ALK, NUT, and TRK Do Not Play Relevant Roles in Gastric Cancer—Results of an Immunohistochemical Study in a Large Series
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tissue Microarray Construction
2.3. Immunohistochemistry and In Situ Hybridization
2.4. TCGA Classification
2.5. Next-Generation Sequencing (NGS) of NTRK-Positive Cases
2.6. Fluorescence In Situ Hybridization (FISH) Analysis of NTRK-Positive Cases
3. Results
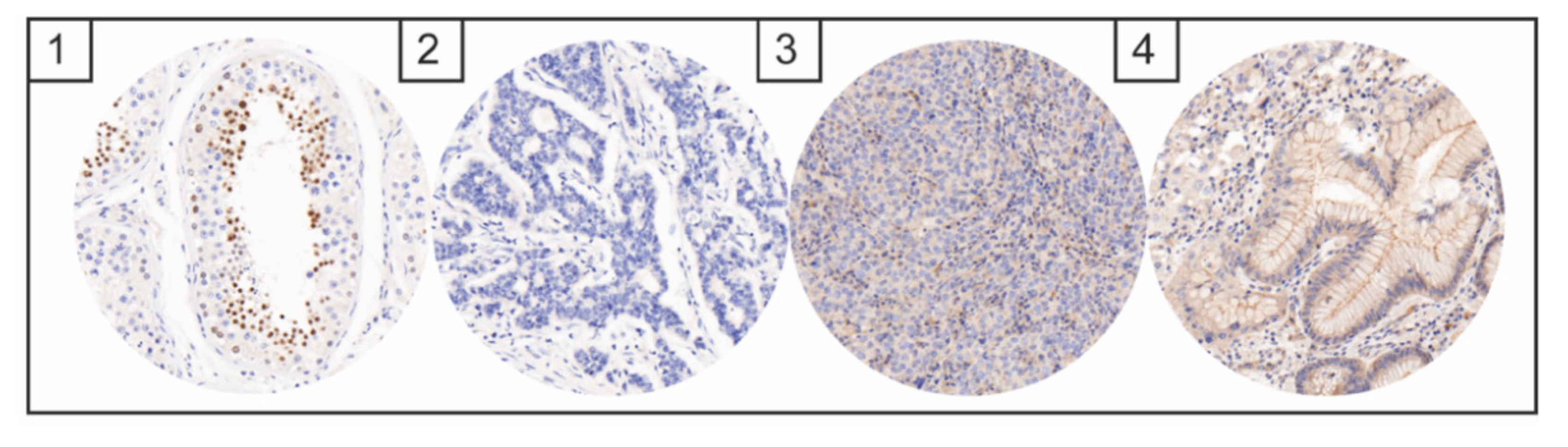
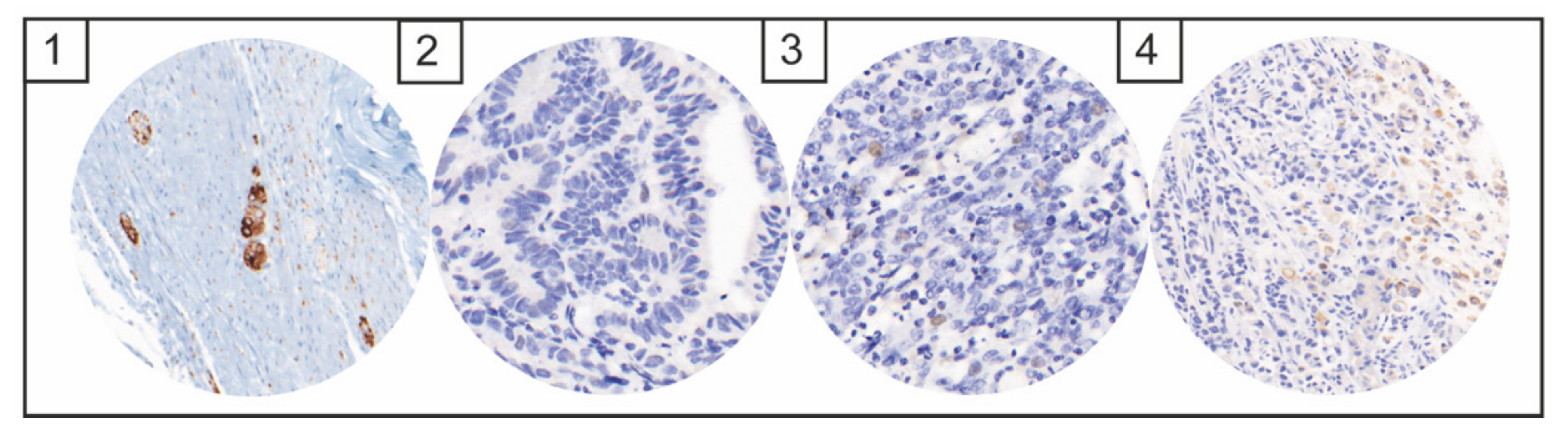
NTRK, ALK, and NUT Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424, Erratum in CA Cancer J. Clin. 2020, 70, 313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Li, N.; Jiang, W.; Hua, Z.; Xia, L.; Wei, Q.; Wang, L. Prognosis significance of HER-2/neu overexpression/amplification in Chinese patients with curatively resected gastric cancer after the ToGA clinical trial. World J. Surg. Oncol. 2012, 10, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böger, C.; Behrens, H.-M.; Mathiak, M.; Krüger, S.; Kalthoff, H.; Röcken, C. PD-L1 is an independent prognostic predictor in gastric cancer of Western patients. Oncotarget 2016, 7, 24269–24283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muro, K.; Chung, H.C.; Shankaran, V.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Golan, T.; Le, D.T.; Burtness, B.; et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): A multicentre, open-label, phase 1b trial. Lancet Oncol. 2016, 17, 717–726. [Google Scholar] [CrossRef]
- Baniak, N.; Senger, J.-L.; Ahmed, S.; Kanthan, S.C.; Kanthan, R. Gastric biomarkers: A global review. World J. Surg. Oncol. 2016, 14, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, P.; Oliveira, M.J.; Beraldi, E.; Mateus, A.R.; Nakajima, T.; Gleave, M.; Yokota, J.; Carneiro, F.; Huntsman, D.; Seruca, R.; et al. Loss of functional E-cadherin renders cells more resistant to the apoptotic agent taxol in vitro. Exp. Cell Res. 2005, 310, 99–104. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Hallberg, B.; Palmer, R.H. The role of the ALK receptor in cancer biology. Ann. Oncol. 2016, 27, iii4–iii15. [Google Scholar] [CrossRef]
- Ducray, S.P.; Natarajan, K.; Garland, G.D.; Turner, S.D.; Egger, G. The Transcriptional Roles of ALK Fusion Proteins in Tumorigenesis. Cancers 2019, 11, 1074. [Google Scholar] [CrossRef] [Green Version]
