Ensembles of Convolutional Neural Networks for Survival Time Estimation of High-Grade Glioma Patients from Multimodal MRI
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
3.1. Data Acquisition and Preparation
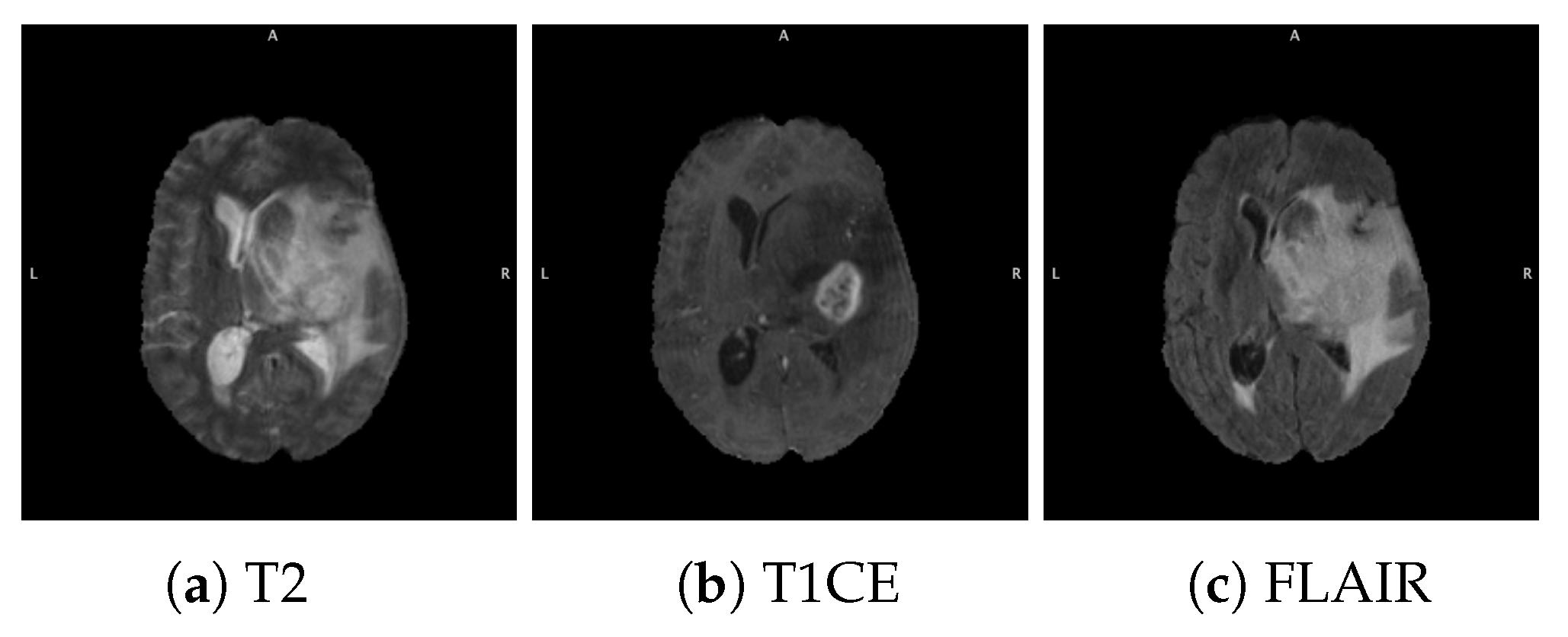
3.1.1. BraTS Dataset
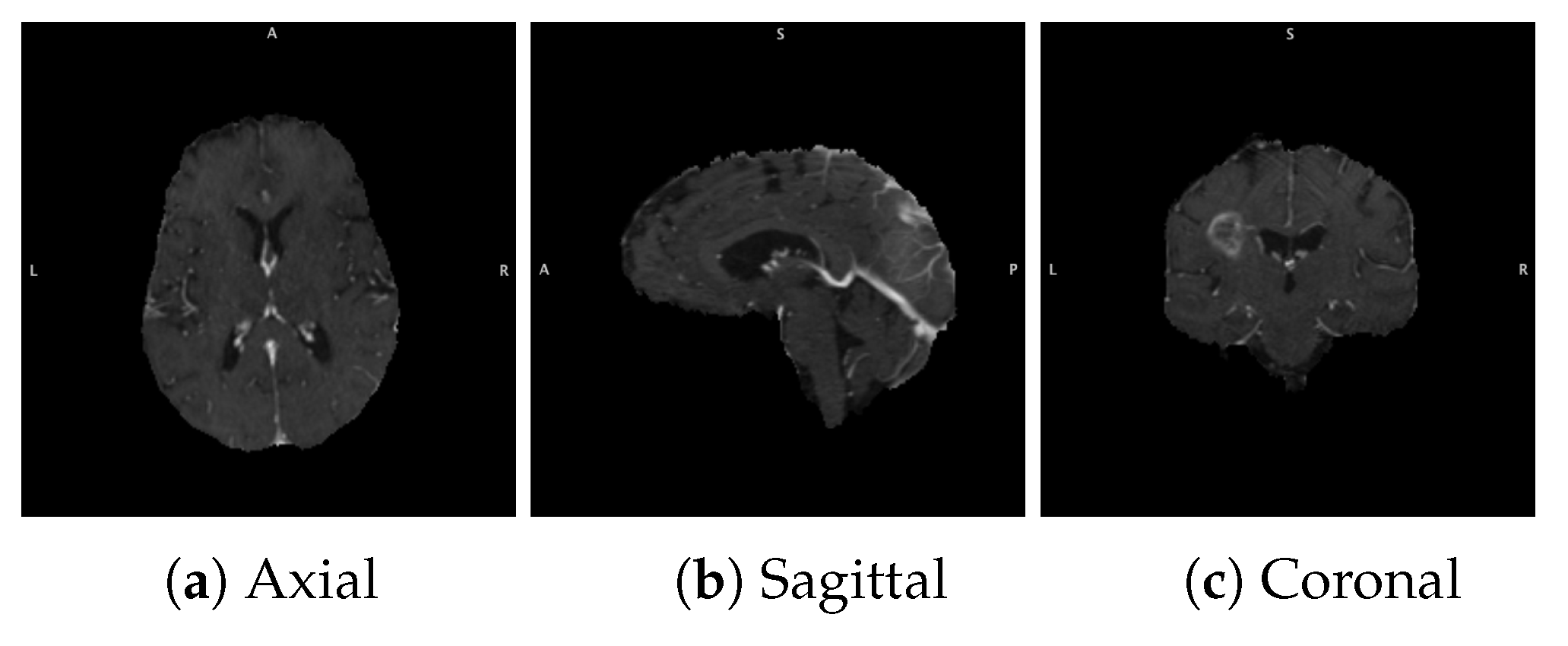
3.1.2. Data Acquisition
3.1.3. Data Pre-Processing and Augmentation
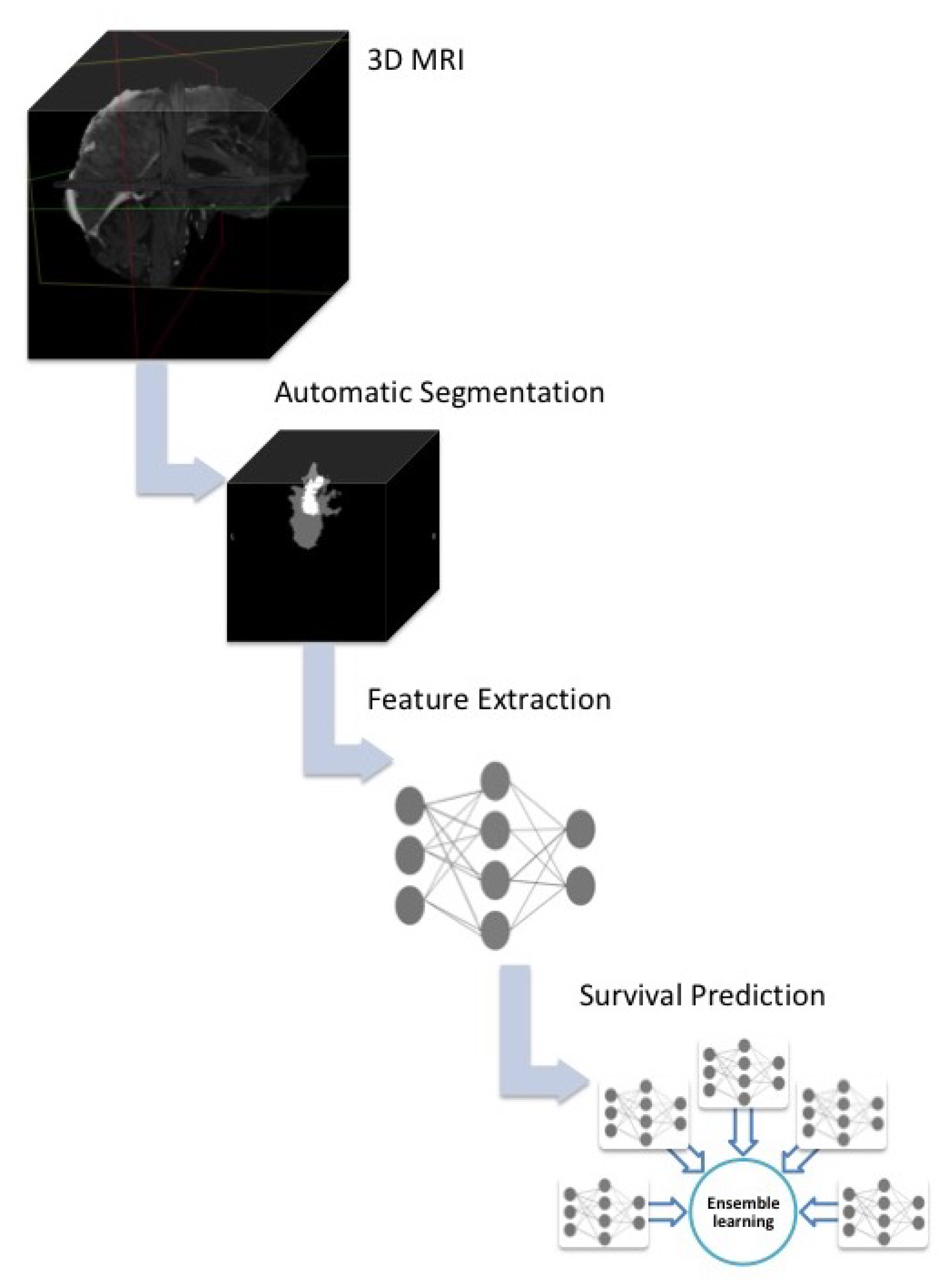
3.2. Overall Survival Prediction System
3.2.1. System Outline
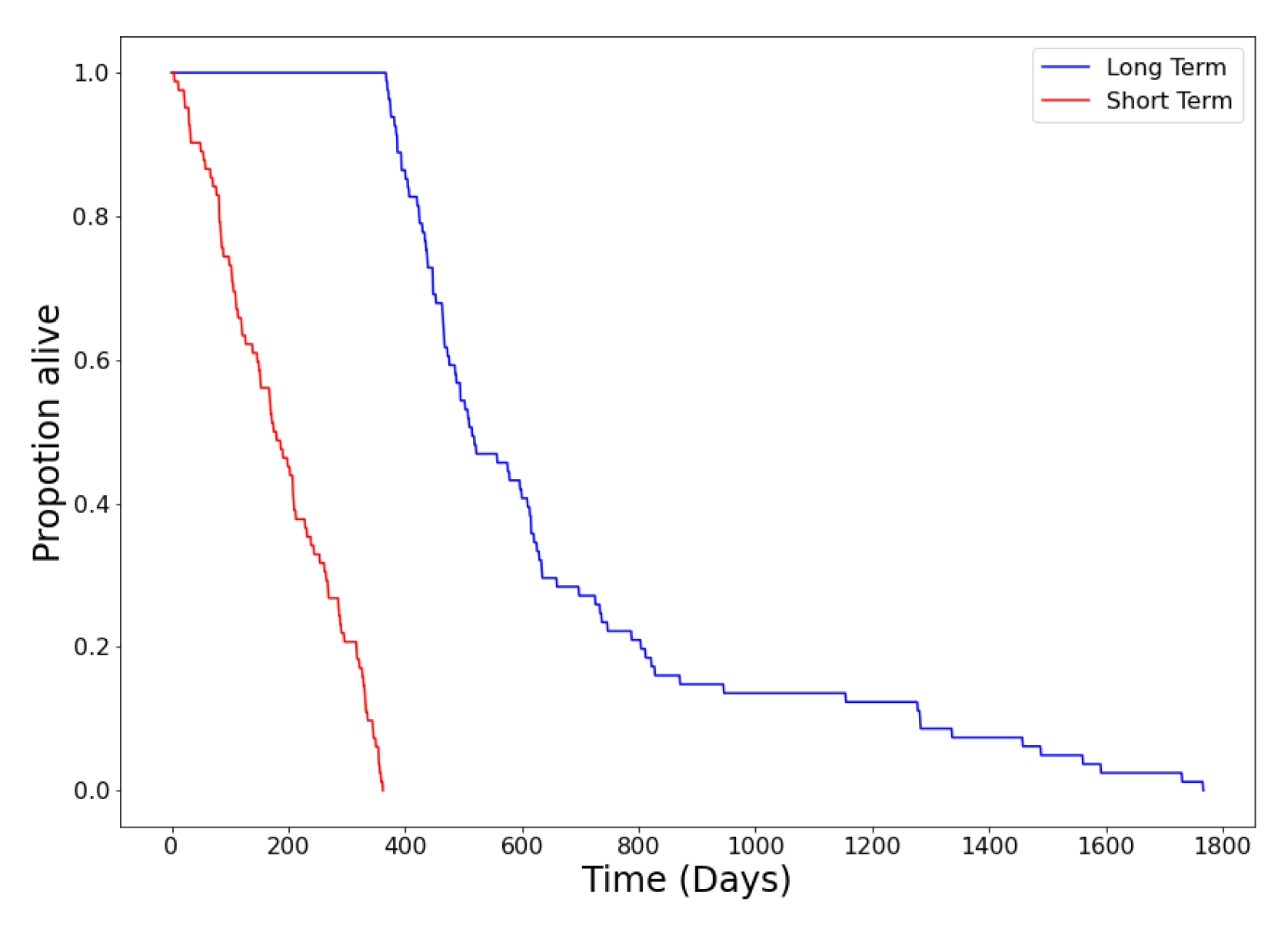
3.2.2. Snapshot Learning for Survival Prediction
3.3. Main CNN Model Construction and Training
4. Experiments and Results
4.1. Parameter Exploration Results
4.1.1. Choice of Snapshots
4.1.2. Ensemble of Convolutional Neural Networks vs. Snapshot Learning
4.1.3. Comparison of 3D T1CE vs. 2D T1CE MRIs
4.2. Combination of Multi-Sequence Data for Survival Prediction
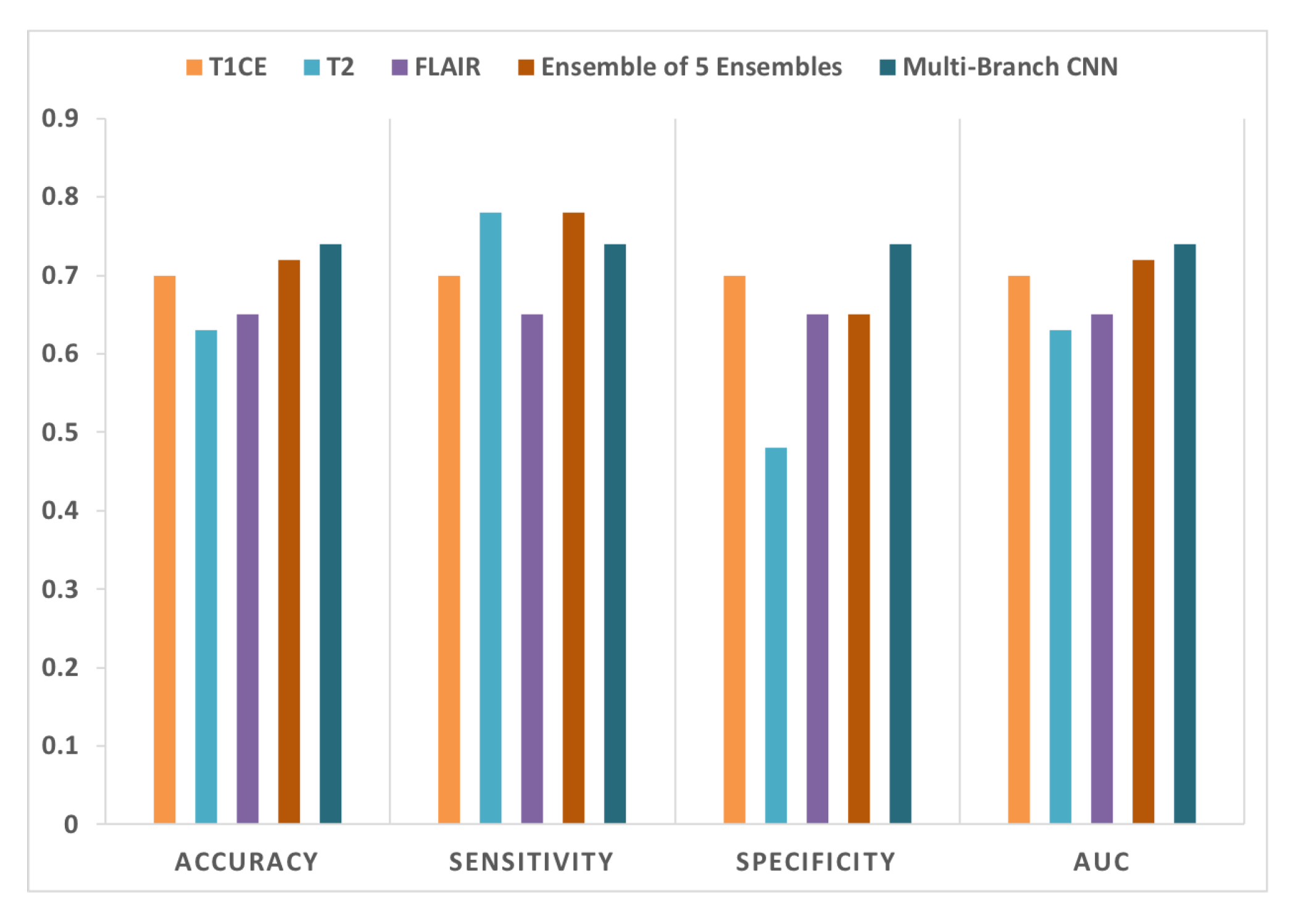
4.2.1. Ensemble of Ensembles Using T1CE, T2, and FLAIR MRIs
4.2.2. Multi-Branch CNN Training with Multi-Sequence 3D MRIs
4.3. Comparison with Other Methods
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncology 2018, 20, iv1–iv86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma. JAMA 2017, 318, 2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Scott, J.; Chaudhury, B.; Hall, L.; Goldgof, D.; Yeom, K.W.; Iv, M.; Ou, Y.; Kalpathy-Cramer, J.; Napel, S.; et al. Radiomics in brain tumor: Image assessment, quantitative feature descriptors, and machine-learning approaches. Am. J. Neuroradiol. 