Scholar] [CrossRef]
- Acharya, U.R.; Joseph, K.P.; Kannathal, N.; Min, L.C.; Suri, J.S. Advances in Cardiac Signal Processing; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Giri, D.; Acharya, U.R.; Martis, R.J.; Sree, S.V.; Lim, T.-C.; VI, T.A.; Suri, J.S. Automated diagnosis of coronary artery disease affected patients using LDA, PCA, ICA and discrete wavelet transform. Knowl.-Based Syst. 2013, 37, 274–282. [Google Scholar] [CrossRef]
- Rajendra Acharya, U.; Joseph, K.P.; Kannathal, N.; Lim, C.M.; Suri, J.S. Heart rate variability: A review. Med. Biol. Eng. Comput. 2006, 44, 1031–1051. [Google Scholar] [CrossRef]
- Hippisley-Cox, J.; Coupland, C.; Brindle, P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: Prospective cohort study. BMJ 2017, 357, j2099. [Google Scholar] [CrossRef]
- D’Agostino Sr, R.B.; Vasan, R.S.; Pencina, M.J.; Wolf, P.A.; Cobain, M.; Massaro, J.M.; Kannel, W.B. General cardiovascular risk profile for use in primary care: The Framingham Heart Study. Circulation 2008, 117, 743–753. [Google Scholar] [CrossRef]
- Conroy, R.M.; Pyörälä, K.; Fitzgerald, A.E.; Sans, S.; Menotti, A.; de Backer, G.; de Bacquer, D.; Ducimetiere, P.; Jousilahti, P.; Keil, U. Estimation of ten-year risk of fatal cardiovascular disease in Europe: The SCORE project. Eur. Heart J. 2003, 24, 987–1003. [Google Scholar] [CrossRef]
- Ridker, P.M.; Buring, J.E.; Rifai, N.; Cook, N.R. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: The Reynolds Risk Score. JAMA 2007, 297, 611–619. [Google Scholar] [CrossRef]
- Goff, D.C.; Lloyd-Jones, D.M.; Bennett, G.; Coady, S.; D’agostino, R.B.; Gibbons, R.; Greenland, P.; Lackland, D.T.; Levy, D.; O’donnell, C.J. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 2014, 63 Pt B, 2935–2959. [Google Scholar] [CrossRef]
- Damman, P.; van’t Hof, A.; Berg, J.T.; Jukema, J.; Appelman, Y.; Liem, A.; de Winter, R. 2015 ESC guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: Comments from the Dutch ACS working group. Neth. Heart J. 2017, 25, 181–185. [Google Scholar] [CrossRef]
- Members, T.F.; Montalescot, G.; Sechtem, U.; Achenbach, S.; Andreotti, F.; Arden, C.; Budaj, A.; Bugiardini, R.; Crea, F.; Cuisset, T. 2013 ESC guidelines on the management of stable coronary artery disease: The Task Force on the management of stable coronary artery disease of the European Society of Cardiology. Eur. Heart J. 2013, 34, 2949–3003. [Google Scholar]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes: The Task Force for the diagnosis and management of chronic coronary syndromes of the European Society of Cardiology (ESC). Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef]
- Anderson, T.J.; Grégoire, J.; Pearson, G.J.; Barry, A.R.; Couture, P.; Dawes, M.; Francis, G.A.; Genest, J., Jr.; Grover, S.; Gupta, M. 2016 Canadian Cardiovascular Society guidelines for the management of dyslipidemia for the prevention of cardiovascular disease in the adult. Can. J. Cardiol. 2016, 32, 1263–1282. [Google Scholar] [CrossRef]
- Anderson, T.J.; Grégoire, J.; Hegele, R.A.; Couture, P.; Mancini, G.J.; McPherson, R.; Francis, G.A.; Poirier, P.; Lau, D.C.; Grover, S. 2012 update of the Canadian Cardiovascular Society guidelines for the diagnosis and treatment of dyslipidemia for the prevention of cardiovascular disease in the adult. Can. J. Cardiol. 2013, 29, 151–167. [Google Scholar] [CrossRef]
- Goldstein, B.A.; Navar, A.M.; Carter, R.E. Moving beyond regression techniques in cardiovascular risk prediction: Applying machine learning to address analytic challenges. Eur. Heart J. 2017, 38, 1805–1814. [Google Scholar] [CrossRef]
- Deyama, J.; Nakamura, T.; Takishima, I.; Fujioka, D.; Kawabata, K.-I.; Obata, J.-E.; Watanabe, K.; Watanabe, Y.; Saito, Y.; Mishina, H. Contrast-enhanced ultrasound imaging of carotid plaque neovascularization is useful for identifying high-risk patients with coronary artery disease. Circ. J. 2013, 77, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Colledanchise, K.N.; Mantella, L.E.; Bullen, M.; Hétu, M.-F.; Abunassar, J.G.; Johri, A.M. Combined femoral and carotid plaque burden identifies obstructive coronary artery disease in women. J. Am. Soc. Echocardiogr. 2020, 33, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.N.; Jamthikar, A.D.; Araki, T.; Gupta, D.; Piga, M.; Saba, L.; Carcassi, C.; Nicolaides, A.; Laird, J.R.; Suri, H.S. Nonlinear model for the carotid artery disease 10-year risk prediction by fusing conventional cardiovascular factors to carotid ultrasound image phenotypes: A Japanese diabetes cohort study. Echocardiography 2019, 36, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Jamthikar, A.; Gupta, D.; Saba, L.; Khanna, N.N.; Viskovic, K.; Mavrogeni, S.; Laird, J.R.; Sattar, N.; Johri, A.M.; Pareek, G. Artificial intelligence framework for predictive cardiovascular and stroke risk assessment models: A narrative review of integrated approaches using carotid ultrasound. Comput. Biol. Med. 2020, 126, 104043. [Google Scholar] [CrossRef] [PubMed]
- Jamthikar, A.D.; Gupta, D.; Johri, A.M.; Mantella, L.E.; Saba, L.; Kolluri, R.; Sharma, A.M.; Viswanathan, V.; Nicolaides, A.; Suri, J.S. Low-cost office-based cardiovascular risk stratification using machine learning and focused carotid ultrasound in an Asian-Indian cohort. J. Med. Syst. 2020, 44, 208. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Agarwal, N.; Cau, R.; Gerosa, C.; Sanfilippo, R.; Porcu, M.; Montisci, R.; Cerrone, G.; Qi, Y.; Balestrieri, A. Review of Imaging biomarkers for the vulnerable carotid plaque. JVS Vasc. Sci. 2021, 2, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Suri, J.S. Multi-Detector CT Imaging: Principles, Head, Neck, and Vascular Systems; CRC Press: Boca Raton, FL, USA, 2013; Volume 1. [Google Scholar]
- Sanches, J.M.; Laine, A.F.; Suri, J.S. Ultrasound Imaging; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Londhe, N.D.; Suri, J.S. Superharmonic imaging for medical ultrasound: A review. J. Med. Syst. 2016, 40, 279. [Google Scholar] [CrossRef]
- Sudeep, P.; Palanisamy, P.; Rajan, J.; Baradaran, H.; Saba, L.; Gupta, A.; Suri, J.S. Speckle reduction in medical ultrasound images using an unbiased non-local means method. Biomed. Signal Process. Control 2016, 28, 1–8. [Google Scholar] [CrossRef]
- Khalifa, F.; Beache, G.M.; Gimel’farb, G.; Suri, J.S.; El-Baz, A.S. State-of-the-art medical image registration methodologies: A survey. In Multi Modality State-of-the-Art Medical Image Segmentation and Registration Methodologies; Springer: Berlin/Heidelberg, Germany, 2011; pp. 235–280. [Google Scholar]
- Roumeliotis, S.; Liakopoulos, V.; Roumeliotis, A.; Stamou, A.; Panagoutsos, S.; D’Arrigo, G.; Tripepi, G. Prognostic Factors of Fatal and Nonfatal Cardiovascular Events in Patients with Type 2 Diabetes: The Role of Renal Function Biomarkers. Clin. Diabetes 2021, 39, 188–196. [Google Scholar] [CrossRef]
- van den Munckhof, I.C.; Jones, H.; Hopman, M.T.; de Graaf, J.; Nyakayiru, J.; van Dijk, B.; Eijsvogels, T.M.; Thijssen, D.H. Relation between age and carotid artery intima-medial thickness: A systematic review. Clin. Cardiol. 2018, 41, 698–704. [Google Scholar] [CrossRef]
- Ho, S.S.Y. Current status of carotid ultrasound in atherosclerosis. Quant. Imaging Med. Surg. 2016, 6, 285–296. [Google Scholar] [CrossRef]
- Touboul, P.-J.; Hennerici, M.; Meairs, S.; Adams, H.; Amarenco, P.; Bornstein, N.; Csiba, L.; Desvarieux, M.; Ebrahim, S.; Hernandez, R.H. Mannheim carotid intima-media thickness and plaque consensus (2004–2006–2011). Cerebrovasc. Dis. 2012, 34, 290–296. [Google Scholar] [CrossRef]
- Stein, J.; Korcarz, C.; Post, W. Use of carotid ultrasound to identify subclinical vascular disease and evaluate cardiovascular disease risk: Summary and discussion of the American Society of Echocardiography consensus statement. Prev. Cardiol. 2009, 12, 34–38. [Google Scholar] [CrossRef]
- Ikeda, N.; Araki, T.; Sugi, K.; Nakamura, M.; Deidda, M.; Molinari, F.; Meiburger, K.M.; Acharya, U.R.; Saba, L.; Bassareo, P.P. Ankle–brachial index and its link to automated carotid ultrasound measurement of intima–media thickness variability in 500 Japanese coronary artery disease patients. Curr. Atheroscler. Rep. 2014, 16, 393. [Google Scholar] [CrossRef]
- Naqvi, T.Z.; Lee, M.-S. Carotid intima-media thickness and plaque in cardiovascular risk assessment. JACC Cardiovasc. Imaging 2014, 7, 1025–1038. [Google Scholar] [CrossRef]
- Santos-Neto, P.J.; Sena-Santos, E.H.; Meireles, D.P.; Bittencourt, M.S.; Santos, I.S.; Bensenor, I.M.; Lotufo, P.A. Association of carotid plaques and common carotid intima-media thickness with modifiable cardiovascular risk factors. J. Stroke Cerebrovasc. Dis. 2021, 30, 105671. [Google Scholar] [CrossRef]
- Gooty, V.D.; Sinaiko, A.R.; Ryder, J.R.; Dengel, D.R.; Jacobs, D.R., Jr.; Steinberger, J. Association between carotid intima media thickness, age, and cardiovascular risk factors in children and adolescents. Metab. Syndr. Relat. Disord. 2018, 16, 122–126. [Google Scholar] [CrossRef]
- Johri, A.M.; Mantella, L.E.; Jamthikar, A.D.; Saba, L.; Laird, J.R.; Suri, J.S. Role of artificial intelligence in cardiovascular risk prediction and outcomes: Comparison of machine-learning and conventional statistical approaches for the analysis of carotid ultrasound features and intra-plaque neovascularization. Int. J. Cardiovasc. Imaging 2021, 37, 3145–3156. [Google Scholar] [CrossRef]
- Johri, A.M.; Behl, P.; Hétu, M.F.; Haqqi, M.; Ewart, P.; Day, A.G.; Parfrey, B.; Matangi, M.F. Carotid ultrasound maximum plaque height—A sensitive imaging biomarker for the assessment of significant coronary artery disease. Echocardiography 2016, 33, 281–289. [Google Scholar] [CrossRef]
- Mantella, L.E.; Colledanchise, K.; Bullen, M.; Hétu, M.-F.; Day, A.G.; McLellan, C.S.; Johri, A.M. Handheld versus conventional vascular ultrasound for assessing carotid artery plaque. Int. J. Cardiol. 2019, 278, 295–299. [Google Scholar] [CrossRef]
- Saba, L.; Ikeda, N.; Deidda, M.; Araki, T.; Molinari, F.; Meiburger, K.M.; Acharya, U.R.; Nagashima, Y.; Mercuro, G.; Nakano, M. Association of automated carotid IMT measurement and HbA1c in Japanese patients with coronary artery disease. Diabetes Res. Clin. Pract. 2013, 100, 348–353. [Google Scholar] [CrossRef]
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Nicolaides, A.N.; Suri, J.S. Automated deep learning-based paradigm for high-risk plaque detection in B-mode common carotid ultrasound scans: An asymptomatic Japanese cohort study. Int. Angiol. J. Int. Union Angiol. 2021, 41, 9–23. [Google Scholar] [CrossRef]
- Mitchell, C.; Korcarz, C.E.; Gepner, A.D.; Kaufman, J.D.; Post, W.; Tracy, R.; Gassett, A.J.; Ma, N.; McClelland, R.L.; Stein, J.H. Ultrasound carotid plaque features, cardiovascular disease risk factors and events: The Multi-Ethnic Study of Atherosclerosis. Atherosclerosis 2018, 276, 195–202. [Google Scholar] [CrossRef]
- Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Cuadrado-Godia, E.; Laird, J.R.; Marinhoe, R.T.; Sanches, J.M.; Nicolaides, A. State-of-the-art review on deep learning in medical imaging. Front. Biosci. 2019, 24, 392–426. [Google Scholar]
- Saba, L.; Jain, P.K.; Suri, H.S.; Ikeda, N.; Araki, T.; Singh, B.K.; Nicolaides, A.; Shafique, S.; Gupta, A.; Laird, J.R. Plaque tissue morphology-based stroke risk stratification using carotid ultrasound: A polling-based PCA learning paradigm. J. Med. Syst. 2017, 41, 98. [Google Scholar] [CrossRef]
- Motwani, M.; Dey, D.; Berman, D.S.; Germano, G.; Achenbach, S.; Al-Mallah, M.H.; Andreini, D.; Budoff, M.J.; Cademartiri, F.; Callister, T.Q. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: A 5-year multicentre prospective registry analysis. Eur. Heart J. 2017, 38, 500–507. [Google Scholar] [CrossRef]
- Alaa, A.M.; Bolton, T.; di Angelantonio, E.; Rudd, J.H.; Van der Schaar, M. Cardiovascular disease risk prediction using automated machine learning: A prospective study of 423,604 UK Biobank participants. PLoS ONE 2019, 14, e0213653. [Google Scholar]
- Kakadiaris, I.A.; Vrigkas, M.; Yen, A.A.; Kuznetsova, T.; Budoff, M.; Naghavi, M. Machine learning outperforms ACC/AHA CVD risk calculator in MESA. J. Am. Heart Assoc. 2018, 7, e009476. [Google Scholar] [CrossRef]
- Cawley, G.C.; Talbot, N.L. On over-fitting in model selection and subsequent selection bias in performance evaluation. J. Mach. Learn. Res. 2010, 11, 2079–2107. [Google Scholar]
- Alalawi, H.H.; Manal, S.A. Detection of Cardiovascular Disease using Machine Learning Classification Models. Int. J. Eng. Res. Technol. ISSN 2021, 10, 2278-0181. [Google Scholar]
- Chauhan, Y.J. Cardiovascular Disease Prediction using Classification Algorithms of Machine Learning. Int. J. Sci. Res. ISSN 2018, 2319–7064. [Google Scholar]
- Choi, E.; Schuetz, A.; Stewart, W.F.; Sun, J. Using recurrent neural network models for early detection of heart failure onset. J. Am. Med. Inform. Assoc. 2017, 24, 361–370. [Google Scholar] [CrossRef]