- Clinical Lung Cancer Genome Project; Network Genomic Medicine. A genomics-based classification of human lung tumors. Sci. Transl. Med. 2013, 5, 209ra153. [Google Scholar]
- Schrank, Z.; Chhabra, G.; Lin, L.; Iderzorig, T.; Osude, C.; Khan, N.; Kuckovic, A.; Singh, S.; Miller, R.J.; Puri, N. Current Molecular-Targeted Therapies in NSCLC and Their Mechanism of Resistance. Cancers 2018, 10, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alese, O.B.; El-Rayes, B.F.; Sica, G.; Zhang, G.; Alexis, D.; La Rosa, F.G.; Varella-Garcia, M.; Chen, Z.; Rossi, M.R.; Adsay, V.; et al. Anaplastic lymphoma kinase (ALK) gene alteration in signet ring cell carcinoma of the gastrointestinal tract. Ther. Adv. Med. Oncol. 2015, 7, 56–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chon, H.J.; Kim, H.R.; Shin, E.; Kim, C.; Heo, S.J.; Lee, C.-K.; Park, J.K.; Noh, S.H.; Chung, H.; Rha, S.Y. The Clinicopathologic Features and Prognostic Impact of ALK Positivity in Patients with Resected Gastric Cancer. Ann. Surg. Oncol. 2015, 22, 3938–3945. [Google Scholar] [CrossRef]
- Ying, J.; Lin, C.; Wu, J.; Guo, L.; Qiu, T.; Ling, Y.; Shan, L.; Zhou, H.; Zhao, D.; Wang, J.; et al. Anaplastic Lymphoma Kinase Rearrangement in Digestive Tract Cancer: Implication for Targeted Therapy in Chinese Population. PLoS ONE 2015, 10, e0144731. [Google Scholar] [CrossRef] [Green Version]
- Okamura, R.; Boichard, A.; Kato, S.; Sicklick, J.K.; Bazhenova, L.; Kurzrock, R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis. Oncol. 2018, 2, 1–20. [Google Scholar] [CrossRef]
- Chetty, R. Neurotrophic tropomyosin or t yrosine receptor kinase (NTRK) genes. J. Clin. Pathol. 2019, 72, 187–190. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Doebele, R.C.; Davis, L.E.; Vaishnavi, A.; Le, A.T.; Estrada-Bernal, A.; Keysar, S.B.; Jimeno, A.; Varella-Garcia, M.; Aisner, D.L.; Li, Y.; et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer Discov. 2015, 5, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 2016, 1, e000023. [Google Scholar] [CrossRef] [Green Version]
- Märkl, B.; Hirschbühl, K.; Dhillon, C. NTRK-Fusions–A new kid on the block. Pathol. Res. Pract. 2019, 215, 152572. [Google Scholar] [CrossRef]
- Shinozaki-Ushiku, A.; Ishikawa, S.; Komura, D.; Seto, Y.; Aburatani, H.; Ushiku, T. The first case of gastric carcinoma with NTRK rearrangement: Identification of a novel ATP1B–NTRK1 fusion. Gastric Cancer 2020, 23, 944–947. [Google Scholar] [CrossRef] [PubMed]
