The Eutopic Endometrium Proteome in Endometriosis Reveals Candidate Markers and Molecular Mechanisms of Physiopathology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Preparation
2.3. Protein Extraction, Digestion, and Liquid Chromatography—Tandem Mass Spectrometry (LC-MS/MS) Analyses
2.4. MS/MS Data Analysis
2.5. Statistical Analysis and Data Mining
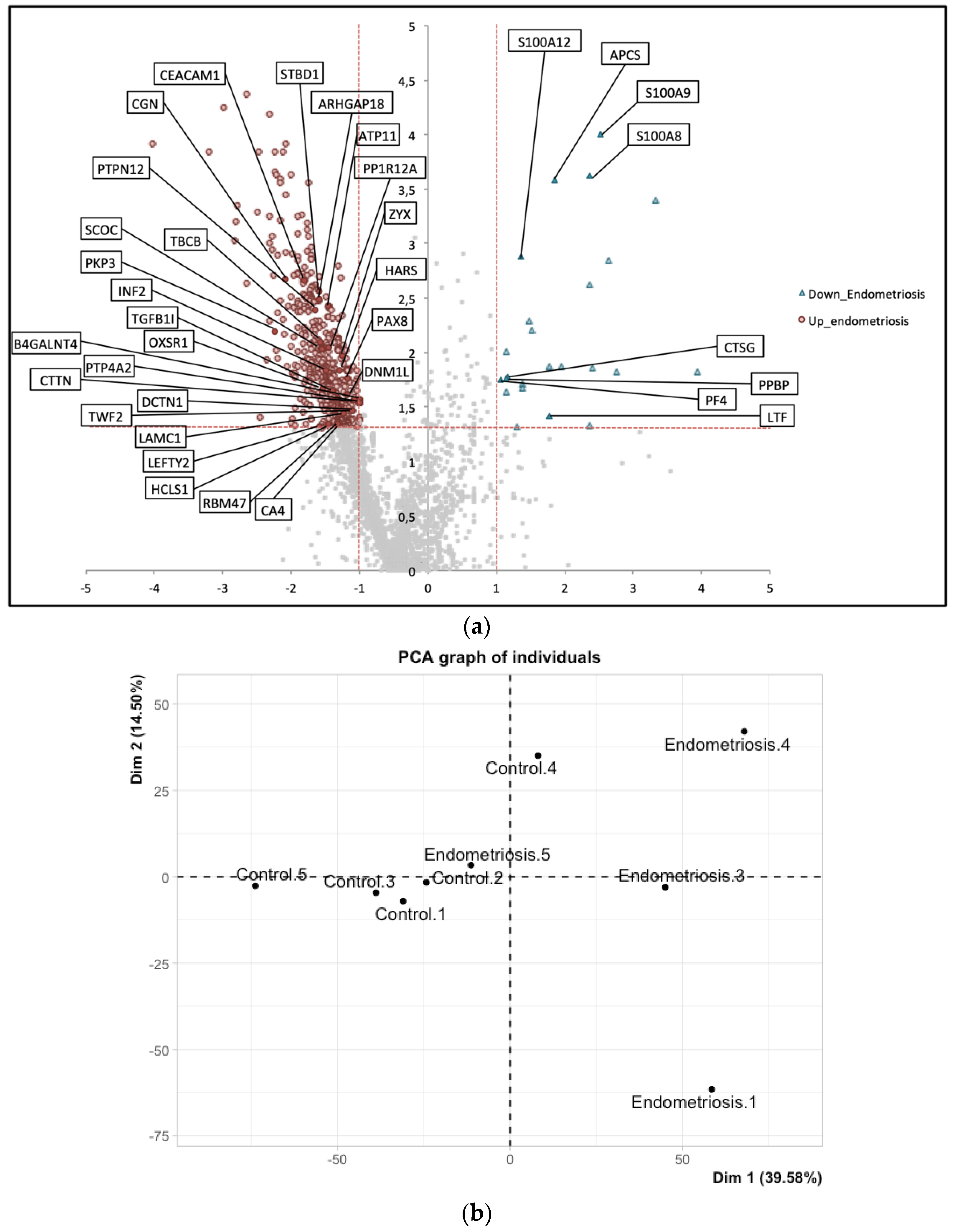
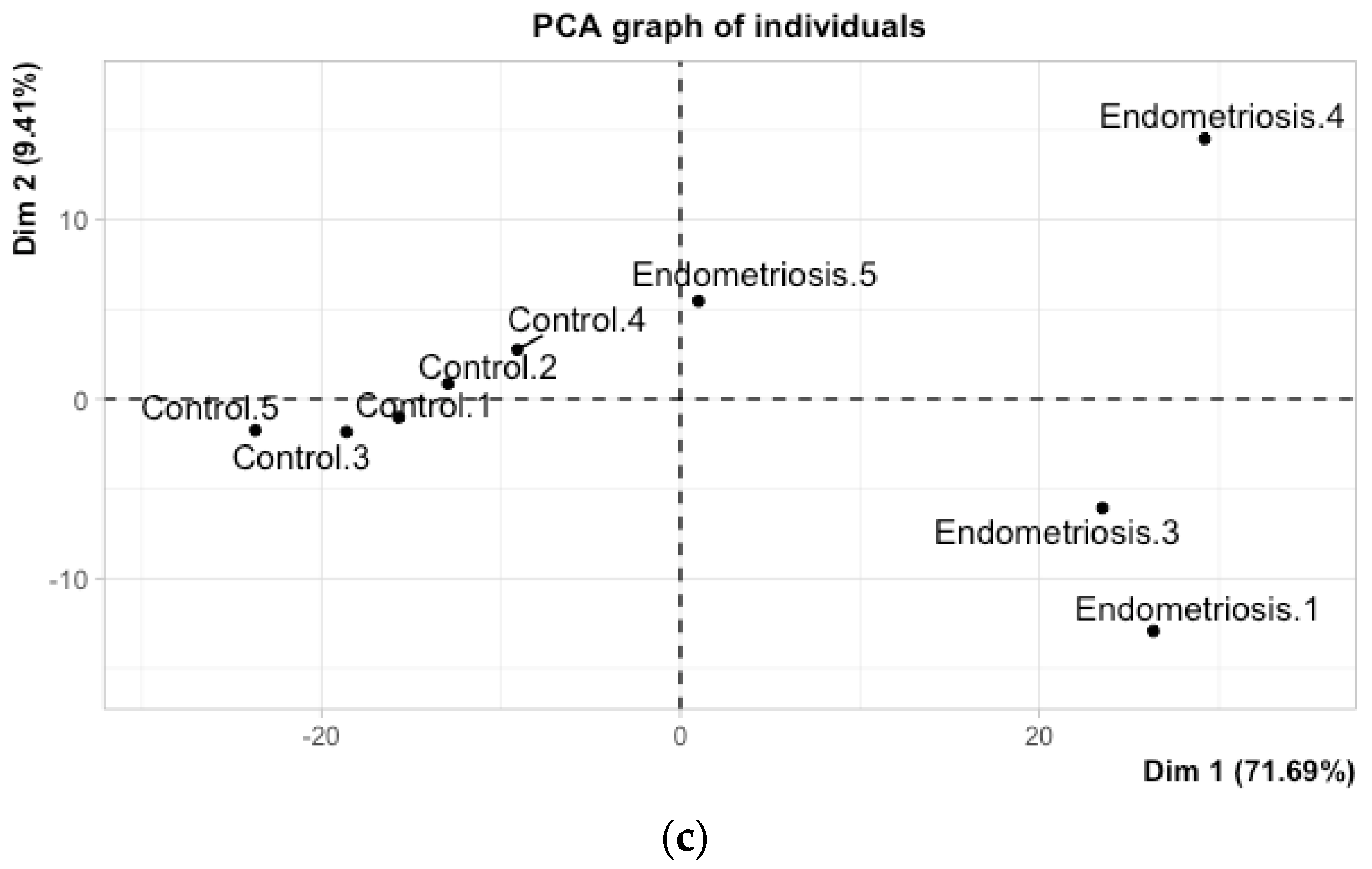
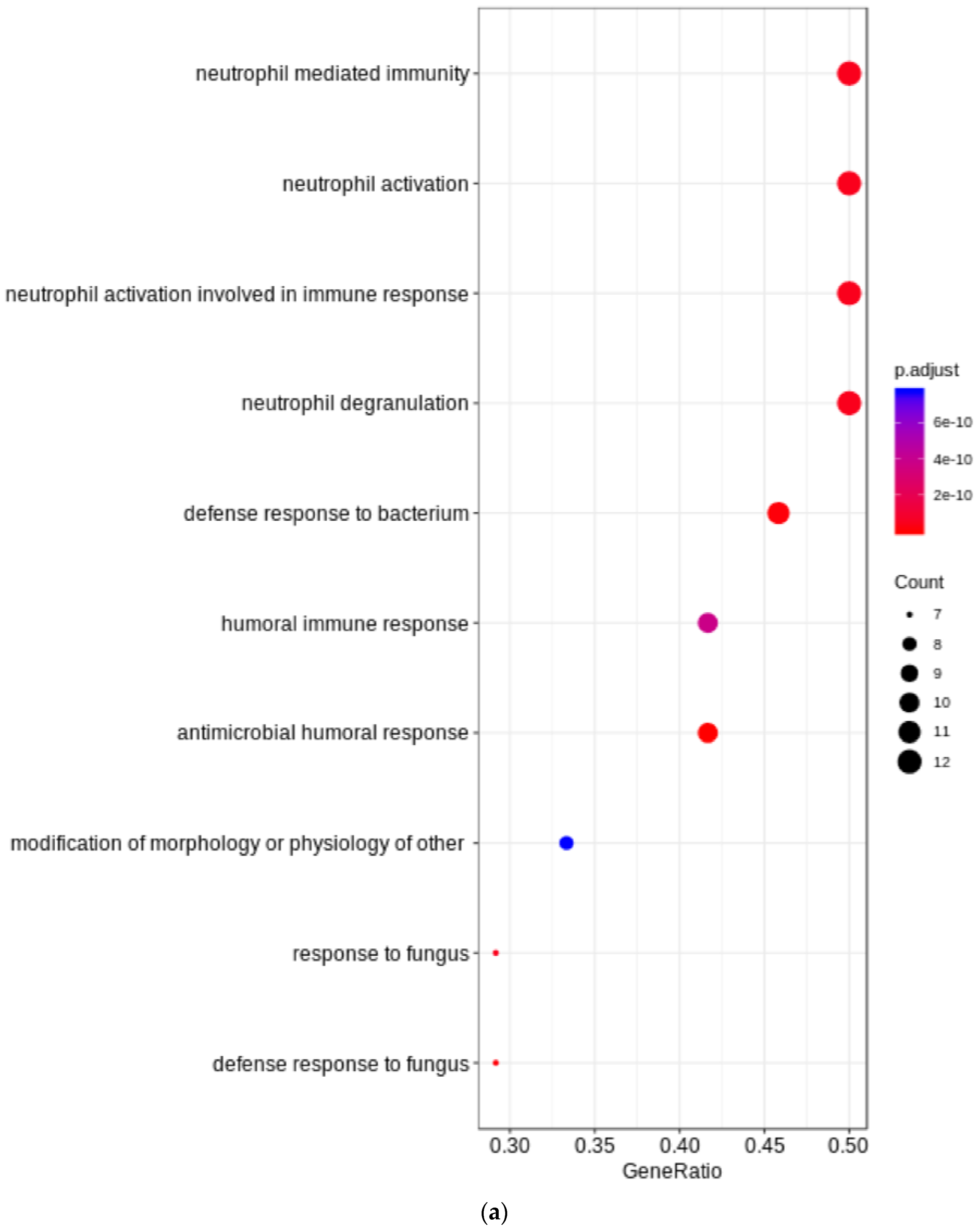
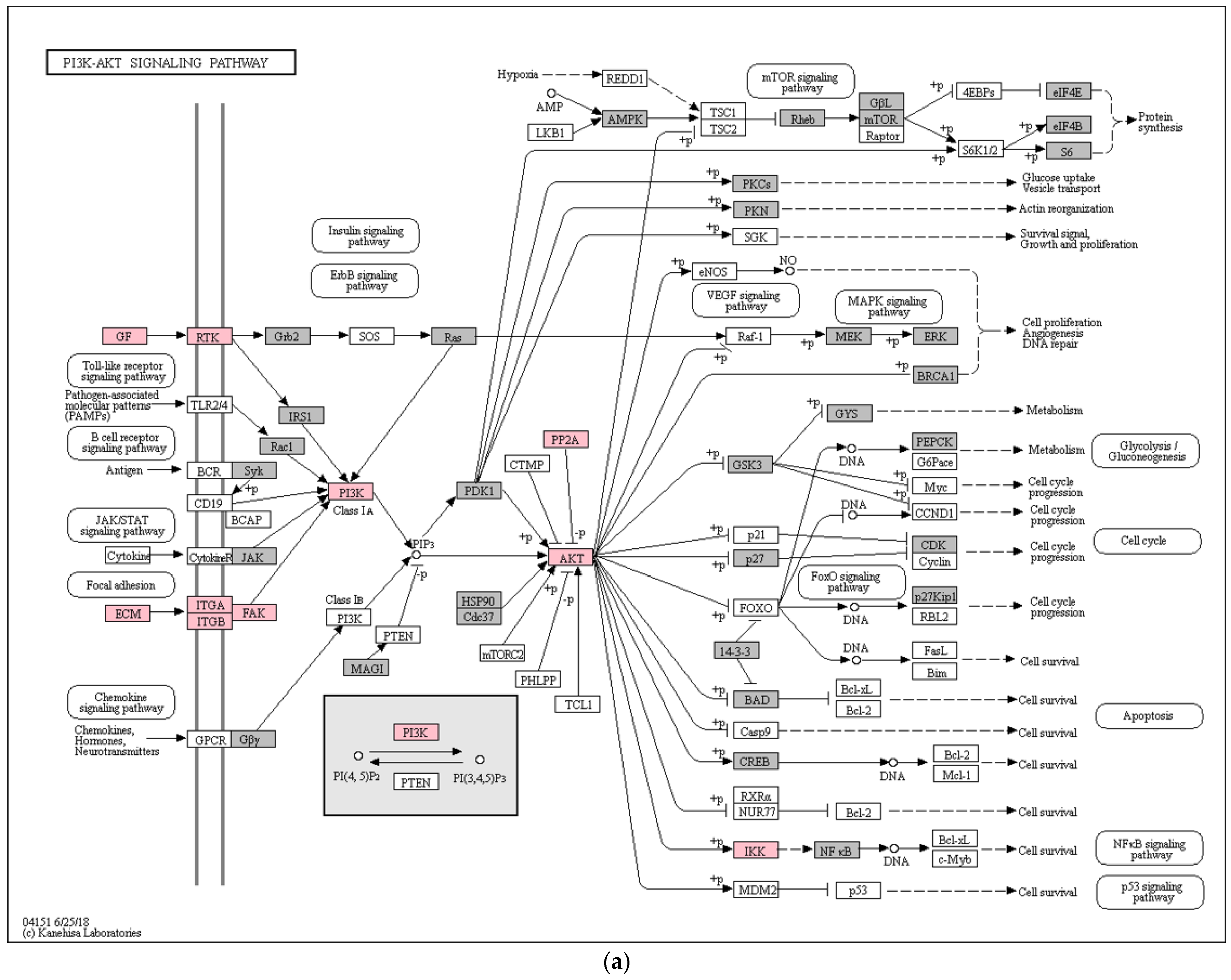
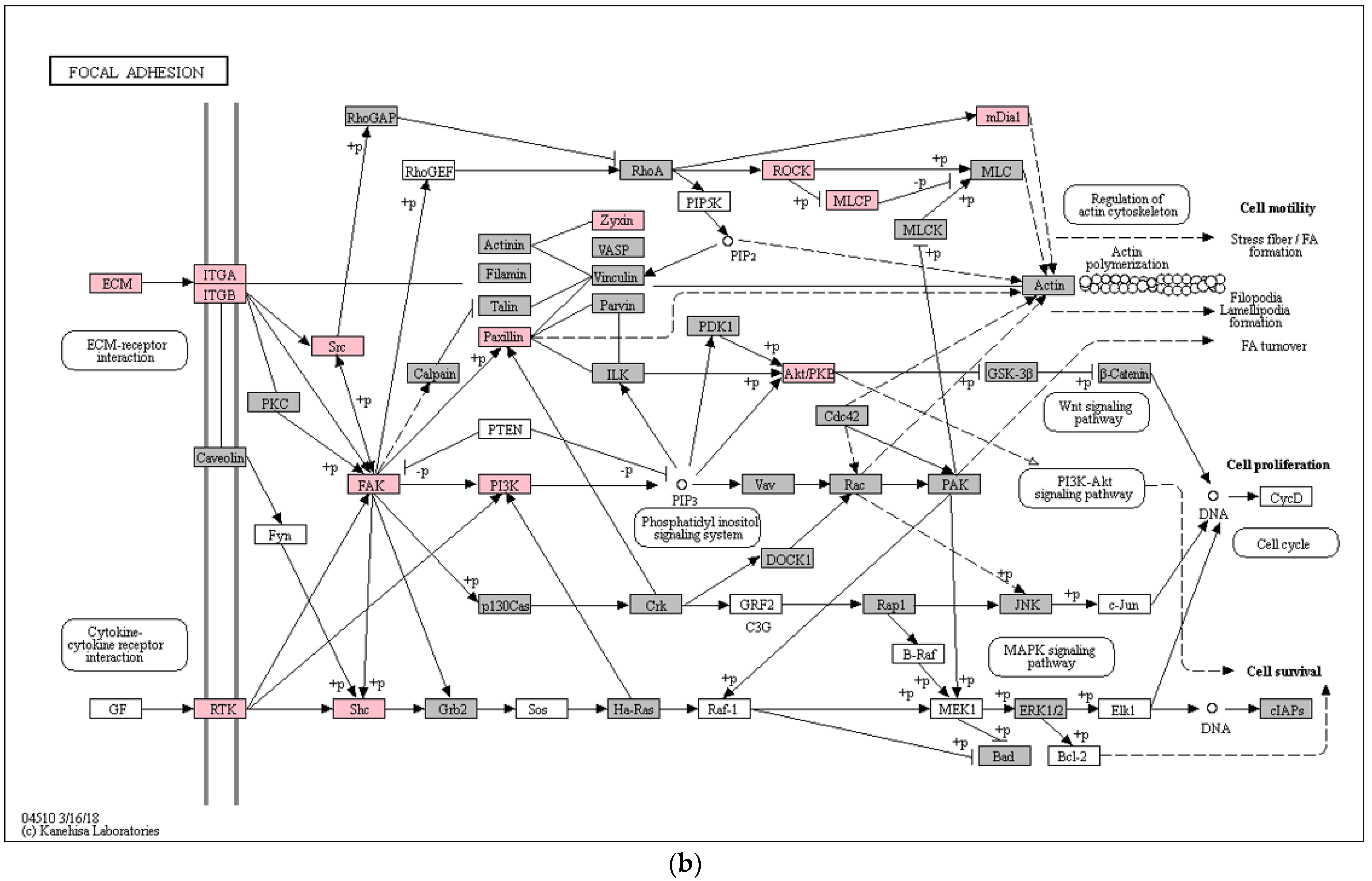
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giudice, L.C.; Kao, L.C. Endometriosis. Lancet 2004, 364, 1789–1799. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Becker, C.M.; Koga, K.; Missmer, S.A.; Taylor, R.N.; Viganò, P. Endometriosis. Nat. Rev. Dis. Primers 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Nisolle, M.; Donnez, J. Peritoneal Endometriosis, Ovarian Endometriosis, and Adenomyotic Nodules of the Rectovaginal Septum Are Three Different Entities. Fertil. Steril. 1997, 68, 585–596. [Google Scholar] [CrossRef]
- Nezhat, C.; Nezhat, F.; Nezhat, C. Endometriosis: Ancient Disease, Ancient Treatments. Fertil. Steril. 2012, 98, S1–S62. [Google Scholar] [CrossRef]
- Vinatier, D.; Orazi, G.; Cosson, M.; Dufour, P. Theories of Endometriosis. Eur. J. Obs. Gynecol. Reprod. Biol. 2001, 96, 21–34. [Google Scholar] [CrossRef]
- Vercellini, P.; Viganò, P.; Somigliana, E.; Fedele, L. Endometriosis: Pathogenesis and Treatment. Nat. Rev. Endocrinol. 2014, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Bulun, S.E. Endometriosis. N. Engl. J. Med. 2009, 360, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.A. Ovarian Hematomas of Endometrial Type (Perforating Hemorrhagic Cysts of the Ovary) and Implantation Adenomas of Endometrial Type. Boston Med. Surg. J. 1922, 186, 445–456. [Google Scholar] [CrossRef]
- Sampson, J.A. Metastatic or Embolic Endometriosis, Due to the Menstrual Dissemination of Endometrial Tissue into the Venous Circulation. Am. J. Pathol. 1927, 3, 93–110.43. [Google Scholar]
- D’Hooghe, T.M.; Debrock, S. Endometriosis, Retrograde Menstruation and Peritoneal Inflammation in Women and in Baboons. Hum. Reprod. Update 2002, 8, 84–88. [Google Scholar] [CrossRef]
- Koninckx, P.R.; Ussia, A.; Adamyan, L.; Wattiez, A.; Gomel, V.; Martin, D.C. Pathogenesis of Endometriosis: The Genetic/Epigenetic Theory. Fertil. Steril. 2019, 111, 327–340. [Google Scholar] [CrossRef] [Green Version]
- Ramin-Wright, A.; Schwartz, A.S.K.; Geraedts, K.; Rauchfuss, M.; Wölfler, M.M.; Haeberlin, F.; von Orelli, S.; Eberhard, M.; Imthurn, B.; Imesch, P.; et al. Fatigue—A Symptom in Endometriosis. Hum. Reprod. 2018, 33, 1459–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballard, K.; Lowton, K.; Wright, J. What’s the Delay? A Qualitative Study of Women’s Experiences of Reaching a Diagnosis of Endometriosis. Fertil. Steril. 2006, 86, 1296–1301. [Google Scholar] [CrossRef] [PubMed]