- Nayan, N.A.; Hamid, H.A.; Suboh, M.Z.; Abdullah, N.; Jaafar, R.; Yusof, N.A.M.; Hamid, M.A.; Zubiri, N.F.; Arifin, A.S.K.; Daud, S.M.A. Cardiovascular Disease Prediction from Electrocardiogram by using Machine Learning Method: A Snapshot from the Subjects of the Malaysian Cohort. Int. J. Online Biomed. Eng. 2020, 16, 2626–8493. [Google Scholar]
- Pasanisi, S.; Paiano, R. A hybrid information mining approach for knowledge discovery in cardiovascular disease (CVD). Information 2018, 9, 90. [Google Scholar] [CrossRef]
- Sánchez-Cabo, F.; Rossello, X.; Fuster, V.; Benito, F.; Manzano, J.P.; Silla, J.C.; Fernández-Alvira, J.M.; Oliva, B.; Fernández-Friera, L.; López-Melgar, B. Machine learning improves cardiovascular risk definition for young, asymptomatic individuals. J. Am. Coll. Cardiol. 2020, 76, 1674–1685. [Google Scholar] [CrossRef]
- Buddi, S.; Taylor, T.; Borges, C.; Nelson, R. SVM multi-classification of T2D/CVD patients using biomarker features. In Proceedings of the 2011 10th International Conference on Machine Learning and Applications and Workshops, Honolulu, HI, USA, 18–21 December 2011; pp. 338–341. [Google Scholar]
- Chao, H.; Shan, H.; Homayounieh, F.; Singh, R.; Khera, R.D.; Guo, H.; Su, T.; Wang, G.; Kalra, M.K.; Yan, P. Deep learning predicts cardiovascular disease risks from lung cancer screening low dose computed tomography. Nat. Commun. 2021, 12, 2963. [Google Scholar] [CrossRef]
- Devi, R.; Tyagi, H.K.; Kumar, D. A novel multi-class approach for early-stage prediction of sudden cardiac death. Biocybern. Biomed. Eng. 2019, 39, 586–598. [Google Scholar] [CrossRef]
- Emaus, M.J.; Išgum, I.; van Velzen, S.G.; van den Bongard, H.D.; Gernaat, S.A.; Lessmann, N.; Sattler, M.G.; Teske, A.J.; Penninkhof, J.; Meijer, H. Bragatston study protocol: A multicentre cohort study on automated quantification of cardiovascular calcifications on radiotherapy planning CT scans for cardiovascular risk prediction in patients with breast cancer. BMJ Open 2019, 9, e028752. [Google Scholar] [CrossRef]
- Hedman, Å.K.; Hage, C.; Sharma, A.; Brosnan, M.J.; Buckbinder, L.; Gan, L.-M.; Shah, S.J.; Linde, C.M.; Donal, E.; Daubert, J.-C. Identification of novel pheno-groups in heart failure with preserved ejection fraction using machine learning. Heart 2020, 106, 342–349. [Google Scholar] [CrossRef]
- Hussein, A.F.; Hashim, S.J.; Rokhani, F.Z.; Wan Adnan, W.A. An Automated High-Accuracy Detection Scheme for Myocardial Ischemia Based on Multi-Lead Long-Interval ECG and Choi-Williams Time-Frequency Analysis Incorporating a Multi-Class SVM Classifier. Sensors 2021, 21, 2311. [Google Scholar] [CrossRef]
- Jamthikar, A.D.; Gupta, D.; Mantella, L.E.; Saba, L.; Laird, J.R.; Johri, A.M.; Suri, J.S. Multiclass machine learning vs. conventional calculators for stroke/CVD risk assessment using carotid plaque predictors with coronary angiography scores as gold standard: A 500 participants study. Int. J. Cardiovasc. Imaging 2021, 37, 1171–1187. [Google Scholar] [CrossRef]
- Khan, M.U.; Ali, S.Z.-e.-Z.; Ishtiaq, A.; Habib, K.; Gul, T.; Samer, A. Classification of Multi-Class Cardiovascular Disorders using Ensemble Classifier and Impulsive Domain Analysis. In Proceedings of the 2021 Mohammad Ali Jinnah University International Conference on Computing (MAJICC), Karachi, Pakistan, 15–17 July 2021; pp. 1–8. [Google Scholar]
- Krupa, B.N.; Bharathi, K.; Gaonkar, M.; Karun, S.; Nath, S.; Ali, M. Multiclass Classification of APG Signals using ELM for CVD Risk Identification: A Real-Time Application. In Proceedings of the 16th International Conference on Biomedical Engineering, Singapore, 7–10 December 2016; Springer: Singapore, 2017; pp. 32–37. [Google Scholar]
- Lui, H.W.; Chow, K.L. Multiclass classification of myocardial infarction with convolutional and recurrent neural networks for portable ECG devices. Inform. Med. Unlocked 2018, 13, 26–33. [Google Scholar] [CrossRef]
- Nakanishi, R.; Slomka, P.J.; Rios, R.; Betancur, J.; Blaha, M.J.; Nasir, K.; Miedema, M.D.; Rumberger, J.A.; Gransar, H.; Shaw, L.J. Machine learning adds to clinical and CAC assessments in predicting 10-year CHD and CVD deaths. Cardiovasc. Imaging 2021, 14, 615–625. [Google Scholar] [CrossRef]
- Ni, J.; Jiang, Y.; Zhai, S.; Chen, Y.; Li, S.; Amei, A.; Tran, D.-M.T.; Zhai, L.; Kuang, Y. Multi-class Cardiovascular Disease Detection and Classification from 12-Lead ECG Signals Using an Inception Residual Network. In Proceedings of the 2021 IEEE 45th Annual Computers, Software, and Applications Conference (COMPSAC), Madrid, Spain, 12–16 July 2021; pp. 1532–1537. [Google Scholar]
- Wiharto, W.; Kusnanto, H.; Herianto, H. Performance analysis of multiclass support vector machine classification for diagnosis of coronary heart diseases. arXiv 2015, arXiv:1511.02352. [Google Scholar] [CrossRef]
- Ambale-Venkatesh, B.; Yang, X.; Wu, C.O.; Liu, K.; Hundley, W.G.; McClelland, R.; Gomes, A.S.; Folsom, A.R.; Shea, S.; Guallar, E. Cardiovascular event prediction by machine learning: The multi-ethnic study of atherosclerosis. Circ. Res. 2017, 121, 1092–1101. [Google Scholar] [CrossRef]
- Jamthikar, A.; Gupta, D.; Johri, A.M.; Mantella, L.E.; Saba, L.; Suri, J.S. A machine learning framework for risk prediction of multi-label cardiovascular events based on focused carotid plaque B-Mode ultrasound: A Canadian study. Comput. Biol. Med. 2021, 140, 105102. [Google Scholar] [CrossRef]
- Kumar, P.; Sharma, R.; Misra, S.; Kumar, A.; Nath, M.; Nair, P.; Vibha, D.; Srivastava, A.K.; Prasad, K. CIMT as a risk factor for stroke subtype: A systematic review. Eur. J. Clin. Investig. 2020, 50, e13348. [Google Scholar] [CrossRef]
- Mehrang, S.; Lahdenoja, O.; Kaisti, M.; Tadi, M.J.; Hurnanen, T.; Airola, A.; Knuutila, T.; Jaakkola, J.; Jaakkola, S.; Vasankari, T. Classification of Atrial Fibrillation and Acute Decompensated Heart Failure Using Smartphone Mechanocardiography: A Multilabel Learning Approach. IEEE Sens. J. 2020, 20, 7957–7968. [Google Scholar] [CrossRef]
- Mohamed, M.; Farah, M.-C.; Fahed, A. Multi-label classification and evidential approach in diseases diagnoses using physiological signals. In Proceedings of the 2020 IEEE 5th Middle East and Africa Conference on Biomedical Engineering (MECBME), Amman, Jordan, 27–29 October 2020. [Google Scholar]
- Nigam, P. Applying Deep Learning to ICD-9 Multi-Label Classification from Medical Records; Technical Report; Stanford University: Stanford, CA, USA, 2016. [Google Scholar]
- Zamzmi, G.; Hsu, L.-Y.; Li, W.; Sachdev, V.; Antani, S. Harnessing machine intelligence in automatic echocardiogram analysis: Current status, limitations, and future directions. IEEE Rev. Biomed. Eng. 2020, 14, 181–203. [Google Scholar] [CrossRef]
- Zeng, X.; Hu, Y.; Shu, L.; Li, J.; Duan, H.; Shu, Q.; Li, H. Explainable machine-learning predictions for complications after pediatric congenital heart surgery. Sci. Rep. 2021, 11, 17244. [Google Scholar] [CrossRef]
- Abdar, M.; Książek, W.; Acharya, U.R.; Tan, R.-S.; Makarenkov, V.; Pławiak, P. A new machine learning technique for an accurate diagnosis of coronary artery disease. Comput. Methods Programs Biomed. 2019, 179, 104992. [Google Scholar] [CrossRef] [PubMed]
- Baccouche, A.; Garcia-Zapirain, B.; Castillo Olea, C.; Elmaghraby, A. Ensemble deep learning models for heart disease classification: A case study from Mexico. Information 2020, 11, 207. [Google Scholar] [CrossRef]
- Chu, H.; Chen, L.; Yang, X.; Qiu, X.; Qiao, Z.; Song, X.; Zhao, E.; Zhou, J.; Zhang, W.; Mehmood, A. Roles of anxiety and depression in predicting cardiovascular disease among patients with type 2 diabetes mellitus: A machine learning approach. Front. Psychol. 2021, 12, 645418. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Tafti, A.P.; Ngufor, C.; Zhang, P.; Xiao, P.; Dai, M.; Liu, H.; Noseworthy, P.; Chen, M.; Friedman, P.A. Using ensemble of ensemble machine learning methods to predict outcomes of cardiac resynchronization. J. Cardiovasc. Electrophysiol. 2021, 32, 2504–2514. [Google Scholar] [CrossRef]
- Esfahani, H.A.; Ghazanfari, M. Cardiovascular disease detection using a new ensemble classifier. In Proceedings of the 2017 IEEE 4th International Conference on Knowledge-Based Engineering and Innovation (KBEI), Tehran, Iran, 22 December 2017; pp. 1011–1014. [Google Scholar]
- Gibson, W.J.; Nafee, T.; Travis, R.; Yee, M.; Kerneis, M.; Ohman, M.; Gibson, C.M. Machine learning versus traditional risk stratification methods in acute coronary syndrome: A pooled randomized clinical trial analysis. J. Thromb. 2020, 49, 1–9. [Google Scholar] [CrossRef]
- Gao, X.-Y.; Amin Ali, A.; Shaban Hassan, H.; Anwar, E.M. Improving the Accuracy for Analyzing Heart Diseases Prediction Based on the Ensemble Method. Complexity 2021, 2021, 6663455. [Google Scholar] [CrossRef]
- Gao, L.; Ding, Y. Disease prediction via Bayesian hyperparameter optimization and ensemble learning. BMC Res. Notes 2020, 13, 205. [Google Scholar] [CrossRef]
- Ghosh, P.; Azam, S.; Jonkman, M.; Karim, A.; Shamrat, F.J.M.; Ignatious, E.; Shultana, S.; Beeravolu, A.R.; De Boer, F. Efficient Prediction of Cardiovascular Disease Using Machine Learning Algorithms with Relief and LASSO Feature Selection Techniques. IEEE Access 2021, 9, 19304–19326. [Google Scholar] [CrossRef]
- Hosni, M.; Carrillo de Gea, J.M.; Idri, A.; El Bajta, M.; Fernandez Aleman, J.L.; García-Mateos, G.; Abnane, I. A systematic mapping study for ensemble classification methods in cardiovascular disease. Artif. Intell. Rev. 2021, 54, 2827–2861. [Google Scholar] [CrossRef]
- Mustafa, J.; Awan, A.A.; Khalid, M.S.; Nisar, S. Ensemble approach for developing a smart heart disease prediction system using classification algorithms. Res. Rep. Clin. Cardiol. 2018, 9, 33. [Google Scholar]
- Jamthikar, A.D.; Gupta, D.; Mantella, L.E.; Saba, L.; Johri, A.M.; Suri, J.S. Ensemble Machine Learning and its Validation for Prediction of Coronary Artery Disease and Acute Coronary Syndrome using Focused Carotid Ultrasound. IEEE Trans. Instrum. Meas. 2021, 43, 2503810. [Google Scholar] [CrossRef]
- Prakash, V.J.; Karthikeyan, N. Enhanced Evolutionary Feature Selection and Ensemble Method for Cardiovascular Disease Prediction. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 389–412. [Google Scholar] [CrossRef]
- Liu, N.; Li, X.; Qi, E.; Xu, M.; Li, L.; Gao, B. A novel Ensemble Learning Paradigm for Medical Diagnosis with Imbalanced Data. IEEE Access 2020, 8, 171263–171280. [Google Scholar] [CrossRef]
- Miao, K.H.; Miao, J.H.; Miao, G.J. Diagnosing coronary heart disease using ensemble machine learning. Int. J. Adv. Comput. Sci. Appl. 2016, 7, 1–12. [Google Scholar]
- Mienye, I.D.; Sun, Y.; Wang, Z. An improved ensemble learning approach for the prediction of heart disease risk. Inform. Med. Unlocked 2020, 20, 100402. [Google Scholar] [CrossRef]
- Negassa, A.; Ahmed, S.; Zolty, R.; Patel, S.R. Prediction Model Using Machine Learning for Mortality in Patients with Heart Failure. Am. J. Cardiol. 2021, 153, 86–93. [Google Scholar] [CrossRef]
- Pławiak, P.; Acharya, U.R. Novel deep genetic ensemble of classifiers for arrhythmia detection using ECG signals. Neural Comput. 2020, 32, 11137–11161. [Google Scholar] [CrossRef]
- Reddy, K.V.V.; Elamvazuthi, I.; Aziz, A.A.; Paramasivam, S.; Chua, H.N.; Pranavanand, S. Heart Disease Risk Prediction Using Machine Learning Classifiers with Attribute Evaluators. Appl. Sci. 2021, 11, 8352. [Google Scholar] [CrossRef]
- Rousset, A.; Dellamonica, D.; Menuet, R.; Lira Pineda, A.; Sabatine, M.S.; Giugliano, R.P.; Trichelair, P.; Zaslavskiy, M.; Ricci, L. Can machine learning bring cardiovascular risk assessment to the next level? A methodological study using FOURIER trial data. Eur. Heart J. Digit. Health 2021, 093, 93. [Google Scholar] [CrossRef]
- Sherly, S.I. An Ensemble Basedheart Disease Predictionusing Gradient Boosting Decision Tree. Turk. J. Comput. Math. Educ. 2021, 12, 3648–3660. [Google Scholar]
- Sherazi, S.W.A.; Bae, J.-W.; Lee, J.Y. A soft voting ensemble classifier for early prediction and diagnosis of occurrences of major adverse cardiovascular events for STEMI and NSTEMI during 2-year follow-up in patients with acute coronary syndrome. PLoS ONE 2021, 16, e0249338. [Google Scholar] [CrossRef]
- Tan, C.; Chen, H.; Xia, C. The prediction of cardiovascular disease based on trace element contents in hair and a classifier of boosting decision stumps. Biol. Trace Elem. Res. 2009, 129, 9–19. [Google Scholar] [CrossRef]
- Uddin, M.N.; Halder, R.K. An Ensemble Method Based Multilayer Dynamic System to Predict Cardiovascular Disease Using Machine Learning Approach. Inform. Med. Unlocked 2021, 24, 100584. [Google Scholar] [CrossRef]
- Velusamy, D.; Ramasamy, K. Ensemble of heterogeneous classifiers for diagnosis and prediction of coronary artery disease with reduced feature subset. Comput. Methods Programs Biomed. 2021, 198, 105770. [Google Scholar] [CrossRef]
- Wankhede, J.; Sambandam, P.; Kumar, M. Effective prediction of heart disease using hybrid ensemble deep learning and tunicate swarm algorithm. J. Biomol. Struct. Dyn. 2021, 128, 1–12. [Google Scholar] [CrossRef]