2018, 39, 208–216. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation ({OSDI} 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems; Curran Associates Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Bernal, J.; Kushibar, K.; Asfaw, D.S.; Valverde, S.; Oliver, A.; Martí, R.; Lladó, X. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: A review. Artif. Intell. Med. 2019, 95, 64–81. [Google Scholar] [CrossRef] [Green Version]
- Walid, M. Prognostic Factors for Long-Term Survival after Glioblastoma. Perm. J. 2008, 12, 45–48. [Google Scholar] [CrossRef] [Green Version]
- Bangalore Yogananda, C.G.; Shah, B.R.; Vejdani-Jahromi, M.; Nalawade, S.S.; Murugesan, G.K.; Yu, F.F.; Pinho, M.C.; Wagner, B.C.; Mickey, B.; Patel, T.R.; et al. A novel fully automated MRI-based deep-learning method for classification of IDH mutation status in brain gliomas. Neuro-Oncology 2020, 22, 402–411. [Google Scholar]
- Amian, M.; Soltaninejad, M. Multi-Resolution 3D CNN for MRI Brain Tumor Segmentation and Survival Prediction. arXiv 2019, arXiv:1911.08388. [Google Scholar]
- Agravat, R.; Raval, M.S. Brain Tumor Segmentation and Survival Prediction. arXiv 2019, arXiv:1909.09399. [Google Scholar]