- Hudelist, G.; Fritzer, N.; Thomas, A.; Niehues, C.; Oppelt, P.; Haas, D.; Tammaa, A.; Salzer, H. Diagnostic Delay for Endometriosis in Austria and Germany: Causes and Possible Consequences. Hum. Reprod. 2012, 27, 3412–3416. [Google Scholar] [CrossRef]
- Staal, A.H.J.; van der Zanden, M.; Nap, A.W. Diagnostic Delay of Endometriosis in the Netherlands. Gynecol. Obs. Investig. 2016, 81, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Fassbender, A.; Burney, R.O.; O, D.F.; D’Hooghe, T.; Giudice, L. Update on Biomarkers for the Detection of Endometriosis. Biomed. Res. Int. 2015, 2015, 130854. [Google Scholar] [CrossRef] [Green Version]
- Rogers, P.A.W.; Adamson, G.D.; Al-Jefout, M.; Becker, C.M.; D’Hooghe, T.M.; Dunselman, G.A.J.; Fazleabas, A.; Giudice, L.C.; Horne, A.W.; Hull, M.L.; et al. Research Priorities for Endometriosis. Reprod. Sci. 2017, 24, 202–226. [Google Scholar] [CrossRef] [Green Version]
- Bergman-Larsson, J.; Gustafsson, S.; Méar, L.; Huvila, J.; Tolf, A.; Olovsson, M.; Pontén, F.; Edqvist, P.-H.D. Combined Expression of HOXA11 and CD10 Identifies Endometriosis versus Normal Tissue and Tumors. Ann. Diagn. Pathol. 2022, 56, 151870. [Google Scholar] [CrossRef]
- Bedaiwy, M.A.; Falcone, T. Laboratory Testing for Endometriosis. Clin. Chim. Acta 2004, 340, 41–56. [Google Scholar] [CrossRef]
- May, K.E.; Conduit-Hulbert, S.A.; Villar, J.; Kirtley, S.; Kennedy, S.H.; Becker, C.M. Peripheral Biomarkers of Endometriosis: A Systematic Review. Hum. Reprod. Update 2010, 16, 651–674. [Google Scholar] [CrossRef]
- May, K.E.; Villar, J.; Kirtley, S.; Kennedy, S.H.; Becker, C.M. Endometrial Alterations in Endometriosis: A Systematic Review of Putative Biomarkers. Hum. Reprod. Update 2011, 17, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Acién, P.; Velasco, I. Endometriosis: A Disease That Remains Enigmatic. ISRN Obs. Gynecol. 2013, 2013, 242149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, D.; Hull, M.L.; Fraser, I.; Miller, L.; Bossuyt, P.M.M.; Johnson, N.; Nisenblat, V. Endometrial Biomarkers for the Non-Invasive Diagnosis of Endometriosis. Cochrane Database Syst. Rev. 2016, 4, CD012165. [Google Scholar] [CrossRef] [PubMed]
- Nisenblat, V.; Bossuyt, P.M.M.; Shaikh, R.; Farquhar, C.; Jordan, V.; Scheffers, C.S.; Mol, B.W.J.; Johnson, N.; Hull, M.L. Blood Biomarkers for the Non-Invasive Diagnosis of Endometriosis. Cochrane Database Syst. Rev. 2016, CD012179. [Google Scholar] [CrossRef] [Green Version]