- Yadav, D.C.; Pal, S. Analysis of Heart Disease Using Parallel and Sequential ensemble Methods with Feature Selection Techniques: Heart Disease Prediction. Int. J. Big Data Anal. Healthc. 2021, 6, 40–56. [Google Scholar] [CrossRef]
- Ye, C.; Fu, T.; Hao, S.; Zhang, Y.; Wang, O.; Jin, B.; Xia, M.; Liu, M.; Zhou, X.; Wu, Q. Prediction of incident hypertension within the next year: Prospective study using statewide electronic health records and machine learning. J. Med. Internet Res. 2018, 20, e22. [Google Scholar] [CrossRef]
- Yekkala, I.; Dixit, S.; Jabbar, M. Prediction of heart disease using ensemble learning and Particle Swarm Optimization. In Proceedings of the 2017 International Conference on Smart Technologies for Smart Nation (SmartTechCon), Bengaluru, India, 17–19 August 2017; pp. 691–698. [Google Scholar]
- Zarkogianni, K.; Athanasiou, M.; Thanopoulou, A.C.; Nikita, K.S. Comparison of machine learning approaches toward assessing the risk of developing cardiovascular disease as a long-term diabetes complication. IEEE J. Biomed. Health Inform. 2017, 22, 1637–1647. [Google Scholar] [CrossRef]
- Zhenya, Q.; Zhang, Z. A hybrid cost-sensitive ensemble for heart disease prediction. BMC Med. Inform. Decis. Mak. 2021, 21, 73. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD’16), San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K. Xgboost: Extreme gradient boosting. R Package Version 0.4-2 2015, 1, 1–4. [Google Scholar]
- Hansson, G.K.; Libby, P.; Tabas, I. Inflammation and plaque vulnerability. J. Intern. Med. 2015, 278, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, K.; Zhao, R.; Hu, J.; Hao, Z.; Wang, F.; Lu, Y.; Liu, F.; Zhang, Y. Inflammatory biomarkers of coronary heart disease. Front. Biosci. 2017, 22, 504–515. [Google Scholar]
- Libby, P.; Ridker, P.M.; Hansson, G.K.; Leducq Transatlantic Network on Atherothrombosis. Inflammation in atherosclerosis: From pathophysiology to practice. J. Am. Coll. Cardiol. 2009, 54, 2129–2138. [Google Scholar] [CrossRef] [PubMed]
- Ross, R. Cell biology of atherosclerosis. Annu. Rev. Physiol. 1995, 57, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Tabas, I.; Williams, K.J.; Borén, J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: Update and therapeutic implications. Circulation 2007, 116, 1832–1844. [Google Scholar] [CrossRef]
- Virmani, R.; Burke, A.P.; Kolodgie, F.D.; Farb, A. Pathology of the thin-cap fibroatheroma: A type of vulnerable plaque. J. Interv. Cardiol. 2003, 16, 267–272. [Google Scholar] [CrossRef]
- Burke, A.P.; Kolodgie, F.D.; Farb, A.; Weber, D.; Virmani, R. Morphological predictors of arterial remodeling in coronary atherosclerosis. Circulation 2002, 105, 297–303. [Google Scholar] [CrossRef]
- Patel, A.K.; Suri, H.S.; Singh, J.; Kumar, D.; Shafique, S.; Nicolaides, A.; Jain, S.K.; Saba, L.; Gupta, A.; Laird, J.R. A review on atherosclerotic biology, wall stiffness, physics of elasticity, and its ultrasound-based measurement. Curr. Atheroscler. Rep. 2016, 18, 83. [Google Scholar] [CrossRef]
- Arroyo, L.H.; Lee, R.T. Mechanisms of plaque rupture: Mechanical and biologic interactions. Cardiovasc. Res. 1999, 41, 369–375. [Google Scholar] [CrossRef]
- Teng, Z.; Zhang, Y.; Huang, Y.; Feng, J.; Yuan, J.; Lu, Q.; Sutcliffe, M.P.; Brown, A.J.; Jing, Z.; Gillard, J.H. Material properties of components in human carotid atherosclerotic plaques: A uniaxial extension study. Acta Biomater. 2014, 10, 5055–5063. [Google Scholar] [CrossRef]
- Kumar, P.R.; Priya, M. Classification of atherosclerotic and non-atherosclerotic individuals using multiclass support vector machine. Technol. Health Care 2014, 22, 583–595. [Google Scholar] [CrossRef]
- Herr, J.E.; Hétu, M.-F.; Li, T.Y.; Ewart, P.; Johri, A.M. Presence of calcium-like tissue composition in carotid plaque is indicative of significant coronary artery disease in high-risk patients. J. Am. Soc. Echocardiogr. 2019, 32, 633–642. [Google Scholar] [CrossRef]
- Jeong, B.; Cho, H.; Kim, J.; Kwon, S.K.; Hong, S.; Lee, C.; Kim, T.; Park, M.S.; Hong, S.; Heo, T.-Y. Comparison between statistical models and machine learning methods on classification for highly imbalanced multiclass kidney data. Diagnostics 2020, 10, 415. [Google Scholar] [CrossRef]
- Rim, T.H.; Lee, C.J.; Tham, Y.-C.; Cheung, N.; Yu, M.; Lee, G.; Kim, Y.; Ting, D.S.; Chong, C.C.Y.; Choi, Y.S. Deep-learning-based cardiovascular risk stratification using coronary artery calcium scores predicted from retinal photographs. Lancet Digit. Health 2021, 3, e306–e316. [Google Scholar] [CrossRef]
- Tandel, G.S.; Balestrieri, A.; Jujaray, T.; Khanna, N.N.; Saba, L.; Suri, J.S. Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput. Biol. Med. 2020, 122, 103804. [Google Scholar] [CrossRef]
- Mercan, C.; Aksoy, S.; Mercan, E.; Shapiro, L.G.; Weaver, D.L.; Elmore, J.G. Multi-instance multi-label learning for multi-class classification of whole slide breast histopathology images. IEEE Trans. Med. Imaging 2017, 37, 316–325. [Google Scholar] [CrossRef]
- Unnikrishnan, P.; Kumar, D.K.; Poosapadi Arjunan, S.; Kumar, H.; Mitchell, P.; Kawasaki, R. Development of health parameter model for risk prediction of CVD using SVM. Comput. Math. Methods Med. 2016, 2016, 3016245. [Google Scholar] [CrossRef]
- Cheng, B.; Liu, M.; Zhang, D.; Shen, D. Robust multi-label transfer feature learning for early diagnosis of Alzheimer’s disease. Brain Imaging Behav. 2019, 13, 138–153. [Google Scholar] [CrossRef]
- Nikhar, S.; Karandikar, A. Prediction of heart disease using machine learning algorithms. Int. J. Adv. Eng. Manag. Sci. 2016, 2, 239484. [Google Scholar]
- Rosengren, A.; Hawken, S.; Ôunpuu, S.; Sliwa, K.; Zubaid, M.; Almahmeed, W.A.; Blackett, K.N.; Sitthi-Amorn, C.; Sato, H.; Yusuf, S. Association of psychosocial risk factors with risk of acute myocardial infarction in 11,119 cases and 13,648 controls from 52 countries (the INTERHEART study): Case-control study. Lancet 2004, 364, 953–962. [Google Scholar] [CrossRef]
- Yusuf, S.; Hawken, S.; Ôunpuu, S.; Dans, T.; Avezum, A.; Lanas, F.; McQueen, M.; Budaj, A.; Pais, P.; Varigos, J. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): Case-control study. Lancet 2004, 364, 937–952. [Google Scholar] [CrossRef]
- Shrivastava, V.K.; Londhe, N.D.; Sonawane, R.S.; Suri, J.S. Reliable and accurate psoriasis disease classification in dermatology images using comprehensive feature space in machine learning paradigm. Expert Syst. Appl. 2015, 42, 6184–6195. [Google Scholar] [CrossRef]
- Araki, T.; Ikeda, N.; Shukla, D.; Jain, P.K.; Londhe, N.D.; Shrivastava, V.K.; Banchhor, S.K.; Saba, L.; Nicolaides, A.; Shafique, S.; et al. PCA-based polling strategy in machine learning framework for coronary artery disease risk assessment in intravascular ultrasound: A link between carotid and coronary grayscale plaque morphology. Comput. Methods Programs Biomed. 2016, 128, 137–158. [Google Scholar] [CrossRef]
- Acharya, U.R.; Mookiah, M.R.K.; Sree, S.V.; Yanti, R.; Martis, R.; Saba, L.; Molinari, F.; Guerriero, S.; Suri, J.S. Evolutionary algorithm-based classifier parameter tuning for automatic ovarian cancer tissue characterization and classification. Ultraschall Der Med.-Eur. J. Ultrasound 2014, 35, 237–245. [Google Scholar]
- Olier, I.; Ortega-Martorell, S.; Pieroni, M.; Lip, G.Y. How machine learning is impacting research in atrial fibrillation: Implications for risk prediction and future management. Cardiovasc. Res. 2021, 117, 1700–1717. [Google Scholar] [CrossRef]
- Angelaki, E.; Marketou, M.E.; Barmparis, G.D.; Patrianakos, A.; Vardas, P.E.; Parthenakis, F.; Tsironis, G.P. Detection of abnormal left ventricular geometry in patients without cardiovascular disease through machine learning: An ECG-based approach. J. Clin. Hypertens. 2021, 23, 935–945. [Google Scholar] [CrossRef]
- Shen, Y.; Yang, Y.; Parish, S.; Chen, Z.; Clarke, R.; Clifton, D.A. Risk prediction for cardiovascular disease using ECG data in the China Kadoorie Biobank. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016. [Google Scholar]
- Chang, A.; Cadaret, L.M.; Liu, K. Machine learning in electrocardiography and echocardiography: Technological advances in clinical cardiology. Curr. Cardiol. Rep. 2020, 22, 161. [Google Scholar] [CrossRef]
- Bos, J.M.; Attia, Z.I.; Albert, D.E.; Noseworthy, P.A.; Friedman, P.A.; Ackerman, M.J. Use of artificial intelligence and deep neural networks in evaluation of patients with electrocardiographically concealed long QT syndrome from the surface 12-lead electrocardiogram. JAMA Cardiol. 2021, 6, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Paragliola, G.; Coronato, A. An hybrid ECG-based deep network for the early identification of high-risk to major cardiovascular events for hypertension patients. J. Biomed. Inform. 2021, 113, 103648. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, R.; Wang, X.; Shen, S.; Zhou, B.; Wang, Z. Multiscale Residual Network Based on Channel Spatial Attention Mechanism for Multilabel ECG Classification. J. Healthc. Eng. 2021, 2021, 6630643. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, Z.-X.; Lu, P.; Guo, R.; Yan, H.-X.; Xu, W.-J.; Wang, Y.-Q.; Xia, C.-M. Classifying syndromes in Chinese medicine using multi-label learning algorithm with relevant features for each label. Chin. J. Integr. Med. 2016, 22, 867–871. [Google Scholar] [CrossRef]
- Longato, E.; Fadini, G.P.; Sparacino, G.; Avogaro, A.; Tramontan, L.; Di Camillo, B. A deep learning approach to predict diabetes’ cardiovascular complications from administrative claims. IEEE J. Biomed. Health Inform. 2021, 25, 3608–3617. [Google Scholar] [CrossRef]
- Fang, J.; Xu, Y.; Zhao, Y.; Yan, Y.; Liu, J.; Liu, J. Weighing features of lung and heart regions for thoracic disease classification. BMC Med. Imaging 2021, 21, 99. [Google Scholar] [CrossRef]
- Cen, L.-P.; Ji, J.; Lin, J.-W.; Ju, S.-T.; Lin, H.-J.; Li, T.-P.; Wang, Y.; Yang, J.-F.; Liu, Y.-F.; Tan, S. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat. Commun. 2021, 12, 4828. [Google Scholar] [CrossRef]
- Deng, F.; Zhou, H.; Lin, Y.; Heim, J.A.; Shen, L.; Li, Y.; Zhang, L. Predict multicategory causes of death in lung cancer patients using clinicopathologic factors. Comput. Biol. Med. 2021, 129, 104161. [Google Scholar] [CrossRef]
- Wagenaar, D.J.; Chen, J.A. Nuclear imaging of vulnerable plaque: Contrast improvements through multi-labeling of nanoparticles. In Proceedings of the IEEE Nuclear Science Symposium Conference Record, Fajardo, PR, USA, 23–29 October 2005; p. 5. [Google Scholar]
- Jie, M.; Hong, Z. Image classification algorithm based on LTS-HD multi instance multi label RBF. In Proceedings of the 2017 12th IEEE Conference on Industrial Electronics and Applications (ICIEA), Siem Reap, Cambodia, 18–20 June 2017; pp. 190–194. [Google Scholar]
- Wu, Q.; Tan, M.; Song, H.; Chen, J.; Ng, M.K. ML-FOREST: A multi-label tree ensemble method for multi-label classification. IEEE Trans. Knowl. Data Eng. 2016, 28, 2665–2680. [Google Scholar] [CrossRef]
- Li, G.-Z.; He, Z.; Shao, F.-F.; Ou, A.-H.; Lin, X.-Z. Patient classification of hypertension in Traditional Chinese Medicine using multi-label learning techniques. BMC Med. Genom. 2015, 8, S4. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, C.; Zhao, Y.; Yan, C. Multi-label ECG signal classification based on ensemble classifier. IEEE Access 2020, 8, 117986–117996. [Google Scholar] [CrossRef]
- Rezaei Ravari, M.; Eftekhari, M.; Saberi Movahed, F. ML-CK-ELM: An efficient multi-layer extreme learning machine using combined kernels for multi-label classification. Sci. Iran. 2020, 27, 3005–3018. [Google Scholar] [CrossRef]
- Rezaei-Ravari, M.; Eftekhari, M.; Saberi-Movahed, F. Regularizing extreme learning machine by dual locally linear embedding manifold learning for training multi-label neural network classifiers. Eng. Appl. Artif. Intell. 2021, 97, 104062. [Google Scholar] [CrossRef]
- Baghel, N.; Dutta, M.K.; Burget, R.J.C.M.; Biomedicine, P.i. Automatic diagnosis of multiple cardiac diseases from PCG signals using convolutional neural network. Comput. Methods Programs Biomed. 2020, 197, 105750. [Google Scholar] [CrossRef]
- Cuadrado-Godia, E.; Dwivedi, P.; Sharma, S.; Santiago, A.O.; Gonzalez, J.R.; Balcells, M.; Laird, J.; Turk, M.; Suri, H.S.; Nicolaides, A.; et al. Cerebral small vessel disease: A review focusing on pathophysiology, biomarkers, and machine learning strategies. J. Stroke 2018, 20, 302–320. [Google Scholar] [CrossRef]
- Huang, W.; Ying, T.W.; Chin, W.L.C.; Baskaran, L.; Marcus, O.E.H.; Yeo, K.K.; Kiong, N.S. Application of ensemble machine learning algorithms on lifestyle factors and wearables for cardiovascular risk prediction. Sci. Rep. 2022, 12, 1033. [Google Scholar] [CrossRef]
- Brownless, J. Nested Cross-Validation for Machine Learning with Python. 2020. Available online: https://machinelearningmastery.com/nested-cross-validation-for-machine-learning-with-python/ (accessed on 20 February 2022).