- French, C.A. NUT Carcinoma: Clinicopathologic features, pathogenesis, and treatment. Pathol. Int. 2018, 68, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Dickson, B.C.; Sung, Y.-S.; Rosenblum, M.K.; Reuter, V.E.; Harb, M.; Wunder, J.S.; Swanson, D.; Antonescu, C.R. NUTM1 Gene Fusions Characterize a Subset of Undifferentiated Soft Tissue and Visceral Tumors. Am. J. Surg. Pathol. 2018, 42, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Siewert, J.R.; Stein, H.J. Classification of adenocarcinoma of the oesophagogastric junction. Br. J. Surg. 2003, 85, 1457–1459. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Mueller, J.D.; Schulmacher, C.; Ott, K.; Fink, U.; Busch, R.; Bottcher, K.; Siewert, J.R.; Hofler, H. Histomorphology and grading of regression in gastric carcinoma treated with neoadjuvant chemotherapy. Cancer 2003, 98, 1521–1530. [Google Scholar] [CrossRef]
- Songun, I.; Putter, H.; Kranenbarg, E.M.-K.; Sasako, M.; van de Velde, C.J. Surgical treatment of gastric cancer: 15-year follow-up results of the randomised nationwide Dutch D1D2 trial. Lancet Oncol. 2010, 11, 439–449. [Google Scholar] [CrossRef]
- Shuster, J.J. Median follow-up in clinical trials. J. Clin. Oncol. 1991, 9, 191–192. [Google Scholar] [CrossRef]
- Ahn, S.; Lee, S.-J.; Kim, Y.; Kim, A.; Shin, N.; Choi, K.U.; Lee, C.-H.; Huh, G.Y.; Kim, K.-M.; Setia, N.; et al. High-throughput Protein and mRNA Expression–based Classification of Gastric Cancers Can Identify Clinically Distinct Subtypes, Concordant With Recent Molecular Classifications. Am. J. Surg. Pathol. 2017, 41, 106–115. [Google Scholar] [CrossRef]
- Setia, N.; Agoston, A.T.; Han, H.S.; Mullen, J.T.; Duda, D.G.; Clark, J.W.; Deshpande, V.; Mino-Kenudson, M.; Srivastava, A.; Lennerz, J.K.; et al. A protein and mRNA expression-based classification of gastric cancer. Mod. Pathol. 2016, 29, 772–784. [Google Scholar] [CrossRef]
- Bronsert, P.; Kohler, I.; Timme, S.; Kiefer, S.; Werner, M.; Schilling, O.; Vashist, Y.; Makowiec, F.; Brabletz, T.; Hopt, U.T.; et al. Prognostic significance of Zinc finger E-box binding homeobox 1 (ZEB1) expression in cancer cells and cancer-associated fibroblasts in pancreatic head cancer. Surgery 2014, 156, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Köbel, M.; Piskorz, A.M.; Lee, S.; Lui, S.; LePage, C.; Marass, F.; Rosenfeld, N.; Mes Masson, A.-M.; Brenton, J.D. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J. Pathol. Clin. Res. 2016, 2, 247–258. [Google Scholar] [CrossRef]