- Ferrero, S.; Gillott, D.J.; Remorgida, V.; Ragni, N.; Venturini, P.L.; Grudzinskas, J.G. Proteomics Technologies in Endometriosis. Expert. Rev. Proteom. 2008, 5, 705–714. [Google Scholar] [CrossRef]
- Siva, A.B.; Srivastava, P.; Shivaji, S. Understanding the Pathogenesis of Endometriosis through Proteomics: Recent Advances and Future Prospects. Proteom. Clin. Appl. 2014, 8, 86–98. [Google Scholar] [CrossRef]
- Ferrero, S. Proteomics in the Diagnosis of Endometriosis: Opportunities and Challenges. Proteom. Clin. Appl. 2019, 13, e1800183. [Google Scholar] [CrossRef]
- Ferrero, S.; Gillott, D.J.; Remorgida, V.; Anserini, P.; Leung, K.-Y.; Ragni, N.; Grudzinskas, J.G. Proteomic Analysis of Peritoneal Fluid in Women with Endometriosis. J. Proteome Res. 2007, 6, 3402–3411. [Google Scholar] [CrossRef]
- Ten Have, S.; Fraser, I.; Markham, R.; Lam, A.; Matsumoto, I. Proteomic Analysis of Protein Expression in the Eutopic Endometrium of Women with Endometriosis. Proteom. Clin. Appl. 2007, 1, 1243–1251. [Google Scholar] [CrossRef]
- El-Kasti, M.M.; Wright, C.; Fye, H.K.S.; Roseman, F.; Kessler, B.M.; Becker, C.M. Urinary Peptide Profiling Identifies a Panel of Putative Biomarkers for Diagnosing and Staging Endometriosis. Fertil. Steril. 2011, 95, 1261–1266.e6. [Google Scholar] [CrossRef]
- Kyama, C.M.; Mihalyi, A.; Gevaert, O.; Waelkens, E.; Simsa, P.; Van de Plas, R.; Meuleman, C.; De Moor, B.; D’Hooghe, T.M. Evaluation of Endometrial Biomarkers for Semi-Invasive Diagnosis of Endometriosis. Fertil. Steril. 2011, 95, 1338–1343.e3. [Google Scholar] [CrossRef] [PubMed]
- Fassbender, A.; Verbeeck, N.; Börnigen, D.; Kyama, C.M.; Bokor, A.; Vodolazkaia, A.; Peeraer, K.; Tomassetti, C.; Meuleman, C.; Gevaert, O.; et al. Combined MRNA Microarray and Proteomic Analysis of Eutopic Endometrium of Women with and without Endometriosis. Hum. Reprod. 2012, 27, 2020–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, K.E.; Miroshnychenko, O.; Johansen, E.B.; Niles, R.K.; Sundaram, R.; Kannan, K.; Albertolle, M.; Zhou, Y.; Prasad, N.; Drake, P.M.; et al. Urine, Peritoneal Fluid and Omental Fat Proteomes of Reproductive Age Women: Endometriosis-Related Changes and Associations with Endocrine Disrupting Chemicals. J. Proteom. 2015, 113, 194–205. [Google Scholar] [CrossRef] [Green Version]
- Grande, G.; Vincenzoni, F.; Milardi, D.; Pompa, G.; Ricciardi, D.; Fruscella, E.; Mancini, F.; Pontecorvi, A.; Castagnola, M.; Marana, R. Cervical Mucus Proteome in Endometriosis. Clin. Proteom. 2017, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manousopoulou, A.; Hamdan, M.; Fotopoulos, M.; Garay-Baquero, D.J.; Teng, J.; Garbis, S.D.; Cheong, Y. Integrated Eutopic Endometrium and Non-Depleted Serum Quantitative Proteomic Analysis Identifies Candidate Serological Markers of Endometriosis. Proteom. Clin. Appl. 2019, 13, e1800153. [Google Scholar] [CrossRef] [PubMed]