- Pintelas, P.; Livieris, I.E. Special issue on ensemble learning and applications. Algorithms 2020, 13, 140. [Google Scholar] [CrossRef]
- Lo, H.-Y.; Lin, S.-D.; Wang, H.-M. Generalized k-labelsets ensemble for multi-label and cost-sensitive classification. IEEE Trans. Knowl. Data Eng. 2013, 26, 1679–1691. [Google Scholar]
- Wang, P.; Ge, R.; Xiao, X.; Zhou, M.; Zhou, F. hMuLab: A biomedical hybrid MUlti-LABel classifier based on multiple linear regression. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 14, 1173–1180. [Google Scholar] [CrossRef]
- Guo, L.; Jin, B.; Yu, R.; Yao, C.; Sun, C.; Huang, D. Multi-label classification methods for green computing and application for mobile medical recommendations. IEEE Access 2016, 4, 3201–3209. [Google Scholar] [CrossRef]
- Farooq, A.; Anwar, S.; Awais, M.; Rehman, S. A deep CNN based multi-class classification of Alzheimer’s disease using MRI. In Proceedings of the 2017 IEEE International Conference on Imaging Systems and Techniques (IST), Beijing, China, 18–20 October 2017; pp. 1–6. [Google Scholar]
- Zuluaga, M.A.; Cardoso, M.J.; Ourselin, S. Automatic right ventricle segmentation using multi-label fusion in cardiac MRI. arXiv 2020, arXiv:2004.02317. [Google Scholar]
- Al Hinai, G.; Jammoul, S.; Vajihi, Z.; Afilalo, J. Deep learning analysis of resting electrocardiograms for the detection of myocardial dysfunction, hypertrophy, and ischaemia: A systematic review. Eur. Heart J.-Digit. Health 2021, 2, 416–423. [Google Scholar] [CrossRef]
- Mahinrad, S.; Ferguson, I.; Macfarlane, P.W.; Clark, E.N.; Stott, D.J.; Ford, I.; Mooijaart, S.P.; Trompet, S.; Van Heemst, D.; Jukema, J.W. Spatial QRS-T angle and cognitive decline in older subjects. J. Alzheimer’s Dis. 2019, 67, 279–289. [Google Scholar] [CrossRef]
- Narayan, S.M.; Wang, P.J.; Daubert, J.P. New concepts in sudden cardiac arrest to address an intractable epidemic: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019, 73, 70–88. [Google Scholar] [CrossRef] [PubMed]
- Puvar, P.; Patel, N.; Shah, A.; Solanki, R.; Rana, D. Heart Disease Detection using Ensemble Learning Approach. Int. Res. J. Eng. Technol. (IRJET) 2021, 8, 2395-0072. [Google Scholar]
- Gibaja, E.; Ventura, S. A tutorial on multilabel learning. ACM Comput. Surv. 2015, 47, 1–38. [Google Scholar] [CrossRef]
- Krstinić, D.; Braović, M.; Šerić, L.; Božić-Štulić, D. Multi-label classifier performance evaluation with confusion matrix. Comput. Sci. Inf. Technol. 2020, 10, 1–14. [Google Scholar]
- Tsoumakas, G.; Katakis, I. Multi-label classification: An overview. Int. J. Data Warehous. Min. 2007, 3, 1–13. [Google Scholar] [CrossRef]
- Spolaôr, N.; Cherman, E.A.; Monard, M.C.; Lee, H.D. A comparison of multi-label feature selection methods using the problem transformation approach. Electron. Notes Theor. Comput. Sci. 2013, 292, 135–151. [Google Scholar] [CrossRef]
- Xia, C.; Vonder, M.; Sidorenkov, G.; Ma, R.; Oudkerk, M.; van der Harst, P.; De Deyn, P.P.; Vliegenthart, R. Coronary Artery Calcium and Cognitive Function in Dutch Adults: Cross-Sectional Results of the Population-Based ImaLife Study. J. Am. Heart Assoc. 2021, 10, e018172. [Google Scholar] [CrossRef]
- Ribeiro, A.L.P.; Paixao, G.M.; Gomes, P.R.; Ribeiro, M.H.; Ribeiro, A.H.; Canazart, J.A.; Oliveira, D.M.; Ferreira, M.P.; Lima, E.M.; de Moraes, J.L. Tele-electrocardiography and bigdata: The CODE (Clinical Outcomes in Digital Electrocardiography) study. J. Electrocardiol. 2019, 57, S75–S78. [Google Scholar] [CrossRef]
- Castelyn, G.; Laranjo, L.; Schreier, G.; Gallego, B. Predictive performance and impact of algorithms in remote monitoring of chronic conditions: A systematic review and meta-analysis. Int. J. Med. Inform. 2021, 156, 104620. [Google Scholar] [CrossRef]
- Chugh, S.S.; Reinier, K.; Teodorescu, C.; Evanado, A.; Kehr, E.; Al Samara, M.; Mariani, R.; Gunson, K.; Jui, J. Epidemiology of sudden cardiac death: Clinical and research implications. Prog. Cardiovasc. Dis. 2008, 51, 213–228. [Google Scholar] [CrossRef]
- Masarone, D.; Limongelli, G.; Ammendola, E.; Verrengia, M.; Gravino, R.; Pacileo, G. Risk stratification of sudden cardiac death in patients with heart failure: An update. J. Clin. Med. 2018, 7, 436. [Google Scholar] [CrossRef]
- Palacios-Rubioa, J.; Marina-Breysse, M.; Quintanilla, J.G.; Gil-Perdomo, J.M.; Juárez-Fernández, M.; Garcia-Gonzalez, I.; Rial-Bastón, V.; Corcobado, M.C.; Espinosa, M.C.; Ruiz, F. Early prognostic value of an Algorithm based on spectral Variables of Ventricular fibrillAtion from the EKG of patients with suddEn cardiac death: A multicentre observational study (AWAKE). Arch. Cardiol. México 2018, 88, 460–467. [Google Scholar] [CrossRef]
- Filgueiras-Rama, D.; Calvo, C.J.; Salvador-Montañés, Ó.; Cádenas, R.; Ruiz-Cantador, J.; Armada, E.; Rey, J.R.; Merino, J.; Peinado, R.; Pérez-Castellano, N. Spectral analysis-based risk score enables early prediction of mortality and cerebral performance in patients undergoing therapeutic hypothermia for ventricular fibrillation and comatose status. Int. J. Cardiol. 2015, 186, 250–258. [Google Scholar] [CrossRef]
- Hussein, A.F.; Hashim, S.J.; Aziz, A.F.A.; Rokhani, F.Z.; Adnan, W.A.W. Performance evaluation of time-frequency distributions for ECG signal analysis. J. Med. Syst. 2018, 42, 15. [Google Scholar] [CrossRef]
- Mehrabi, N.; Morstatter, F.; Saxena, N.; Lerman, K.; Galstyan, A. A survey on bias and fairness in machine learning. ACM Comput. Surv. 2021, 54, 1–35. [Google Scholar] [CrossRef]
- Cabrera, Á.A.; Epperson, W.; Hohman, F.; Kahng, M.; Morgenstern, J.; Chau, D.H. FairVis: Visual analytics for discovering intersectional bias in machine learning. In Proceedings of the 2019 IEEE Conference on Visual Analytics Science and Technology (VAST), Vancouver, BC, Canada, 20–25 October 2019. [Google Scholar]
- Schelter, S.; Stoyanovich, J. Taming technical bias in machine learning pipelines. Bull. Tech. Comm. Data Eng. 2020, 43, 39–50. [Google Scholar]
- Suri, J.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Viskovic, K.; Suri, N.; Alizad, A.; El-Baz, A.; Saba, L.; Fatemi, M. Systematic review of artificial intelligence in acute respiratory distress syndrome for COVID-19 lung patients: A biomedical imaging perspective. IEEE J. Biomed. Health Inform. 2021, 25, 4128–4139. [Google Scholar] [CrossRef]
- Buss, V.H.; Leesong, S.; Barr, M.; Varnfield, M.; Harris, M. Primary Prevention of Cardiovascular Disease and Type 2 Diabetes Mellitus Using Mobile Health Technology: Systematic Review of the Literature. J. Med. Internet Res. 2020, 22, e21159. [Google Scholar] [CrossRef]
- Villarreal, V.; Berbey-Alvarez, A. Evaluation of mHealth Applications Related to Cardiovascular Diseases: A Systematic Review. Acta Inform. Med. 2020, 28, 130–137. [Google Scholar] [CrossRef]