- Grosser, B.; Kohlruss, M.; Slotta-Huspenina, J.; Jesinghaus, M.; Pfarr, N.; Steiger, K.; Novotny, A.; Gaida, M.M.; Schmidt, T.; Hapfelmeier, A.; et al. Impact of Tumor Localization and Molecular Subtypes on the Prognostic and Predictive Significance of p53 Expression in Gastric Cancer. Cancers 2020, 12, 1689. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Miyoshi, H.; Hiraoka, K.; Arakawa, F.; Haraguchi, T.; Nakashima, S.; Hashiguchi, T.; Shoda, T.; Hamada, T.; Okawa, T.; et al. Anaplastic lymphoma kinase protein expression, genetic abnormalities, and phosphorylation in soft tissue tumors: Phosphorylation is associated with recurrent metastasis. Oncol. Rep. 2015, 33, 1667–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int. J. Mol. Sci. 2018, 19, 3448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Wekken, A.; Pelgrim, R.; Hart, N. ’T.; Werner, N.; Mastik, M.; Hendriks, L.; van der Heijden, E.; Looijen-Salamon, M.; De Langen, A.; Brekel, J.S.-V.D.; et al. Dichotomous ALK-IHC Is a Better Predictor for ALK Inhibition Outcome than Traditional ALK-FISH in Advanced Non–Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 4251–4258. [Google Scholar] [CrossRef] [Green Version]
- Storck, S.; Kennedy, A.L.; Marcus, K.J.; Teot, L.; Vaughn, J.; Gnekow, A.K.; Märkl, B.; Leuschner, I.; Dubois, S.G.; French, C.A.; et al. Pediatric NUT-midline carcinoma: Therapeutic success employing a sarcoma based multimodal approach. Pediatr. Hematol. Oncol. 2017, 34, 231–237. [Google Scholar] [CrossRef]
- Solomon, J.P.; Linkov, I.; Rosado, A.; Mullaney, K.; Rosen, E.Y.; Frosina, D.; Jungbluth, A.A.; Zehir, A.; Benayed, R.; Drilon, A.; et al. NTRK fusion detection across multiple assays and 33,997 cases: Diagnostic implications and pitfalls. Mod. Pathol. 2020, 33, 38–46. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Lee, S.J.; Li, G.G.; Kim, S.T.; Hong, M.E.; Jang, J.; Yoon, N.; Ahn, S.M.; Murphy, D.; Christiansen, J.; Wei, G.; et al. NTRK1 rearrangement in colorectal cancer patients: Evidence for actionable target using patient-derived tumor cell line. Oncotarget 2015, 6, 39028–39035. [Google Scholar] [CrossRef] [Green Version]

| Variable | n = 477 | % | |
|---|---|---|---|
| Median age (range) (years) | 70.0 (30.0–95.0) | ||
| Median survival (range) (months) | 58.0 (49.9–66.1) | ||
| Sex | female | 312 | 65% |
| male | 165 | 35% | |
| T status | pT1/2 | 159 | 33% |
| pT3/4 | 318 | 67% | |
| N status | negative | 178 | 37% |