- Fowler, P.A.; Tattum, J.; Bhattacharya, S.; Klonisch, T.; Hombach-Klonisch, S.; Gazvani, R.; Lea, R.G.; Miller, I.; Simpson, W.G.; Cash, P. An Investigation of the Effects of Endometriosis on the Proteome of Human Eutopic Endometrium: A Heterogeneous Tissue with a Complex Disease. Proteomics 2007, 7, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Greaves, E.; Critchley, H.O.D.; Horne, A.W.; Saunders, P.T.K. Relevant Human Tissue Resources and Laboratory Models for Use in Endometriosis Research. Acta Obs. Gynecol. Scand. 2017, 96, 644–658. [Google Scholar] [CrossRef]
- Becker, C.M.; Laufer, M.R.; Stratton, P.; Hummelshoj, L.; Missmer, S.A.; Zondervan, K.T.; Adamson, G.D. WERF EPHect Working Group World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: I. Surgical Phenotype Data Collection in Endometriosis Research. Fertil. Steril. 2014, 102, 1213–1222. [Google Scholar] [CrossRef]
- Fassbender, A.; Rahmioglu, N.; Vitonis, A.F.; Viganò, P.; Giudice, L.C.; D’Hooghe, T.M.; Hummelshoj, L.; Adamson, G.D.; Becker, C.M.; Missmer, S.A.; et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: IV. Tissue Collection, Processing, and Storage in Endometriosis Research. Fertil. Steril. 2014, 102, 1244–1253. [Google Scholar] [CrossRef]
- Rahmioglu, N.; Fassbender, A.; Vitonis, A.F.; Tworoger, S.S.; Hummelshoj, L.; D’Hooghe, T.M.; Adamson, G.D.; Giudice, L.C.; Becker, C.M.; Zondervan, K.T.; et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: III. Fluid Biospecimen Collection, Processing, and Storage in Endometriosis Research. Fertil. Steril. 2014, 102, 1233–1243. [Google Scholar] [CrossRef]
- Vitonis, A.F.; Vincent, K.; Rahmioglu, N.; Fassbender, A.; Buck Louis, G.M.; Hummelshoj, L.; Giudice, L.C.; Stratton, P.; Adamson, G.D.; Becker, C.M.; et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: II. Clinical and Covariate Phenotype Data Collection in Endometriosis Research. Fertil. Steril. 2014, 102, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Nisenblat, V.; Farquhar, C.; Fraser, I.; Bossuyt, P.M.M.; Johnson, N.; Hull, M.L. Urinary Biomarkers for the Non-Invasive Diagnosis of Endometriosis. Cochrane Database Syst. Rev. 2015, CD012019. [Google Scholar] [CrossRef] [PubMed]
- Melaine, N.; Com, E.; Bellaud, P.; Guillot, L.; Lagarrigue, M.; Morrice, N.A.; Guével, B.; Lavigne, R.; Velez de la Calle, J.-F.; Dojahn, J.; et al. Deciphering the Dark Proteome: Use of the Testis and Characterization of Two Dark Proteins. J. Proteome Res. 2018, 17, 4197–4210. [Google Scholar] [CrossRef] [PubMed]