- Xiao, Q.; Lu, S.; Wang, Y.; Sun, L.; Wu, Y.; e-Health. Current status of cardiovascular disease-related smartphone apps downloadable in China. Telemedicine 2017, 23, 219–225. [Google Scholar] [CrossRef]
- Saba, L.; Banchhor, S.K.; Araki, T.; Viskovic, K.; Londhe, N.D.; Laird, J.R.; Suri, H.S.; Suri, J.S. Intra-and inter-operator reproducibility of automated cloud-based carotid lumen diameter ultrasound measurement. Indian Heart J. 2018, 70, 649–664. [Google Scholar] [CrossRef]
- Huda, N.; Khan, S.; Abid, R.; Shuvo, S.B.; Labib, M.M.; Hasan, T. A Low-cost, Low-energy Wearable ECG System with Cloud-Based Arrhythmia Detection. In Proceedings of the 2020 IEEE Region 10 Symposium (TENSYMP), Dhaka, Bangladesh, 5–7 June 2020; pp. 1840–1843. [Google Scholar]
- Singh, K.K.; Singh, S.S. An Artificial Intelligence based mobile solution for early detection of valvular heart diseases. In Proceedings of the 2019 IEEE International Conference on Electronics, Computing and Communication Technologies (CONECCT), Bangalore, India, 26–27 July 2019; pp. 1–5. [Google Scholar]
- Spanakis, E.G.; Psaraki, M.; Sakkalis, V. Congestive heart failure risk assessment monitoring through internet of things and mobile personal health systems. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 2925–2928. [Google Scholar]
- Paredes, S.; Henriques, J.; Rocha, T.; de Carvalho, P.; Morais, J.; Santos, L.; Carvalho, R. The lookAfterRisk Project: Dynamic Cardiovascular Risk Assessment based on Remote Monitoring Solutions. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 1122–1125. [Google Scholar]
- Freyer, L.; von Stülpnagel, L.; Spielbichler, P.; Sappler, N.; Wenner, F.; Schreinlechner, M.; Krasniqi, A.; Behroz, A.; Eiffener, E.; Zens, M. Rationale and design of a digital trial using smartphones to detect subclinical atrial fibrillation in a population at risk: The eHealth-based bavarian alternative detection of Atrial Fibrillation (eBRAVE-AF) trial. Am. Heart J. 2021, 241, 26–34. [Google Scholar] [CrossRef]
- Giansanti, D.; Monoscalco, L. A smartphone-based survey in mHealth to investigate the introduction of the artificial intelligence into cardiology. Mhealth 2021, 7, 8. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kang, K. Pit-a-Pat: A smart electrocardiogram system for detecting arrhythmia. Telemed. e-Health 2015, 21, 814–821. [Google Scholar] [CrossRef]
- Lillo-Castellano, J.; Mora-Jimenez, I.; Santiago-Mozos, R.; Chavarría-Asso, F.; Cano-González, A.; García-Alberola, A.; Rojo-Álvarez, J.L. Symmetrical compression distance for arrhythmia discrimination in cloud-based big-data services. IEEE J. Biomed. Health Inform. 2015, 19, 1253–1263. [Google Scholar] [CrossRef]
- Sakellarios, A.; Siogkas, P.; Georga, E.; Tachos, N.; Kigka, V.; Tsompou, P.; Andrikos, I.; Karanasiou, G.S.; Rocchiccioli, S.; Correia, J. A clinical decision support platform for the risk stratification, diagnosis, and prediction of coronary artery disease evolution. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 4556–4559. [Google Scholar]
- Suri, J.S.; Rangayyan, R.M. Breast Imaging, Mammography, and Computer-Aided Diagnosis of Breast Cancer; SPIE: Bellingham, WA, USA, 2006. [Google Scholar]
- Boernama, A.W.D.; Setiawan, N.A.; Wahyunggoro, O. Multiclass classification of brain-computer interface motor imagery system: A systematic literature review. In Proceedings of the 2021 International Conference on Artificial Intelligence and Mechatronics Systems (AIMS), Bandung, Indonesia, 28–30 April 2021; pp. 1–6. [Google Scholar]
- Collins, D.R.; Tompson, A.C.; Onakpoya, I.J.; Roberts, N.; Ward, A.M.; Heneghan, C.J. Global cardiovascular risk assessment in the primary prevention of cardiovascular disease in adults: Systematic review of systematic reviews. BMJ Open 2017, 7, e013650. [Google Scholar] [CrossRef]
- Dissanayake, K.; Md Johar, M.G. Comparative Study on Heart Disease Prediction Using Feature Selection Techniques on Classification Algorithms. Appl. Comput. Intell. Soft Comput. 2021, 2021, 5581806. [Google Scholar] [CrossRef]
- Galar, M.; Fernandez, A.; Barrenechea, E.; Bustince, H.; Herrera, F. A review on ensembles for the class imbalance problem: Bagging-, boosting-, and hybrid-based approaches. IEEE Trans. Syst. Man Cybern. Part C 2011, 42, 463–484. [Google Scholar] [CrossRef]
- Stewart, J.; Manmathan, G.; Wilkinson, P. Primary prevention of cardiovascular disease: A review of contemporary guidance and literature. JRSM Cardiovasc. Dis. 2017, 6, 2048004016687211. [Google Scholar] [CrossRef]
- Mathew, R.M.; Gunasundari, R. A Review on Handling Multiclass Imbalanced Data Classification In Education Domain. In Proceedings of the 2021 International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE), Greater Noida, India, 4–5 March 2021; pp. 752–755. [Google Scholar]
- Uike, D.; Thorat, S. Implementation of Multiclass Algorithm for Sickle Cell Identification and Categorization—A Review. In Proceedings of the 2020 2nd International Conference on Innovative Mechanisms for Industry Applications (ICIMIA), Bangalore, India, 5–7 March 2020; pp. 300–303. [Google Scholar]
- Wang, H.; Liu, X.; Lv, B.; Yang, F.; Hong, Y. Reliable multi-label learning via conformal predictor and random forest for syndrome differentiation of chronic fatigue in traditional Chinese medicine. PLoS ONE 2014, 9, e99565. [Google Scholar] [CrossRef]
- Wiharto, W.; Kusnanto, H.; Herianto, H. Intelligence system for diagnosis level of coronary heart disease with K-star algorithm. Healthc. Inform. Res. 2016, 22, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Boi, A.; Jamthikar, A.D.; Saba, L.; Gupta, D.; Sharma, A.; Loi, B.; Laird, J.R.; Khanna, N.N.; Suri, J.S. A survey on coronary atherosclerotic plaque tissue characterization in intravascular optical coherence tomography. Curr. Atheroscler. Rep. 2018, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, E.; Corciu, A.; Venneri, L.; Faita, F.; Giannarelli, C.; Gemignani, V.; Demi, M. Assessment of cardiovascular risk markers from ultrasound images: System reproducibility. In Proceedings of the 2008 Computers in Cardiology, Bologna, Italy, 14–17 September 2008; pp. 105–108. [Google Scholar]
- Liu, L.; Tang, L. A Survey of Statistical Topic Model for Multi-Label Classification. In Proceedings of the 2018 26th International Conference on Geoinformatics, Kunming, China, 28–30 June 2018; pp. 1–5. [Google Scholar]
- Charte, F. A comprehensive and didactic review on multilabel learning software tools. IEEE Access 2020, 8, 50330–50354. [Google Scholar] [CrossRef]