- Rafi, A.; Ali, J.; Akram, T.; Fiaz, K.; Shahid, A.R.; Raza, B.; Madni, T.M. U-Net Based Glioblastoma Segmentation with Patient’s Overall Survival Prediction. In International Symposium on Intelligent Computing Systems; Springer: Berlin, Germany, 2020; pp. 22–32. [Google Scholar]
- Chato, L.; Latifi, S. Machine learning and deep learning techniques to predict overall survival of brain tumor patients using MRI images. In Proceedings of the 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE), Washington, DC, USA, 23–25 October 2017; pp. 9–14. [Google Scholar]
- Hua, R.; Huo, Q.; Gao, Y.; Sui, H.; Zhang, B.; Sun, Y.; Mo, Z.; Shi, F. Segmenting Brain Tumor Using Cascaded V-Nets in Multimodal MR Images. Front. Comput. Neurosci. 2020, 14, 9. [Google Scholar] [CrossRef]
- McKinley, R.; Rebsamen, M.; Daetwyler, K.; Meier, R.; Radojewski, P.; Wiest, R. Uncertainty-driven refinement of tumor-core segmentation using 3D-to-2D networks with label uncertainty. arXiv 2020, arXiv:2012.06436. [Google Scholar]
- Wang, S.; Dai, C.; Mo, Y.; Angelini, E.; Guo, Y.; Bai, W. Automatic Brain Tumour Segmentation and Biophysics-Guided Survival Prediction. arXiv 2019, arXiv:1911.08483. [Google Scholar]
- Nalepa, J.; Lorenzo, P.R.; Marcinkiewicz, M.; Bobek-Billewicz, B.; Wawrzyniak, P.; Walczak, M.; Kawulok, M.; Dudzik, W.; Kotowski, K.; Burda, I.; et al. Fully-automated deep learning-powered system for DCE-MRI analysis of brain tumors. Artif. Intell. Med. 2020, 102, 101769. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In International MICCAI Brainlesion Workshop; Springer: Berlin, Germany, 2017; pp. 178–190. [Google Scholar]