| positive | 299 | 63% | |
| Distant Metastasis | no | 247 | 52% |
| yes | 197 | 41% | |
| NA | 33 | 7% | |
| Grading | low grade | 162 | 34% |
| high grade | 304 | 64% | |
| NA | 11 | 2% | |
| Lymphovascular invasion | negative | 287 | 60% |
| positive | 190 | 40% | |
| Vascular invasion | negative | 401 | 84% |
| positive | 76 | 16% | |
| Lauren | intestinal | 266 | 56% |
| non-intestinal | 211 | 44% | |
| Localization | proximal (AEG II/III, Cardia) | 124 | 26% |
| non proximal (Fundus/Corpus/Antrum) | 335 | 70% | |
| NA | 18 | 4% | |
| R status | R0 | 403 | 84% |
| R1 | 54 | 11% | |
| Rx | 20 | 4% | |
| TCGA | EBV+ | 25 | 5% |
| MSI | 61 | 13% | |
| GS | 110 | 23% | |
| CIN | 151 | 32% | |
| no classification | 130 | 27% | |
| Death | no | 227 | 48% |
| death | 250 | 52% | |
| Preoperative CTx | no | 347 | 73% |
| yes | 130 | 27% | |
| TRG (n = 130) | 1b | 9 | 7% |
| 2 | 36 | 28% | |
| 3 | 82 | 65% | |
| CTx regimen | Cis/Ox + 5-FU or Cap | 34 | 27% |
| Ox + 5-FU + Doc | 45 | 35% | |
| Cis + 5-FU + Epi | 41 | 32% | |
| Ox + Epi + Cap | 5 | 4% | |
| others | 2 | 2% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glückstein, M.-I.; Dintner, S.; Miller, S.; Vlasenko, D.; Schenkirsch, G.; Agaimy, A.; Märkl, B.; Grosser, B. ALK, NUT, and TRK Do Not Play Relevant Roles in Gastric Cancer—Results of an Immunohistochemical Study in a Large Series. Diagnostics 2022, 12, 429. https://doi.org/10.3390/diagnostics12020429
Glückstein M-I, Dintner S, Miller S, Vlasenko D, Schenkirsch G, Agaimy A, Märkl B, Grosser B. ALK, NUT, and TRK Do Not Play Relevant Roles in Gastric Cancer—Results of an Immunohistochemical Study in a Large Series. Diagnostics. 2022; 12(2):429. https://doi.org/10.3390/diagnostics12020429
Chicago/Turabian StyleGlückstein, Marie-Isabelle, Sebastian Dintner, Silvia Miller, Dmytro Vlasenko, Gerhard Schenkirsch, Abbas Agaimy, Bruno Märkl, and Bianca Grosser. 2022. "ALK, NUT, and TRK Do Not Play Relevant Roles in Gastric Cancer—Results of an Immunohistochemical Study in a Large Series" Diagnostics 12, no. 2: 429. https://doi.org/10.3390/diagnostics12020429
APA StyleGlückstein, M.-I., Dintner, S., Miller, S., Vlasenko, D., Schenkirsch, G., Agaimy, A., Märkl, B., & Grosser, B. (2022). ALK, NUT, and TRK Do Not Play Relevant Roles in Gastric Cancer—Results of an Immunohistochemical Study in a Large Series. Diagnostics, 12(2), 429. https://doi.org/10.3390/diagnostics12020429