- Jumeau, F.; Com, E.; Lane, L.; Duek, P.; Lagarrigue, M.; Lavigne, R.; Guillot, L.; Rondel, K.; Gateau, A.; Melaine, N.; et al. Human Spermatozoa as a Model for Detecting Missing Proteins in the Context of the Chromosome-Centric Human Proteome Project. J. Proteome Res. 2015, 14, 3606–3620. [Google Scholar] [CrossRef]
- Colinet, H.; Pineau, C.; Com, E. Large Scale Phosphoprotein Profiling to Explore Drosophila Cold Acclimation Regulatory Mechanisms. Sci. Rep. 2017, 7, 1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fermin, D.; Basrur, V.; Yocum, A.K.; Nesvizhskii, A.I. Abacus: A Computational Tool for Extracting and Pre-Processing Spectral Count Data for Label-Free Quantitative Proteomic Analysis. Proteomics 2011, 11, 1340–1345. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: http://r.meteo.uni.wroc.pl/web/packages/dplR/vignettes/intro-dplR.pdf (accessed on 16 December 2021).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Combes, F.; Loux, V.; Vandenbrouck, Y. GO Enrichment Analysis for Differential Proteomics Using ProteoRE. Methods Mol. Biol. 2021, 2361, 179–196. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.J.; Yin, X. Signaling Pathways in Endometriosis (Eutopic/Ectopic). In Endometriosis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011; pp. 164–172. ISBN 978-1-4443-9851-9. [Google Scholar]
- Laudanski, P.; Szamatowicz, J.; Kowalczuk, O.; Kuźmicki, M.; Grabowicz, M.; Chyczewski, L. Expression of Selected Tumor Suppressor and Oncogenes in Endometrium of Women with Endometriosis. Hum. Reprod. 2009, 24, 1880–1890. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.H.; Yu, Y.; Luo, L.; Lydon, J.P.; Jeong, J.-W.; Kim, J.J. Activated AKT Pathway Promotes Establishment of Endometriosis. Endocrinology 2014, 155, 1921–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Pavone, M.E.; Lu, Z.; Wei, J.; Kim, J.J. Increased Activation of the PI3K/AKT Pathway Compromises Decidualization of Stromal Cells from Endometriosis. J. Clin. Endocrinol. Metab. 2012, 97, E35–E43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lessey, B.A.; Kim, J.J. Endometrial Receptivity in the Eutopic Endometrium of Women with Endometriosis: It Is Affected, and Let Me Show You Why. Fertil. Steril. 2017, 108, 19–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romer, L.H.; Birukov, K.G.; Garcia, J.G.N. Focal Adhesions: Paradigm for a Signaling Nexus. Circ. Res. 2006, 98, 606–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozhan, E.; Kokcu, A.; Yanik, K.; Gunaydin, M. Investigation of Diagnostic Potentials of Nine Different Biomarkers in Endometriosis. Eur. J. Obs. Gynecol. Reprod. Biol. 2014, 178, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Laudanski, P.; Charkiewicz, R.; Kuzmicki, M.; Szamatowicz, J.; Świątecka, J.; Mroczko, B.; Niklinski, J. Profiling of Selected Angiogenesis-Related Genes in Proliferative Eutopic Endometrium of Women with Endometriosis. Eur. J. Obs. Gynecol. Reprod. Biol. 2014, 172, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Christofolini, D.M.; Mafra, F.A.; Catto, M.C.; Bianco, B.; Barbosa, C.P. New Candidate Genes Associated to Endometriosis. Gynecol. Endocrinol. 2019, 35, 62–65. [Google Scholar] [CrossRef]