- Siblini, W.; Kuntz, P.; Meyer, F. A review on dimensionality reduction for multi-label classification. IEEE Trans. Knowl. Data Eng. 2019, 33, 839–857. [Google Scholar] [CrossRef]
- Indhumathi, M.; Kumar, V.A. Healthcare Management of Major Cardiovascular Disease—A review. In Proceedings of the 2021 6th International Conference on Inventive Computation Technologies (ICICT), Coimbatore, India, 20–22 January 2021; pp. 1230–1235. [Google Scholar]
- Kolli, K.K.; Han, D.; Gransar, H.; Lee, J.H.; Choi, S.-Y.; Chun, E.J.; Jung, H.O.; Sung, J.; Han, H.-W.; Park, S.H. Machine learning algorithm to predict coronary artery calcification in asymptomatic healthy population. In Proceedings of the 2019 IEEE Healthcare Innovations and Point of Care Technologies, (HI-POCT), Bethesda, MD, USA, 20–22 November 2019; pp. 95–98. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Acharya, U.R.; Joseph, K.P.; Kannathal, N.; Min, L.C.; Suri, J.S. Heart rate variability. In Advances in Cardiac Signal Processing; Springer: Berlin/Heidelberg, Germany, 2007; pp. 121–165. [Google Scholar]
- Quang, D.; Chen, Y.; Xie, X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015, 31, 761–763. [Google Scholar] [CrossRef]
- Ding, C.; Chen, P.; Jiao, J. Non-linear effects of the built environment on automobile-involved pedestrian crash frequency: A machine learning approach. Accid. Anal. Prev. 2018, 112, 116–126. [Google Scholar] [CrossRef]
- Eder, S.J.; Nicholson, A.A.; Stefanczyk, M.M.; Pieniak, M.; Martínez-Molina, J.; Pešout, O.; Binter, J.; Smela, P.; Scharnowski, F.; Steyrl, D. Securing your relationship: Quality of intimate relationships during the COVID-19 pandemic can be predicted by attachment style. Front. Psychol. 2021, 3016. [Google Scholar] [CrossRef]
- Milicevic, O.; Salom, I.; Rodic, A.; Markovic, S.; Tumbas, M.; Zigic, D.; Djordjevic, M.; Djordjevic, M. PM2. 5 as a major predictor of COVID-19 basic reproduction number in the USA. Environ. Res. 2021, 201, 111526. [Google Scholar] [CrossRef]
- Soltan, A.A.; Kouchaki, S.; Zhu, T.; Kiyasseh, D.; Taylor, T.; Hussain, Z.B.; Peto, T.; Brent, A.J.; Eyre, D.W.; Clifton, D.A. Rapid triage for COVID-19 using routine clinical data for patients attending hospital: Development and prospective validation of an artificial intelligence screening test. Lancet Digit. Health 2021, 3, e78–e87. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Ang, P.C.A.; Yanti, R.; Suri, J.S. Application of non-linear and wavelet based features for the automated identification of epileptic EEG signals. Int. J. Neural Syst. 2012, 22, 1250002. [Google Scholar] [CrossRef]
- Poddar, M.G.; Kumar, V.; Sharma, Y.P. Automated diagnosis of coronary artery diseased patients by heart rate variability analysis using linear and non-linear methods. J. Med. Eng. Technol. 2015, 39, 331–341. [Google Scholar] [CrossRef]
- Tran, J.; Sharma, D.; Gotlieb, N.; Xu, W.; Bhat, M. Application of machine learning in liver transplantation: A review. Hepatol. Int. 2022, 1–14. [Google Scholar] [CrossRef]
- Shandilya, S.; Ward, K.; Kurz, M.; Najarian, K. Non-linear dynamical signal characterization for prediction of defibrillation success through machine learning. BMC Med. Inform. Decis. Mak. 2012, 12, 116. [Google Scholar] [CrossRef]
- Singh, R.S.; Saini, B.S.; Sunkaria, R.K. Detection of coronary artery disease by reduced features and extreme learning machine. Clujul Med. 2018, 91, 166. [Google Scholar] [CrossRef]
- Shandilya, S.; Kurz, M.C.; Ward, K.R.; Najarian, K. Integration of Attributes from Non-Linear Characterization of Cardiovascular Time-Series for Prediction of Defibrillation Outcomes. PLoS ONE 2016, 11, e0141313. [Google Scholar]
- Hedjazi, N.; Kharboutly, H.; Benali, A.; Dibi, Z. PCA-based selection of distinctive stability criteria and classification of post-stroke pathological postural behaviour. Australas. Phys. Eng. Sci. Med. 2018, 41, 189–199. [Google Scholar] [CrossRef]
- Li, X.; Ling, S.H.; Su, S. A hybrid feature selection and extraction methods for sleep apnea detection using bio-signals. Sensors 2020, 20, 4323. [Google Scholar] [CrossRef]
- Jo, Y.-Y.; Kwon, J.-m.; Jeon, K.-H.; Cho, Y.-H.; Shin, J.-H.; Lee, Y.-J.; Jung, M.-S.; Ban, J.-H.; Kim, K.-H.; Lee, S.Y. Detection and classification of arrhythmia using an explainable deep learning model. J. Electrocardiol. 2021, 67, 124–132. [Google Scholar] [CrossRef]
- Muthulakshmi, M.; Kavitha, G. Deep CNN with LM learning based myocardial ischemia detection in cardiac magnetic resonance images. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 824–827. [Google Scholar]
- Liang, Y.; Yin, S.; Tang, Q.; Zheng, Z.; Elgendi, M.; Chen, Z. Deep learning algorithm classifies heartbeat events based on electrocardiogram signals. Front. Physiol. 2020, 11, 1255. [Google Scholar] [CrossRef]
- Tadesse, G.A.; Javed, H.; Weldemariam, K.; Liu, Y.; Liu, J.; Chen, J.; Zhu, T. DeepMI: Deep multi-lead ECG fusion for identifying myocardial infarction and its occurrence-time. Artif. Intell. Med. 2021, 121, 102192. [Google Scholar] [CrossRef]
- Tadesse, G.A.; Zhu, T.; Liu, Y.; Zhou, Y.; Chen, J.; Tian, M.; Clifton, D. Cardiovascular disease diagnosis using cross-domain transfer learning. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 4262–4265. [Google Scholar]
- Butun, E.; Yildirim, O.; Talo, M.; Tan, R.-S.; Acharya, U.R. 1D-CADCapsNet: One dimensional deep capsule networks for coronary artery disease detection using ECG signals. Phys. Med. 2020, 70, 39–48. [Google Scholar] [CrossRef]
- Wang, T.; Lu, C.; Sun, Y.; Yang, M.; Liu, C.; Ou, C. Automatic ECG classification using continuous wavelet transform and convolutional neural network. Entropy 2021, 23, 119. [Google Scholar] [CrossRef]
- Wang, R.; Fan, J.; Li, Y. Deep multi-scale fusion neural network for multi-class arrhythmia detection. IEEE J. Biomed. Health Inform. 2020, 24, 2461–2472. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Wang, K.; Liu, J.; He, R.; Yuan, Y.; Zhang, H. Automatic Multi-Label ECG Classification with Category Imbalance and Cost-Sensitive Thresholding. Biosensors 2021, 11, 453. [Google Scholar] [CrossRef]
- El-Baz, A.; Gimel’farb, G.; Suri, J.S. Stochastic Modeling for Medical Image Analysis; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Lloyd-Jones, D.M. Cardiovascular risk prediction: Basic concepts, current status, and future directions. Circulation 2010, 121, 1768–1777. [Google Scholar] [CrossRef]
- Farzadfar, F. Cardiovascular disease risk prediction models: Challenges and perspectives. Lancet Glob. Health 2019, 7, e1288–e1289. [Google Scholar] [CrossRef]
- Kuppili, V.; Biswas, M.; Sreekumar, A.; Suri, H.S.; Saba, L.; Edla, D.R.; Marinhoe, R.T.; Sanches, J.M.; Suri, J.S. Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. J. Med. Syst. 2017, 41, 152. [Google Scholar] [CrossRef]