- Chollet, F.; Keras. 2015. Available online: https://github.com/fchollet/keras (accessed on 14 December 2021).
- Huang, G.; Li, Y.; Pleiss, G.; Liu, Z.; Hopcroft, J.E.; Weinberger, K.Q. Snapshot ensembles: Train 1, get m for free. arXiv 2017, arXiv:1704.00109. [Google Scholar]
- Ahmed, K.B.; Hall, L.O.; Liu, R.; Gatenby, R.A.; Goldgof, D.B. Neuroimaging Based Survival Time Prediction of GBM Patients Using CNNs from Small Data. In Proceedings of the 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC), Bari, Italy, 6–9 October 2019; pp. 1331–1335. [Google Scholar]
- Kickingereder, P.; Burth, S.; Wick, A.; Götz, M.; Eidel, O.; Schlemmer, H.P.; Maier-Hein, K.H.; Wick, W.; Bendszus, M.; Radbruch, A.; et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology 2016, 280, 880–889. [Google Scholar] [CrossRef]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Vega, J.E.V.; Brat, D.J.; Cooper, L.A. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2017, 115, 201717139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.; Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kang, S.G.; Kim, E.H.; Kim, S.H.; Lee, S.K. Radiomic MRI Phenotyping of Glioblastoma: Improving Survival Prediction. Radiology 2018, 289, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, M.; Rathore, S.; Nasrallah, M. Analysis of microscopic images via deep neural networks can predict outcome and IDH and 1p/19q codeletion status in gliomas. J. Neuropathol. Exp. Neurol. 