- Mu, L.; Ma, Y.-Y. Expression of Focal Adhesion Kinase in Endometrial Stromal Cells of Women with Endometriosis Was Adjusted by Ovarian Steroid Hormones. Int. J. Clin. Exp. Pathol. 2015, 8, 1810–1815. [Google Scholar]
- Mu, L.; Zheng, W.; Wang, L.; Chen, X.-J.; Zhang, X.; Yang, J.-H. Alteration of Focal Adhesion Kinase Expression in Eutopic Endometrium of Women with Endometriosis. Fertil. Steril. 2008, 89, 529–537. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Aguilar, J.; Molloy, M.P. Targeted Mass Spectrometry of S100 Proteins. Methods Mol. Biol. 2019, 1929, 663–678. [Google Scholar] [CrossRef]
- Ryckman, C.; Vandal, K.; Rouleau, P.; Talbot, M.; Tessier, P.A. Proinflammatory Activities of S100: Proteins S100A8, S100A9, and S100A8/A9 Induce Neutrophil Chemotaxis and Adhesion. J. Immunol. 2003, 170, 3233–3242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrero, S.; Gillott, D.J.; Remorgida, V.; Anserini, P.; Ragni, N.; Grudzinskas, J.G. Peritoneal Fluid Proteome in Women with Different ASRM Stages of Endometriosis. Gynecol. Endocrinol. 2008, 24, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Symons, L.K.; Miller, J.E.; Kay, V.R.; Marks, R.M.; Liblik, K.; Koti, M.; Tayade, C. The Immunopathophysiology of Endometriosis. Trends Mol. Med. 2018, 24, 748–762. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, L.; Drissennek, L.; Antoine, Y.; Tiers, L.; Hirtz, C.; Lehmann, S.; Perrochia, H.; Bissonnette, F.; Kadoch, I.-J.; Haouzi, D.; et al. Human S100A10 Plays a Crucial Role in the Acquisition of the Endometrial Receptivity Phenotype. Cell Adh. Migr. 2016, 10, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.-M.; Lin, X.-N.; Song, T.; Liu, L.; Zhang, S.-Y. Calcium-Binding Protein S100P Is Highly Expressed during the Implantation Window in Human Endometrium. Fertil. Steril. 2010, 94, 1510–1518. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-M.; Ding, G.-L.; Jiang, Y.; Pan, H.-J.; Zhang, D.; Wang, T.-T.; Zhang, R.-J.; Shu, J.; Sheng, J.-Z.; Huang, H.-F. Down-Regulation of S100A11, a Calcium-Binding Protein, in Human Endometrium May Cause Reproductive Failure. J. Clin. Endocrinol. Metab. 2012, 97, 3672–3683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polak, G.; Wertel, I.; Tarkowski, R.; Morawska, D.; Kotarski, J. Decreased Lactoferrin Levels in Peritoneal Fluid of Women with Minimal Endometriosis. Eur. J. Obs. Gynecol. Reprod. Biol. 2007, 131, 93–96. [Google Scholar] [CrossRef]