- Banerjee, A.; Chen, S.; Pasea, L.; Lai, A.G.; Katsoulis, M.; Denaxas, S.; Nafilyan, V.; Williams, B.; Wong, W.K.; Bakhai, A. Excess deaths in people with cardiovascular diseases during the COVID-19 pandemic. Eur. J. Prev. Cardiol. 2021, 28, 1599–1609. [Google Scholar] [CrossRef]
- Magadum, A.; Kishore, R. Cardiovascular manifestations of COVID-19 infection. Cells 2020, 9, 2508. [Google Scholar]
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Laird, J.R.; Nicolaides, A.N.; Suri, J.S. Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study. Diagnostics 2021, 11, 2257. [Google Scholar] [CrossRef] [PubMed]
- Cherman, E.A.; Monard, M.C.; Metz, J. Multi-label problem transformation methods: A case study. CLEI Electron. J. 2011, 14, 4. [Google Scholar] [CrossRef]
- Read, J.; Pfahringer, B.; Holmes, G.; Frank, E. Classifier chains for multi-label classification. Mach. Learn. 2011, 85, 333–359. [Google Scholar] [CrossRef]
- Boutell, M.R.; Luo, J.; Shen, X.; Brown, C.M. Learning multi-label scene classification. Pattern Recognit. 2004, 37, 1757–1771. [Google Scholar] [CrossRef]
- Tsoumakas, G.; Vlahavas, I. Random k-labelsets: An ensemble method for multilabel classification. In Proceedings of European Conference on Machine Learning; Springer: Berlin/Heidelberg, Germany, 2007; pp. 406–417. [Google Scholar]
- Zhang, M.-L.; Zhou, Z.-H. ML-KNN: A lazy learning approach to multi-label learning. Pattern Recognit. 2007, 40, 2038–2048. [Google Scholar] [CrossRef]
- Benites, F.; Sapozhnikova, E. Haram: A hierarchical aram neural network for large-scale text classification. In Proceedings of the 2015 IEEE international Conference on Data Mining Workshop (ICDMW), Atlantic City, NJ, USA, 14–17 November 2015; pp. 847–854. [Google Scholar]
- Naing, N.N. Determination of sample size. Malays. J. Med. Sci. MJMS 2003, 10, 84. [Google Scholar]
- Qualtrics, S.S. Determining Sample Size: How to Ensure You Get the Correct Sample Size; Qualtrics: Seattle, WA, USA, 2019. [Google Scholar]
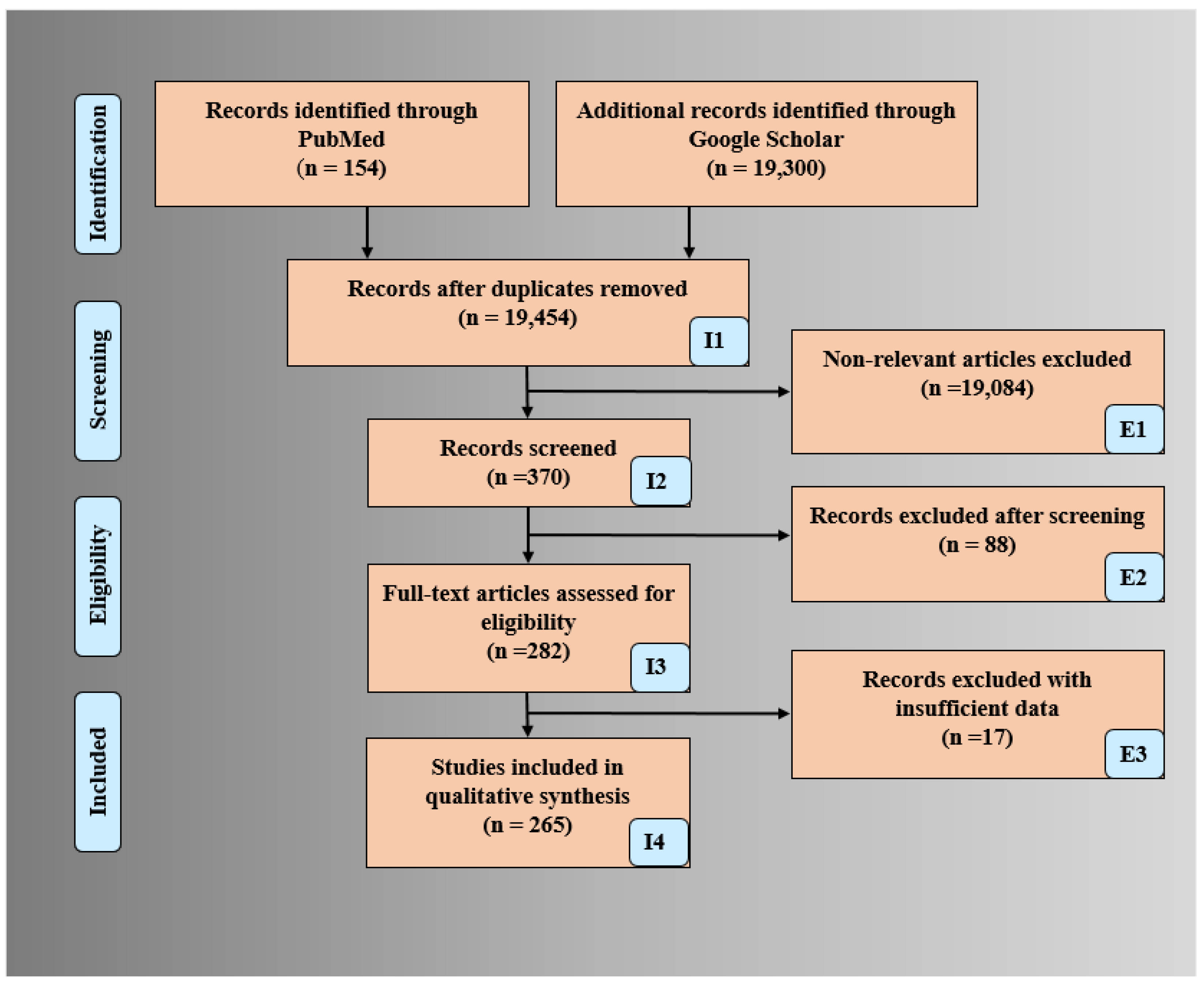
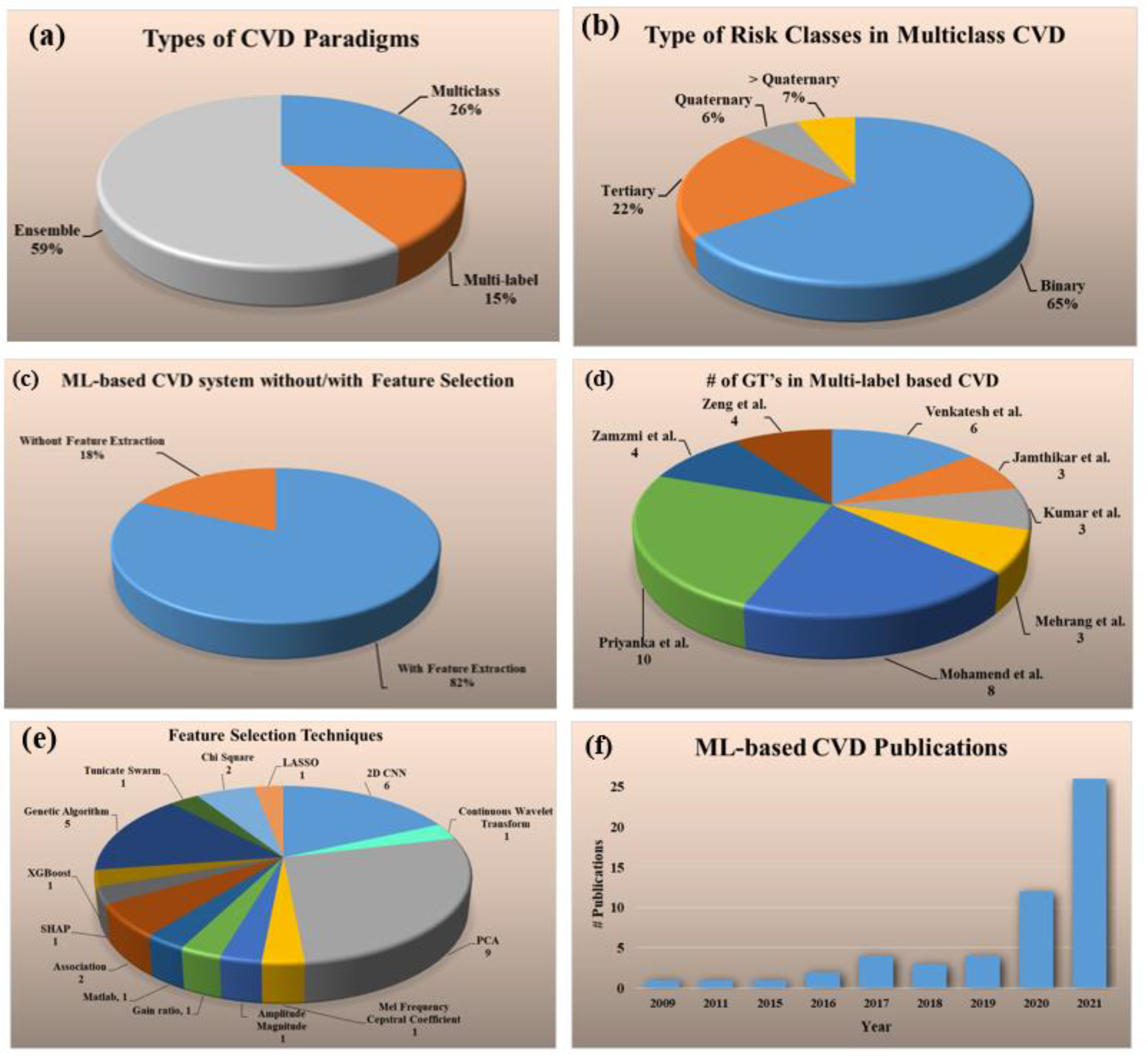
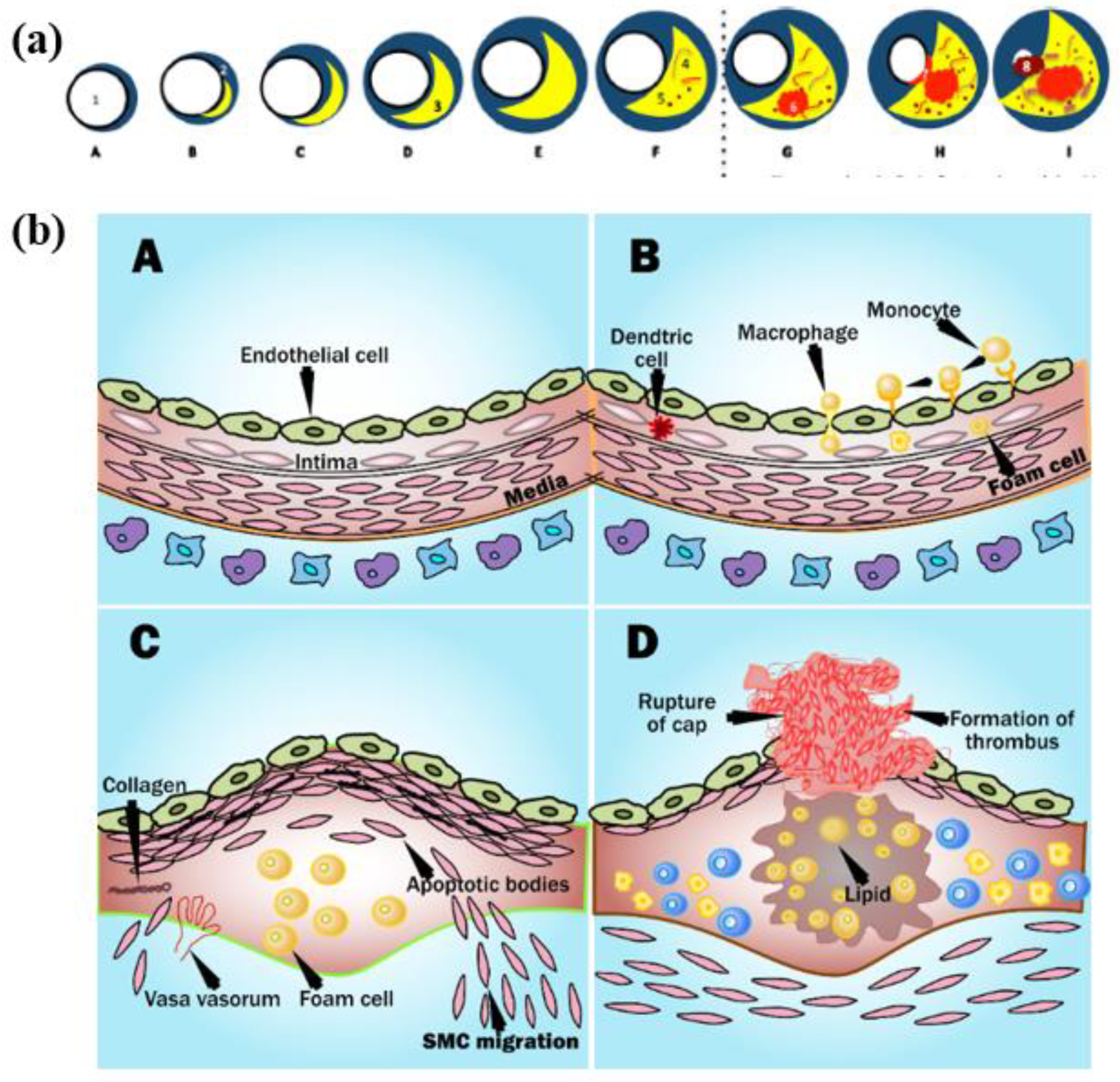
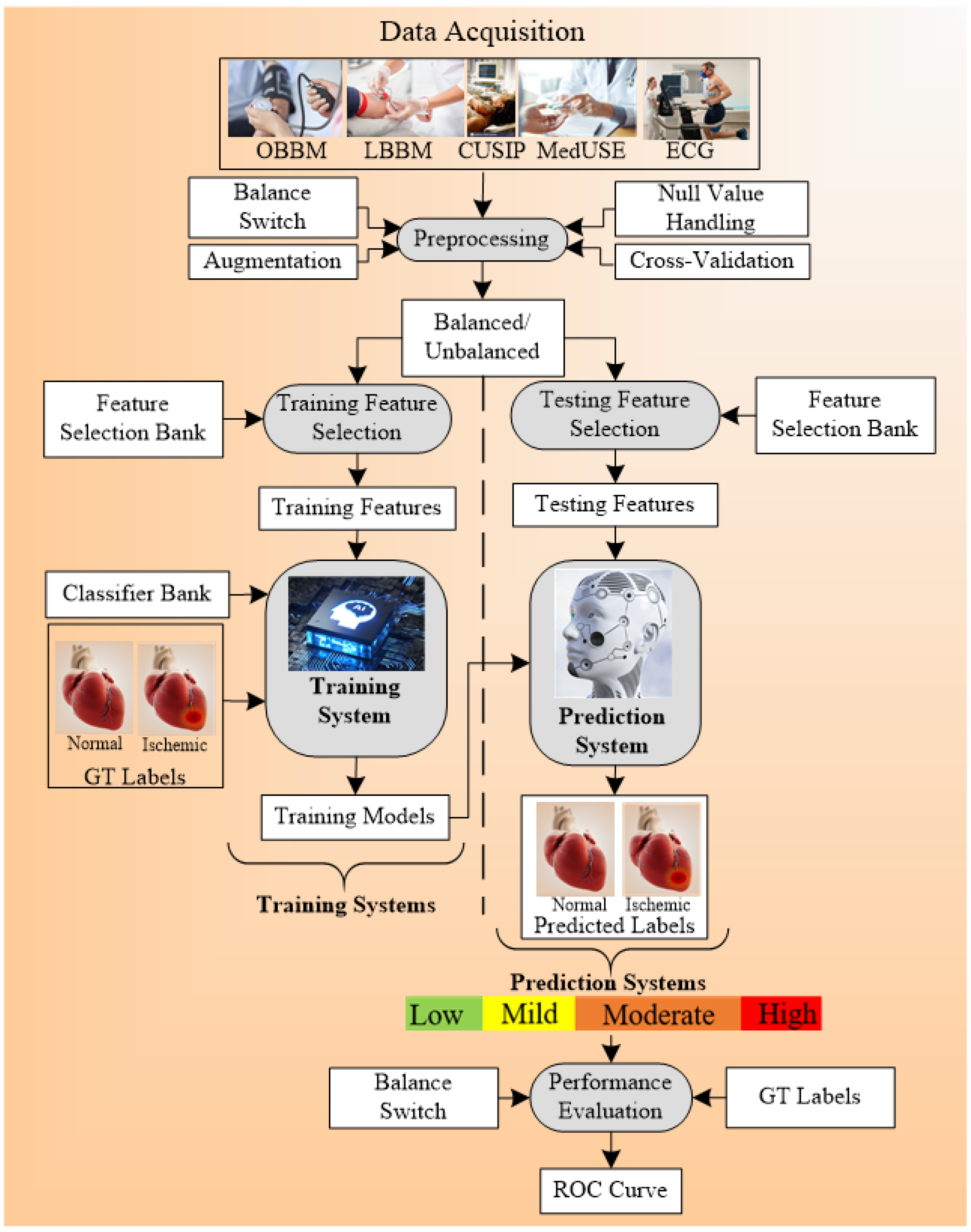
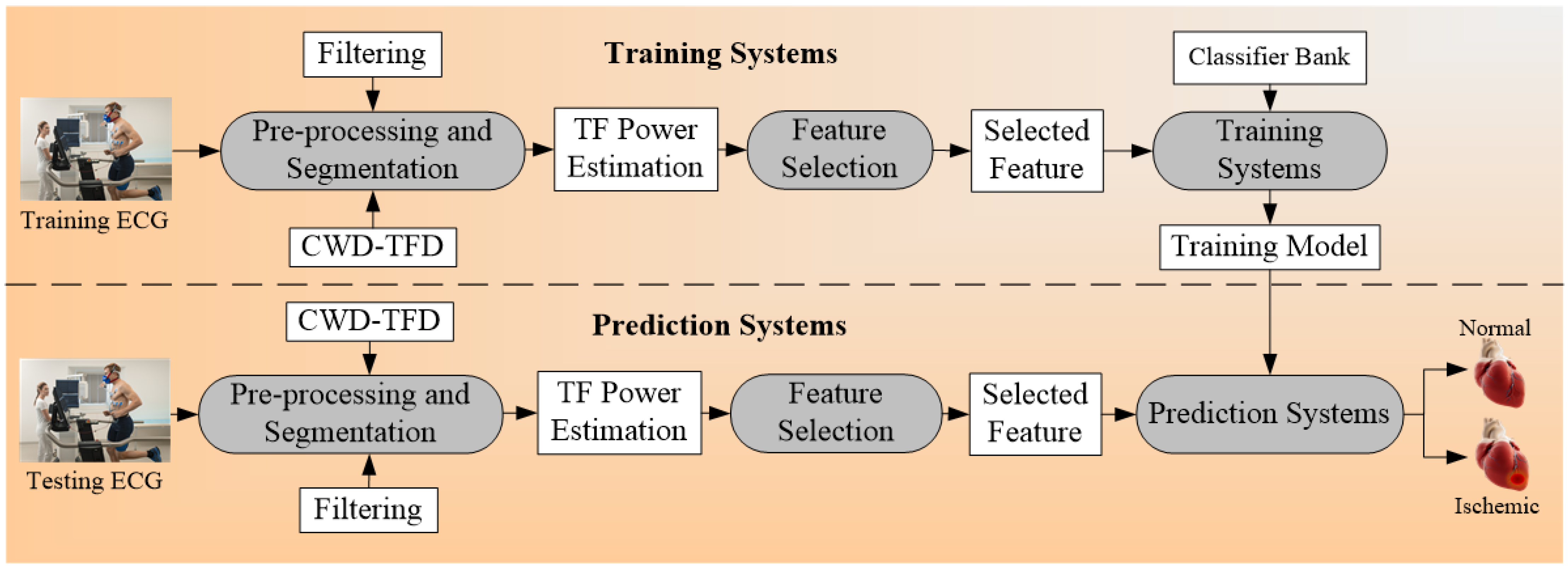
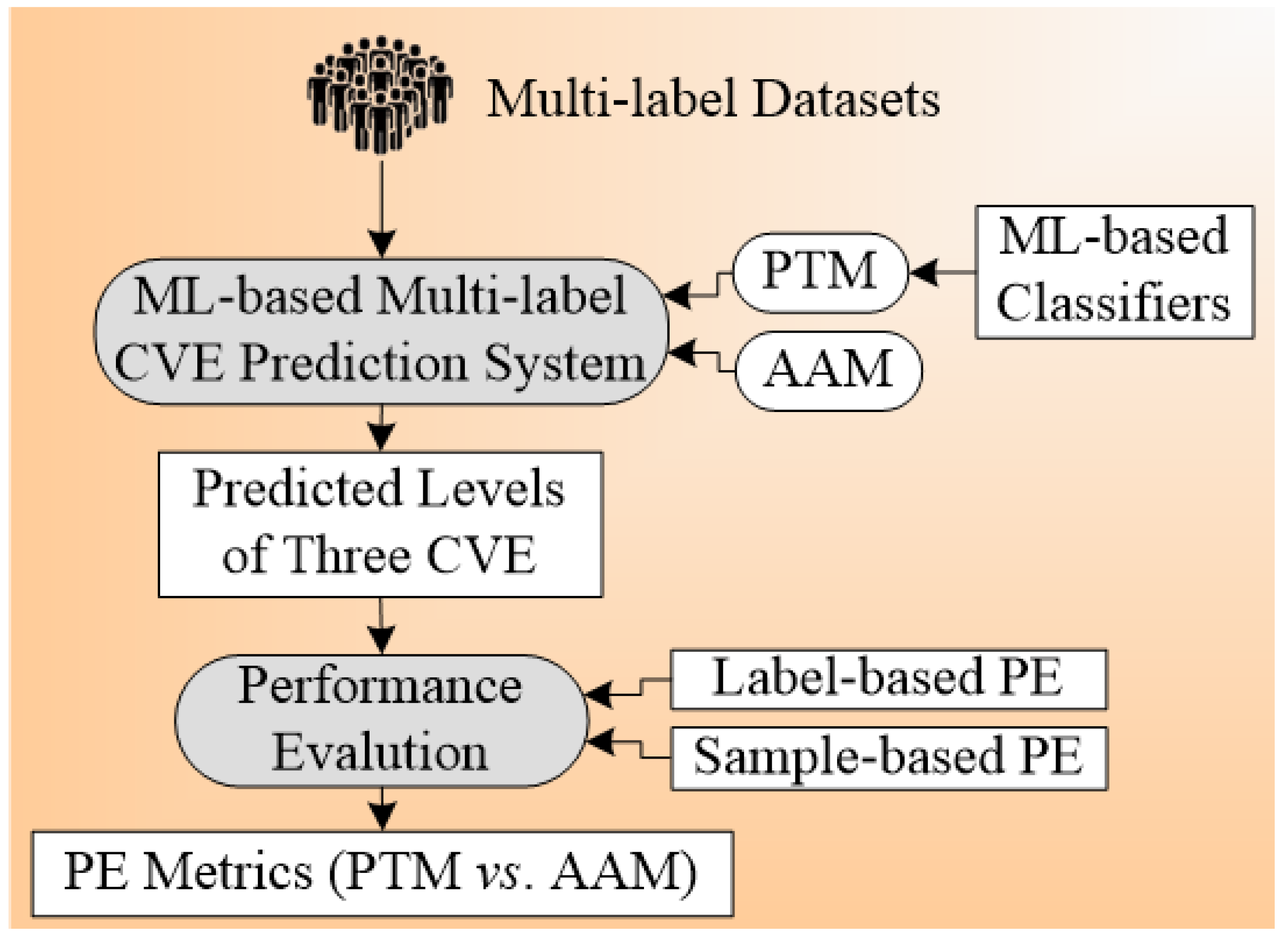

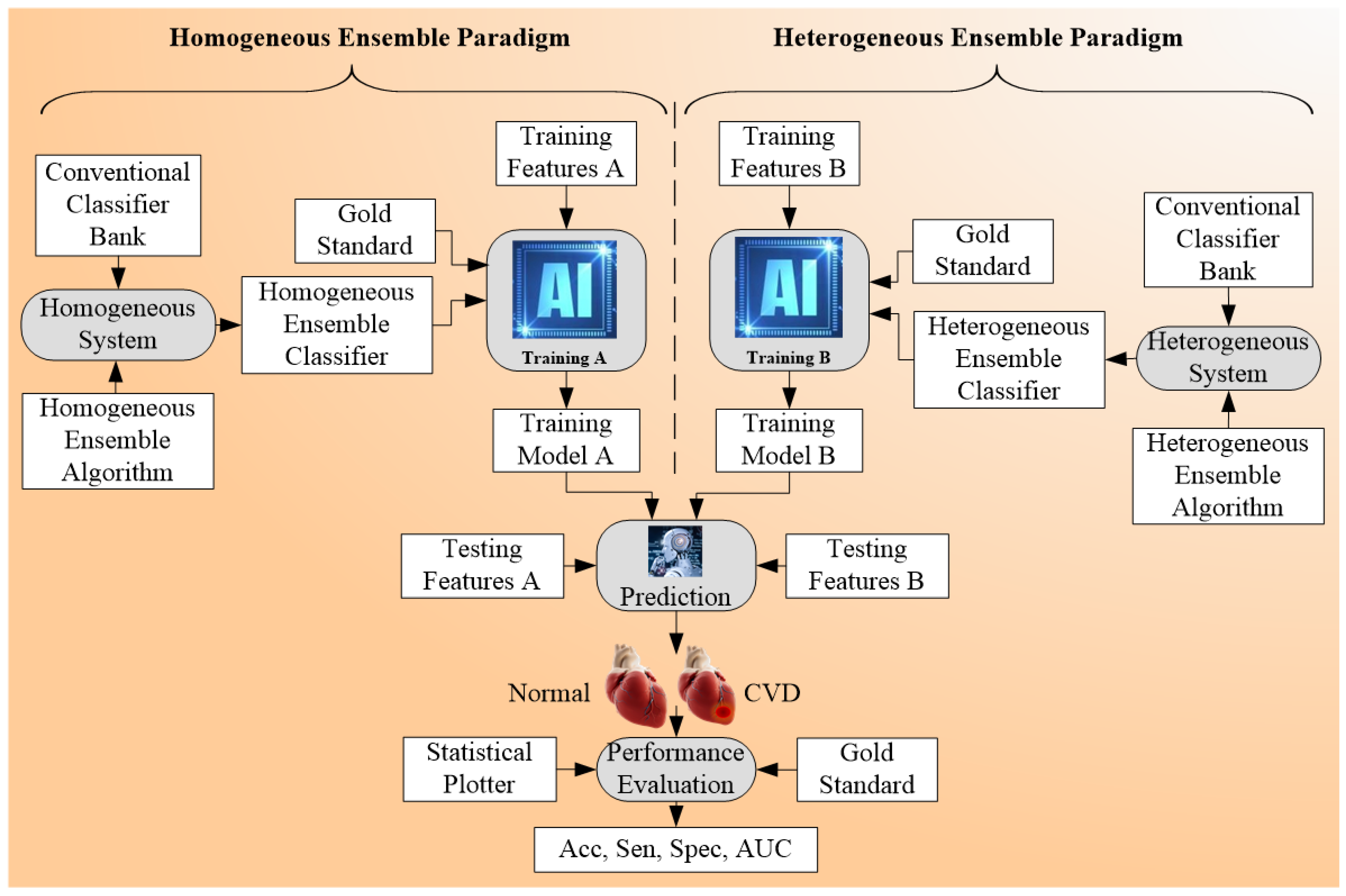
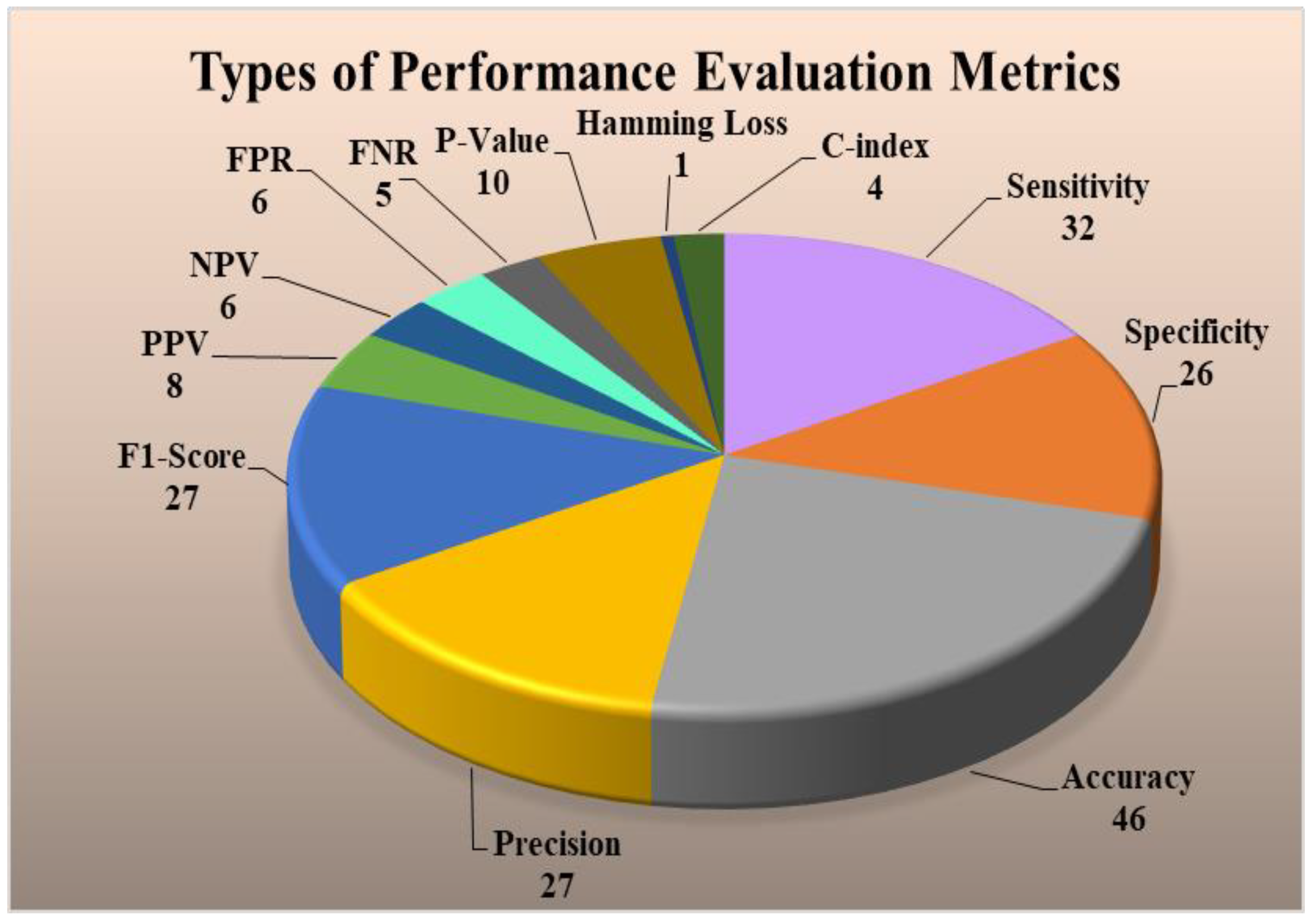
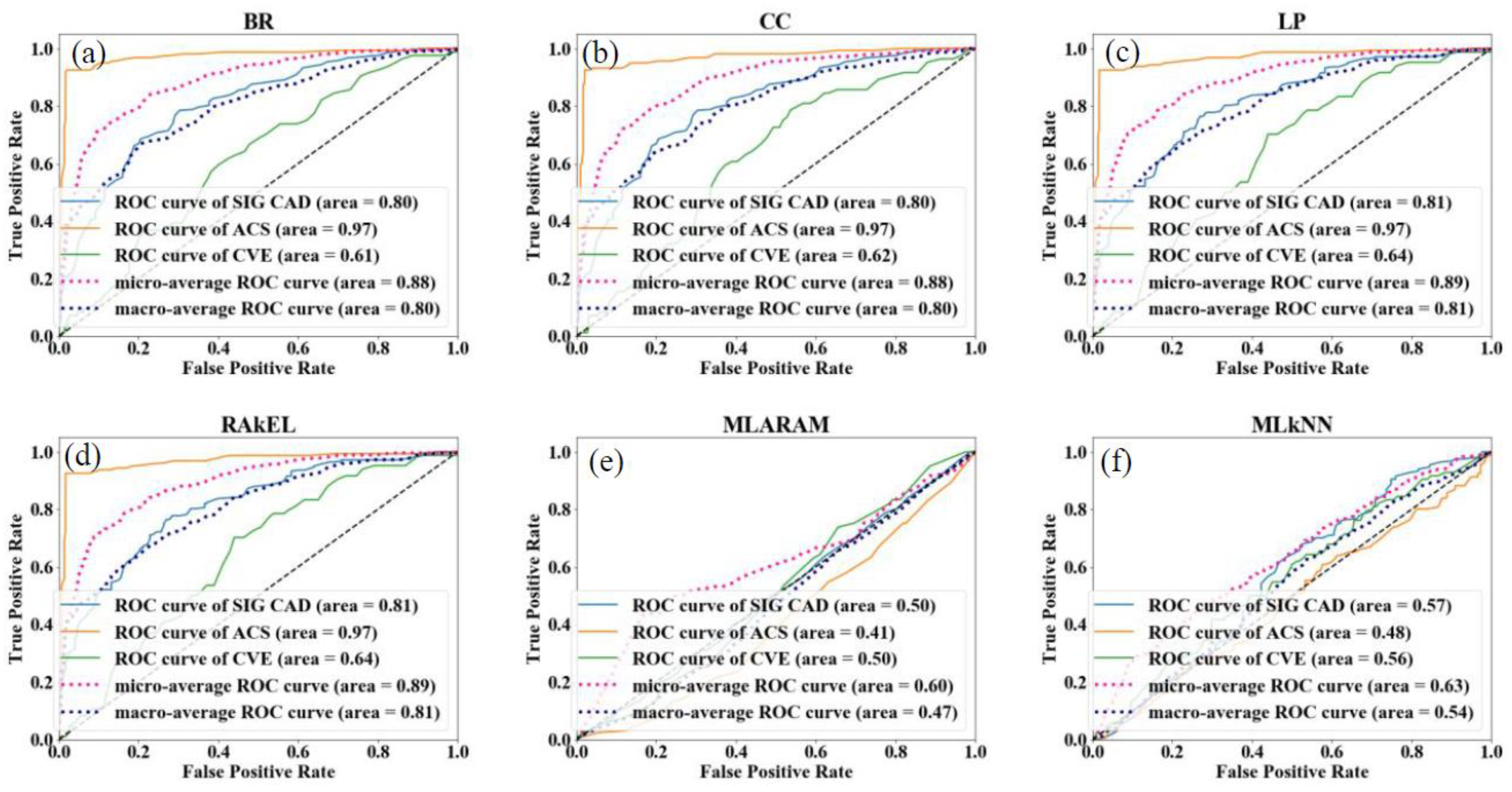
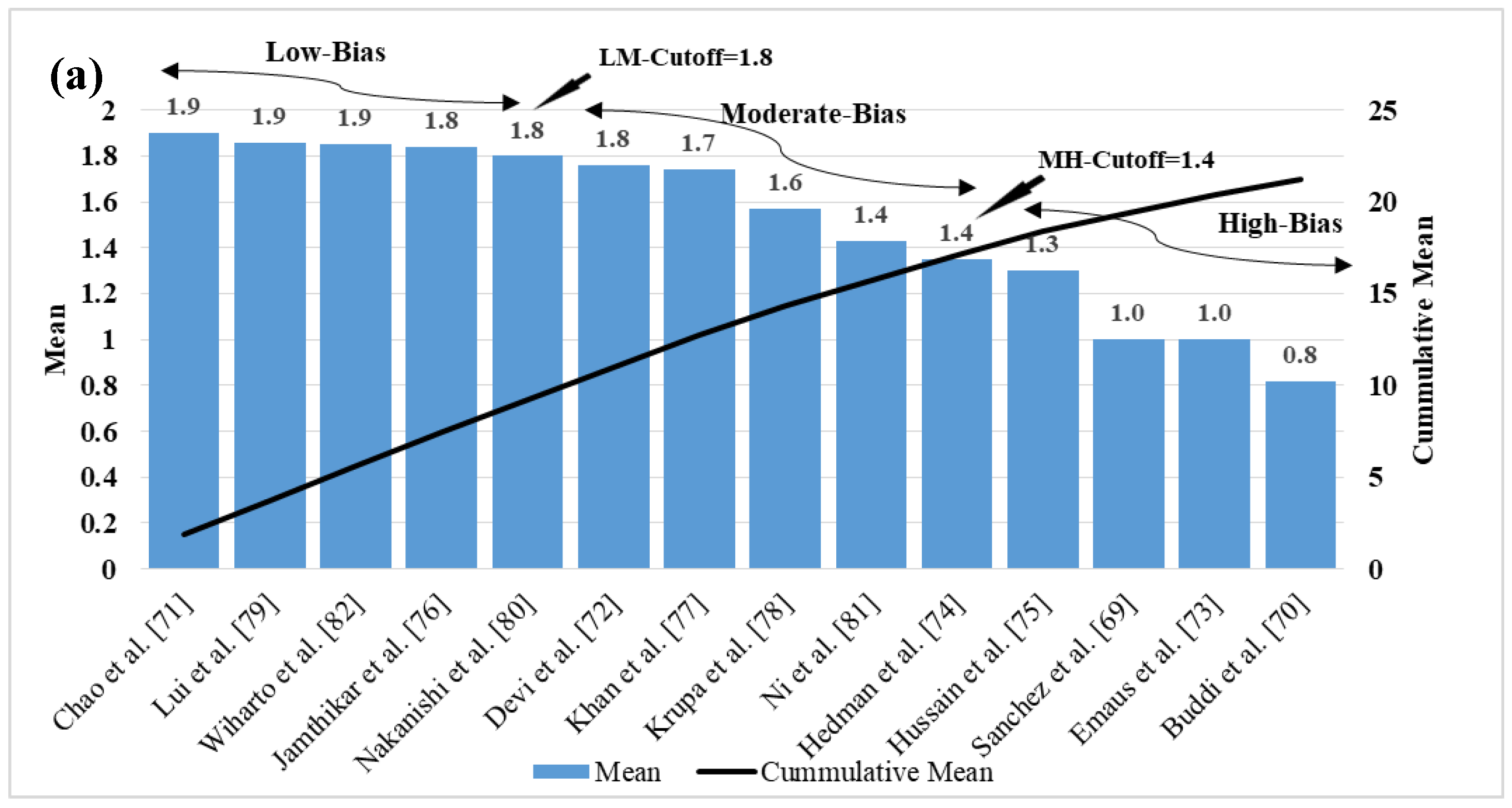
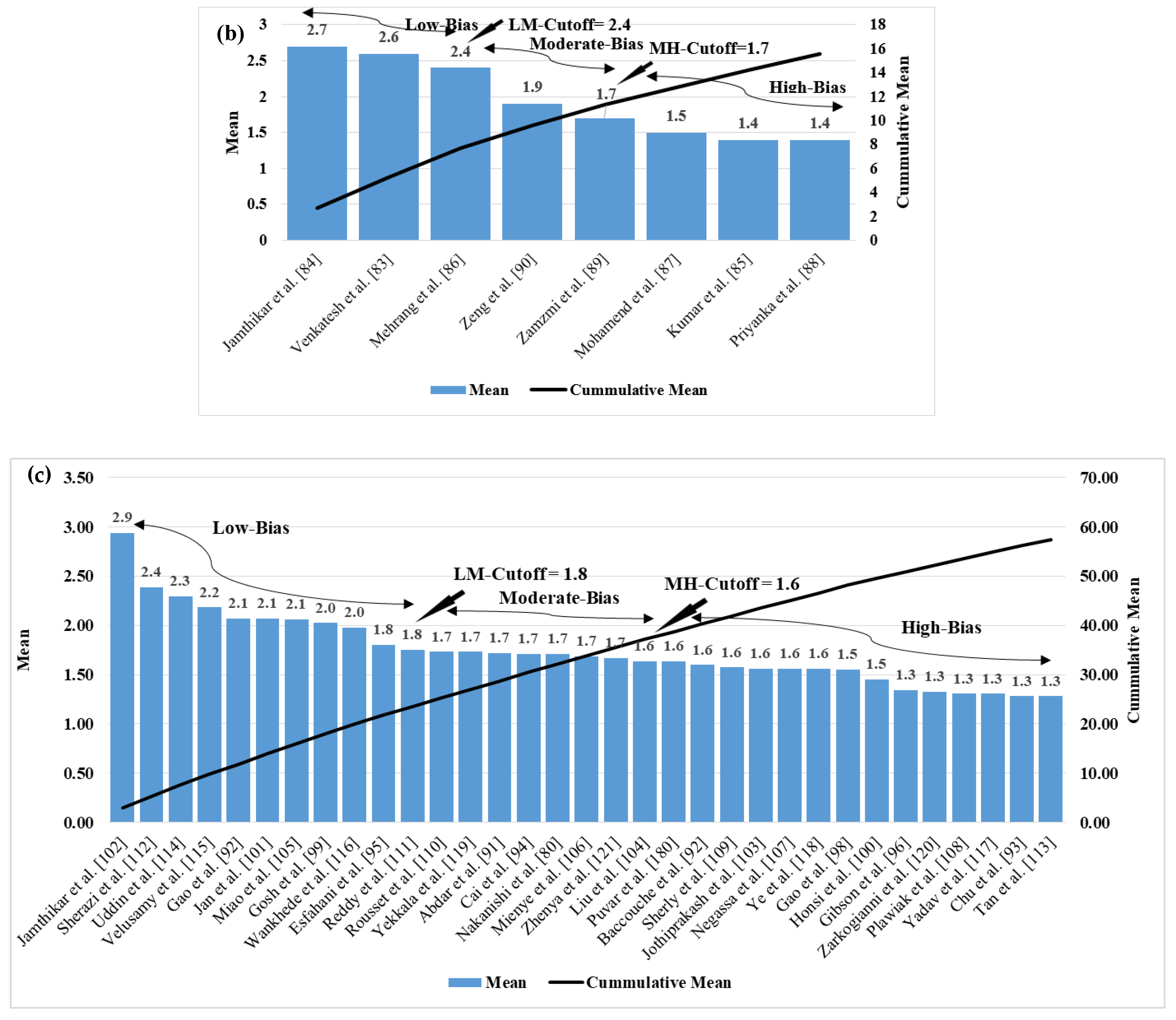

| SN | Studies | Input Covariates | Gold Standard Types | #RC | ML/DL |
|---|---|---|---|---|---|
| 1 | Chao et al. [71] | OBBM, LBBM | CVD Event | K | DL |
| 2 | Lui et al. [79] | ECG parameters | CHC | 3 | ML |
| 3 | Wiharto et al. [82] | OBBM, LBBM, ECG | CHD | 3 | ML |
| 4 | Jamthikar et al. [76] | OBBM, LBBM, CUSIP | CVE | 4 | ML |
| 5 | Nakanishi et al. [80] | OBBM, LBBM, CUSIP | Death | 3 | ML |
| 6 | Devi et al. [72] | ECG Parameters | SCD | 3 | ML |
| 7 | Khan et al. [77] | PCG Signals | CVE | 3 | ML |
| 8 | Krupa et al. [78] | APG signals | BCVD | 3 | ML |
| 9 | Ni et al. [81] | ECG Signals | CVD, No CVD | 4 | DL |
| 10 | Hedman et al. [74] | OBBM, LBBM | Heart Failure | 3 | ML |
| 11 | Hussain et al. [75] | OBBM, LBBM, ECG | MI | 3 | ML |
| 12 | Sanchez et al. [69] | OBBM, LBBM | CAC score | 9 | ML |
| 13 | Emaus et al. [73] | OBBM, CAC (CT) | F/NF CVD | 3 | DL |
| 14 | Buddi et al. [70] | OBBM, LBBM | CVD, Diabetes | 4 | ML |
| SN | Attributes | Multiclass CVD | Multiclass Non-CVD |
|---|---|---|---|
| 1 | Ground truth types | CVE [69,70,71,72,73,76,77,78,79,81,82], HF [74], MI [75], Death [80] | AD, NC, MCI, PMCI vs. SMCI [141], Proliferation, NP [139], ADH, DCS, IC [137,138,142] |
| 2 | Covariates types for the ML design | OBBM [69,70,73,74,75,76,80,82], LBBM [69,70,73,74,75,76,80,82], CUSIP [71,72,76,77,78,79,80,81,82], MU [76] | BHI [139], OBBM [137,138,141,142], LBBM [137,138,141,142] |
| 3 | Disease Type | CVD [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | Diabetes [142], Cancer (Breast, Lung, Brain) [138,139], Alzheimer’s [138,141], Retinal [137] |
| 4 | Image Modalities | ECG, CT, US [71,72,76,77,78,79,80,81,82] | EEG, MRI, CT [137,139] |
| 5 | # Classes | 3–9 [69,70,71,72,73,74,75,76,77,78,79,80,81,82] | 5–14 [137,138,139,141,142] |
| 6 | Architecture Type | ML [70,72,76,77,78,79,80,82], DL [71,81] | ML, rMLTFL [141] |
| 7 | Classifiers used | SVM [70,75,76,77], DT, RF, LR, NB, KNN, CNN [71,79] | RetiCAC [137], PCE, SVM, CNN, DT, LR, NB, SVM, KNN, ensemble [138,139] |
| SN | Studies | Input Covariates | Ground Truth | ML/DL |
|---|---|---|---|---|
| 1 | Venkatesh et al. [83] | OBBM, LBBM | Death, Stroke, CHD, CVD, HF, AF | ML |
| 2 | Jamthikar et al. [84] | OBBM, LBBM, CUSIP | CAD, ACS, Composite CVE | ML |
| 3 | Kumar et al. [85] | OBBM, LBBM, ECG | LVD, SVD, ICH | ML |
| 4 | Mehrang et al. [86] | OBBM, LBBM, CUSIP | Non-AFib-Non-ADHF, Afib-Non-ADHF, Afib-ADHF | ML |
| 5 | Mohamend et al. [87] | OBBM, LBBM, CUSIP | SHF, ASHF, CSHF, ACSHF, DHF, ADHF, CDHF, ACDHF | ML |
| 6 | Priyanka et al. [88] | OBBM, LBBM | HT, CHF, AF, CA, AKF, Dia-TII, HL, ARF, UTI, ER | ML |
| 7 | Zamzmi et al. [89] | MRI, CT Signals | HF, CAD, DCM, MI | DL |
| 8 | Zeng et al. [90] | OBBM, LBBM | LC, CC, IC, RC | ML |
| SN | Studies | Input Covariates | Ground Truth | ML/DL |
|---|---|---|---|---|
| 1 | Abdar et al. [91] | OBBM, LBBM | CAD | ML |
| 2 | Baccouche et al. [92] | OBBM, LBBM | HHD, IHD, MHD, VHD | DL |
| 3 | Chu et al. [93] | OBBM, LBBM, ECG | CVD, Dia | ML |
| 4 | Cai et al. [94] | OBBM, LBBM | CR | ML |
| 5 | Esfahani et al. [95] | OBBM, LBBM | CVD | ML |
| 6 | Gibson et al. [96] | OBBM, LBBM | ACS | ML |
| 7 | Gao et al. [97] | OBBM, LBBM, ECG | CVD, BC | ML |
| 8 | Gao et al. [98] | OBBM, LBBM | CVD | ML |
| 9 | Gosh et al. [99] | OBBM, LBBM, ECG | CVD | ML |
| 10 | Honsi et al. [100] | OBBM, LBBM | CVD | ML |
| 11 | Jan et al. [101] | OBBM, LBBM, ECG | HD | ML |
| 12 | Jamthikar et al. [102] | OBBM, LBBM, CUSIP | CAD, ACS | ML |
| 13 | Jothiprakash et al. [103] | OBBM, LBBM | CVD | ML |
| 14 | Liu et al. [104] | OBBM, LBBM | CA | ML |
| 15 | Miao et al. [105] | OBBM, LBBM, ECG | CHD | ML |
| 16 | Mienye et al. [106] | OBBM, LBBM | HD | ML |
| 17 | Negassa et al. [107] | OBBM, LBBM | HF | ML |
| 18 | Nakanishi et al. [80] | OBBM, LBBM, CT | Death | ML |
| 19 | Plawiak et al. [108] | OBBM, LBBM, ECG | Arrhythmia | DL |
| 20 | Puvar et al. [180] | OBBM, LBBM, ECG | HD | ML |
| 21 | Reddy et al. [109] | OBBM, LBBM | HD | ML |
| 22 | Rousset et al. [110] | OBBM, LBBM | CVD | ML |
| 23 | Sherly et al. [111] | OBBM, LBBM, ECG | HD | ML |
| 24 | Sherazi et al. [112] | OBBM, LBBM | CVE | ML |
| 25 | Tan et al. [113] | OBBM, LBBM | CVD | ML |