- Riley, C.F.; Moen, M.H.; Videm, V. Inflammatory Markers in Endometriosis: Reduced Peritoneal Neutrophil Response in Minimal Endometriosis. Acta Obs. Gynecol. Scand. 2007, 86, 877–881. [Google Scholar] [CrossRef]
- Laudanski, P.; Gorodkiewicz, E.; Ramotowska, B.; Charkiewicz, R.; Kuzmicki, M.; Szamatowicz, J. Determination of Cathepsins B, D and G Concentration in Eutopic Proliferative Endometrium of Women with Endometriosis by the Surface Plasmon Resonance Imaging (SPRI) Technique. Eur. J. Obs. Gynecol. Reprod. Biol. 2013, 169, 80–83. [Google Scholar] [CrossRef]
- Grzywa, R.; Gorodkiewicz, E.; Burchacka, E.; Lesner, A.; Laudański, P.; Łukaszewski, Z.; Sieńczyk, M. Determination of Cathepsin G in Endometrial Tissue Using a Surface Plasmon Resonance Imaging Biosensor with Tailored Phosphonic Inhibitor. Eur. J. Obs. Gynecol. Reprod. Biol. 2014, 182, 38–42. [Google Scholar] [CrossRef] [PubMed]


| Inclusion Criterion | Exclusion Criterion | |
|---|---|---|
| ♀ ⟪endometriosis⟫ N = 4 | Endometriosis (with symptoms) Histologic confirmation of endometriosis No hormonal treatment Laparoscopy between day 19 and 24 of menstrual cycle Presence of superficial and active lesions with angiogenesis around Presence of characteristic lesions in Pouch of Douglas | Adenomyosis No symptom Hormonal treatment Other day of menstrual cylce Hydrosalpinx |
| ♀ ⟪control⟫ N = 5 | Underwent surgery for infertility No pain symptom No hormonal treatment Laparoscopy between day 19 and 24 of menstrual cycle No visible lesion with ultrasound No cyst | Hydrosalpinx Endometriosis or other disease with effect on endometrium Hormonal treatment Other day of menstrual cycle |
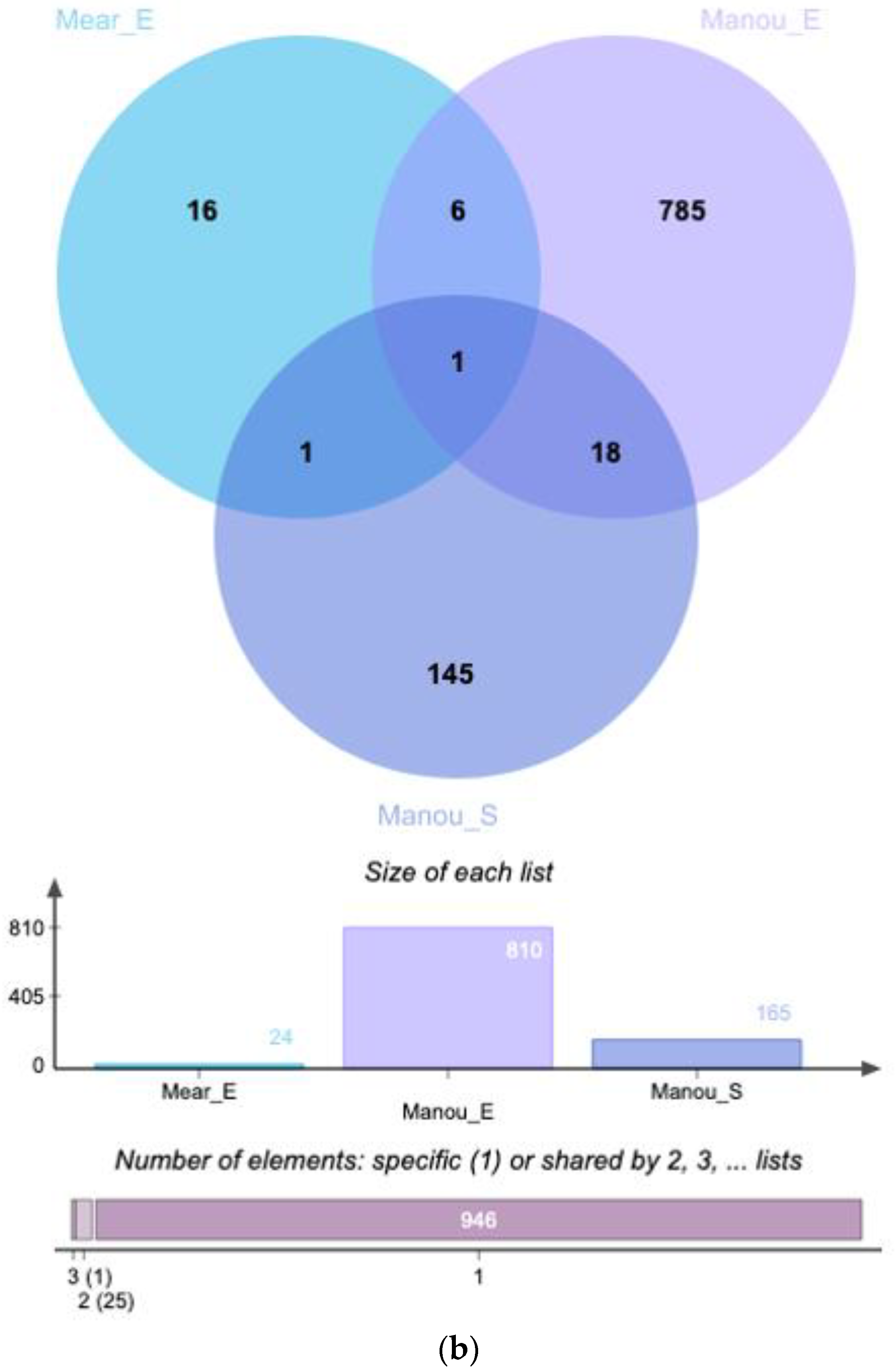
| UniProt iD. | Protein Names | Gene Names | Méar et al. Endometrium (Secretory) | Manousopoulou et al. Endometrium (Proliferative) | Manousopoulou et al. Serum |
|---|---|---|---|---|---|
| Q9Y446 | Plakophilin-3 | PKP3 | up | up | - |
| A0AV96 | RNA-binding protein 47 | RBM47 | up | up | - |
| Q9P2M7 | Cingulin | CGN | up | up | - |
| O95210 | Starch-binding domain-containing protein 1 | STBD1 | up | up | - |
| Q9UIL1 | Short coiled-coil protein | SCOC | up | up | - |
| O00292 | Left-right determination factor 2 | LEFTY2 | up | up | - |
| P13688 | Carcinoembryonic antigen-related cell adhesion molecule 1 | CEACAM1 | up | up | - |
| P22748 | Carbonic anhydrase 4 | CA4 | up | up | - |
| Q06710 | Paired box protein Pax-8 | PAX8 | up | up | - |
| Q12974 | Protein tyrosine phosphatase type IVA 2 | PTP4A2 | up | up | - |
| Q76KP1 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase 1 | B4GALNT4 | up | up | - |
| P11047 | Laminin subunit gamma-1 | LAMC1 | up | - | up |
| Q14203 | Dynactin subunit 1 | DCTN1 | up | - | up |
| Q6IBS0 | Twinfilin-2 | TWF2 | up | - | up |
| O00429 | Dynamin-1-like protein | DNM1L | up | - | up |
| Q15942 | Zyxin | ZYX | up | - | up |
| Q14247 | Src substrate cortactin | CTTN | up | - | up |
| P12081 | Histidine—tRNA ligase, cytoplasmic | HARS | up | - | up |
| O14974 | Protein phosphatase 1 regulatory subunit 12A | PPP1R12A | up | - | up |
| Q27J81 | Inverted formin-2 | INF2 | up | - | up |
| P14317 | Hematopoietic lineage cell-specific protein | HCLS1 | up | - | up |
| O43294 | Transforming growth factor beta-1-induced transcript 1 protein | TGFB1I1 | up | - | up |
| Q99426 | Tubulin-folding cofactor B | TBCB | up | - | up |
| O95747 | Serine/threonine-protein kinase OSR1 | OXSR1 | up | - | up |
| Q05209 | Tyrosine-protein phosphatase non-receptor type 12 | PTPN12 | up | - | up |
| Q8N392 | Rho GTPase-activating protein 18 | ARHGAP18 | up | - | up |
| P98196 | Probable phospholipid-transporting ATPase IH | ATP11A | up | - | up |
| P08311 | Cathepsin G | CTSG | down | - | down |
| P06702 | Protein S100-A9 | S100A9 | down | down | - |
| P05109 | Protein S100-A8 | S100A8 | down | down | - |
| P02775 | Platelet basic protein | PPBP | down | down | - |
| P80511 | Protein S100-A12 | S100A12 | down | down | - |
| P02743 | Serum amyloid P-component | APCS | down | down | - |
| P02776 | Platelet factor 4 | PF4 | down | down | - |
| P02788 | Lactotransferrin | LTF | down | down | down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méar, L.; Com, E.; Fathallah, K.; Guillot, L.; Lavigne, R.; Guével, B.; Fauconnier, A.; Vialard, F.; Pineau, C. The Eutopic Endometrium Proteome in Endometriosis Reveals Candidate Markers and Molecular Mechanisms of Physiopathology. Diagnostics 2022, 12, 419. https://doi.org/10.3390/diagnostics12020419
Méar L, Com E, Fathallah K, Guillot L, Lavigne R, Guével B, Fauconnier A, Vialard F, Pineau C. The Eutopic Endometrium Proteome in Endometriosis Reveals Candidate Markers and Molecular Mechanisms of Physiopathology. Diagnostics. 2022; 12(2):419. https://doi.org/10.3390/diagnostics12020419
Chicago/Turabian StyleMéar, Loren, Emmanuelle Com, Khadija Fathallah, Laetitia Guillot, Régis Lavigne, Blandine Guével, Arnaud Fauconnier, François Vialard, and Charles Pineau. 2022. "The Eutopic Endometrium Proteome in Endometriosis Reveals Candidate Markers and Molecular Mechanisms of Physiopathology" Diagnostics 12, no. 2: 419. https://doi.org/10.3390/diagnostics12020419
APA StyleMéar, L., Com, E., Fathallah, K., Guillot, L., Lavigne, R., Guével, B., Fauconnier, A., Vialard, F., & Pineau, C. (2022). The Eutopic Endometrium Proteome in Endometriosis Reveals Candidate Markers and Molecular Mechanisms of Physiopathology. Diagnostics, 12(2), 419. https://doi.org/10.3390/diagnostics12020419