| 26 | Uddin et al. [114] | OBBM, LBBM | CVD | ML |
| 27 | Velusamy et al. [115] | OBBM, LBBM | CAD | ML |
| 28 | Wankhede et al. [116] | OBBM, LBBM | HD | DL |
| 29 | Yadav et al. [117] | OBBM, LBBM | HD | ML |
| 30 | Ye et al. [118] | OBBM, LBBM | HYT | ML |
| 31 | Yekkala et al. [119] | OBBM, LBBM | CVD | ML |
| 32 | Zarkogianni et al. [120] | OBBM, LBBM | CVD, Dia | ML |
| 33 | Zhenya et al. [121] | OBBM, LBBM, ECG | HD | ML |
| (a) Multiclass Studies | Sum | Mean | Rank | (c) Ensemble Studies | Sum | Mean | Rank |
| Chao et al. [71] | 78 | 1.9 | 1 | Jamthikar et al. [102] | 120.5 | 2.9 | 1 |
| Lui et al. [79] | 76.5 | 1.9 | 2 | Sherazi et al. [112] | 98 | 2.4 | 2 |
| Wiharto et al. [82] | 76 | 1.9 | 3 | Uddin et al. [114] | 94 | 2.3 | 3 |
| Jamthikar et al. [76] | 75.5 | 1.8 | 4 | Velusamy et al. [115] | 89.5 | 2.2 | 4 |
| Nakanishi et al. [80] | 74 | 1.8 | 5 | Gao et al. [97] | 85 | 2.1 | 5 |
| Devi et al. [72] | 72.5 | 1.8 | 6 | Jan et al. [101] | 85 | 2.1 | 6 |
| Khan et al. [77] | 71.5 | 1.7 | 7 | Miao et al. [105] | 84.5 | 2.1 | 7 |
| Krupa et al. [78] | 64.5 | 1.6 | 8 | Gosh et al. [99] | 83 | 2 | 8 |
| Ni et al. [81] | 59 | 1.4 | 9 | Wankhede et al. [116] | 81 | 2 | 9 |
| Hedman et al. [74] | 55.5 | 1.4 | 10 | Esfahani et al. [95] | 74 | 1.8 | 10 |
| Hussain et al. [75] | 53.5 | 1.3 | 11 | Reddy et al. [111] | 72 | 1.8 | 11 |
| Sanchez et al. [69] | 43 | 1 | 12 | Rousset et al. [110] | 71 | 1.7 | 12 |
| Emaus et al. [73] | 41 | 1 | 13 | Yekkala et al. [119] | 71 | 1.7 | 13 |
| Buddi et al. [70] | 33.5 | 0.8 | 14 | Abdar et al. [91] | 70.5 | 1.7 | 14 |
| (b) Multi-label Studies | Sum | Mean | Rank | Cai et al. [94] | 70 | 1.7 | 15 |
| Jamthikar et al. [84] | 111.5 | 2.7 | 1 | Nakanishi et al. [80] | 70 | 1.7 | 16 |
| Venkatesh et al. [83] | 108 | 2.6 | 2 | Mienye et al. [106] | 69 | 1.7 | 17 |
| Mehrang et al. [86] | 96.5 | 2.4 | 3 | Zhenya et al. [121] | 68.5 | 1.7 | 18 |
| Zeng et al. [90] | 76.5 | 1.9 | 4 | Liu et al. [104] | 67 | 1.6 | 19 |
| Zamzmi et al. [89] | 69.5 | 1.7 | 5 | Puvar et al. [180] | 67 | 1.6 | 20 |
| Mohamend et al. [87] | 60 | 1.5 | 6 | Baccouche et al. [92] | 65.5 | 1.6 | 21 |
| Kumar et al. [85] | 59 | 1.4 | 7 | Sherly et al. [109] | 64.5 | 1.6 | 22 |
| Priyanka et al. [88] | 59 | 1.4 | 8 | Jothiprakash et al. [103] | 64 | 1.6 | 23 |
| Negassa et al. [107] | 64 | 1.6 | 24 | ||||
| Ye et al. [118] | 64 | 1.6 | 25 | ||||
| Gao et al. [98] | 63.5 | 1.5 | 26 | ||||
| Honsi et al. [100] | 59.5 | 1.5 | 27 | ||||
| Gibson et al. [96] | 55 | 1.3 | 28 | ||||
| Zarkogianni et al. [120] | 54.5 | 1.3 | 29 | ||||
| Plawiak et al. [108] | 53.5 | 1.3 | 30 | ||||
| Yadav et al. [117] | 53.5 | 1.3 | 31 | ||||
| Chu et al. [93] | 52.5 | 1.2 | 32 | ||||
| Tan et al. [113] | 52.5 | 1.2 | 33 |
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 | C12 | C13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SN | Author | Yr | JOU | DS | CVD | Domain | ML | CT | CVP | MC | MLB | Ensbl | Summary |
| R1 | Boernama et al. [211] | ’21 | IEEE | 30 | 🗶 | EEG | 🗸 | SVM, NN, LDA, OVO | 🗶 | 🗸 | 🗶 | 🗶 | EEG Classification |
| R2 | Collins et al. [212] | ’16 | BMJ | 122 | 🗸 | BP | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | CVD Meta-analysis |
| R3 | Dissanayake et al. [213] | ’21 | Hindawi | CHDD | 🗸 | 🗶 | 🗸 | RF, SVM, DT, KNN, LR, GNB | K5 | 🗸 | 🗶 | 🗶 | CVD risk |
| R4 | Galar et al. [214] | ’12 | IEEE Tran. | Imb D | 🗶 | 🗶 | 🗸 | SMOTE | K5 | 🗶 | 🗶 | 🗸 | Ensemble Classification |
| R5 | Stewart et al. [215] | ’17 | JRSMCD | 🗶 | 🗸 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | 🗶 | CVD risk |
| R6 | Mathew et al. [216] | ’21 | IEEE | 6 | 🗶 | Edu | 🗸 | Adaboost, KNN, BPSO | K7 | 🗸 | 🗶 | 🗸 | Teaching Quality |
| R7 | Uike et al. [217] | ’21 | IEEE | 8 | 🗶 | SC | 🗸 | XG-Boost | Open | 🗸 | 🗶 | 🗶 | SC Classification |
| R8 | Wang et al. [218] | ’14 | Plos One | 736 | 🗶 | CF | 🗸 | RF, NBC, KNN | K10 | 🗶 | 🗸 | 🗶 | CF Classification |
| R9 | Wiharto et al. [219] | ’16 | HIR | 303 | 🗸 | 🗶 | 🗸 | K-Star | K* | 🗸 | 🗶 | 🗶 | CHD Classification |
| R10 | Boi et al. [220] | ’18 | CAR | 126 | 🗸 | 🗶 | 🗸 | SVM, RF, CNN | 🗶 | 🗶 | 🗶 | 🗸 | OCT-based risk stratification |
| R11 | Jamthikar et al. [35] | ’20 | CBM | 208 | 🗸 | 🗶 | 🗸 | KNN, RF, DT | K10 | 🗸 | 🗶 | 🗶 | CVD risk |
| R12 | Bianchini et al. [221] | ’08 | IEEE | 10 | 🗸 | 🗶 | 🗸 | 🗶 | 🗶 | 🗸 | 🗶 | 🗶 | Cardiovascular Risk Markers |
| R13 | Liu et al. [222] | ’12 | IEEE | 15 | 🗶 | Statistics | 🗸 | LDA | 🗶 | 🗶 | 🗸 | 🗶 | Statistical Classification |
| R14 | Charte et al. [223] | ’20 | IEEE | 🗶 | 🗶 | Software | 🗸 | MULAN | 🗶 | 🗶 | 🗸 | 🗶 | Comparison |
| R15 | Siblini et al. [224] | ’15 | IEEE | 156 | 🗶 | DM | 🗸 | LDA, MDDM | 🗶 | 🗶 | 🗸 | 🗶 | DM Reduction |
| R16 | Indhumathi et al. [225] | ’21 | IEEE | 30 | 🗸 | 🗶 | 🗸 | Probabilistic | 🗶 | 🗶 | 🗶 | 🗶 | CVD Management |
| R17 | Kolli et al. [226] | ’19 | IEEE | 86,155 | 🗸 | 🗶 | 🗸 | LogitBoost | K5 | 🗸 | 🗶 | 🗸 | Coronary Artery Calcification |
| R18 | Proposed Study | ’22 | 🗶 | 265 | 🗸 | 🗶 | 🗸 | 🗶 | 🗶 | 🗸 | 🗸 | 🗸 | CVD risk |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suri, J.S.; Bhagawati, M.; Paul, S.; Protogerou, A.D.; Sfikakis, P.P.; Kitas, G.D.; Khanna, N.N.; Ruzsa, Z.; Sharma, A.M.; Saxena, S.; et al. A Powerful Paradigm for Cardiovascular Risk Stratification Using Multiclass, Multi-Label, and Ensemble-Based Machine Learning Paradigms: A Narrative Review. Diagnostics 2022, 12, 722. https://doi.org/10.3390/diagnostics12030722
Suri JS, Bhagawati M, Paul S, Protogerou AD, Sfikakis PP, Kitas GD, Khanna NN, Ruzsa Z, Sharma AM, Saxena S, et al. A Powerful Paradigm for Cardiovascular Risk Stratification Using Multiclass, Multi-Label, and Ensemble-Based Machine Learning Paradigms: A Narrative Review. Diagnostics. 2022; 12(3):722. https://doi.org/10.3390/diagnostics12030722
Chicago/Turabian StyleSuri, Jasjit S., Mrinalini Bhagawati, Sudip Paul, Athanasios D. Protogerou, Petros P. Sfikakis, George D. Kitas, Narendra N. Khanna, Zoltan Ruzsa, Aditya M. Sharma, Sanjay Saxena, and et al. 2022. "A Powerful Paradigm for Cardiovascular Risk Stratification Using Multiclass, Multi-Label, and Ensemble-Based Machine Learning Paradigms: A Narrative Review" Diagnostics 12, no. 3: 722. https://doi.org/10.3390/diagnostics12030722
APA StyleSuri, J. S., Bhagawati, M., Paul, S., Protogerou, A. D., Sfikakis, P. P., Kitas, G. D., Khanna, N. N., Ruzsa, Z., Sharma, A. M., Saxena, S., Faa, G., Laird, J. R., Johri, A. M., Kalra, M. K., Paraskevas, K. I., & Saba, L. (2022). A Powerful Paradigm for Cardiovascular Risk Stratification Using Multiclass, Multi-Label, and Ensemble-Based Machine Learning Paradigms: A Narrative Review. Diagnostics, 12(3), 722. https://doi.org/10.3390/diagnostics12030722