2019, 78, 553. [Google Scholar]
- Shukla, G.; Bakas, S.; Rathore, S.; Akbari, H.; Sotiras, A.; Davatzikos, C. Radiomic features from multi-institutional glioblastoma MRI offer additive prognostic value to clinical and genomic markers: Focus on TCGA-GBM collection. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, E107–E108. [Google Scholar] [CrossRef]
- Chaddad, A.; Daniel, P.; Desrosiers, C.; Toews, M.; Abdulkarim, B. Novel Radiomic Features Based on Joint Intensity Matrices for Predicting Glioblastoma Patient Survival Time. IEEE J. Biomed. Health Inform. 2019, 23, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.; Chaddad, A.; Iftikhar, M.A.; Bilello, M.; Abdulkadir, A. Combining MRI and Histologic Imaging Features for Predicting Overall Survival in Patients with Glioma. Radiol. Imaging Cancer 2021, 3, e200108. [Google Scholar] [CrossRef] [PubMed]
- Macyszyn, L.; Akbari, H.; Pisapia, J.M.; Da, X.; Attiah, M.; Pigrish, V.; Bi, Y.; Pal, S.; Davuluri, R.V.; Roccograndi, L.; et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-Oncology 2015, 18, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaddad, A.; Daniel, P.; Sabri, S.; Desrosiers, C.; Abdulkarim, B. Integration of radiomic and multi-omic analyses predicts survival of newly diagnosed IDH1 wild-type glioblastoma. Cancers 2019, 11, 1148. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; Van Den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Lamborn, K.R.; Chang, S.M.; Prados, M.D. Prognostic factors for survival of patients with glioblastoma: Recursive partitioning analysis. Neuro-Oncology 2004, 6, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Marina, O.; Suh, J.H.; Reddy, C.A.; Barnett, G.H.; Vogelbaum, M.A.; Peereboom, D.M.; Stevens, G.H.; Elinzano, H.; Chao, S.T. Treatment outcomes for patients with glioblastoma multiforme and a low Karnofsky Performance Scale score on presentation to a tertiary care institution. J. Neurosurg. 2011, 115, 220–229. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, C.; Hentschel, B.; Wick, W.; Capper, D.; Felsberg, J.; Simon, M.; Westphal, M.; Schackert, G.; Meyermann, R.; Pietsch, T.; et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: Implications for classification of gliomas. Acta Neuropathol. 2010, 120, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Baumert, B.G.; Hegi, M.E.; van den Bent, M.J.; von Deimling, A.; Gorlia, T.; Hoang-Xuan, K.; Brandes, A.A.; Kantor, G.; Taphoorn, M.J.; Hassel, M.B.; et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033-26033): A randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016, 17, 1521–1532. [Google Scholar] [CrossRef] [Green Version]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquilanti, E.; Miller, J.; Santagata, S.; Cahill, D.P.; Brastianos, P.K. Updates in prognostic markers for gliomas. Neuro-Oncology 2018, 20, vii17–vii26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellingson, B.M.; Abrey, L.E.; Nelson, S.J.; Kaufmann, T.J.; Garcia, J.; Chinot, O.; Saran, F.; Nishikawa, R.; Henriksson, R.; Mason, W.P.; et al. Validation of postoperative residual contrast-enhancing tumor volume as an independent prognostic factor for overall survival in newly diagnosed glioblastoma. Neuro-Oncology 2018, 20, 1240–1250. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hall, L.O.; Goldgof, D.B.; Zhou, M.; Gatenby, R.A.; Ahmed, K.B. Exploring deep features from brain tumor magnetic resonance images via transfer learning. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 235–242. [Google Scholar]
- Tanaka, Y.; Nariai, T.; Momose, T.; Aoyagi, M.; Maehara, T.; Tomori, T.; Yoshino, Y.; Nagaoka, T.; Ishiwata, K.; Ishii, K.; et al. Glioma surgery using a multimodal navigation system with integrated metabolic images. J. Neurosurg. 2009, 110, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Arbizu, J.; Tejada, S.; Marti-Climent, J.; Diez-Valle, R.; Prieto, E.; Quincoces, G.; Vigil, C.; Idoate, M.; Zubieta, J.; Peñuelas, I.; et al. Quantitative volumetric analysis of gliomas with sequential MRI and 11 C-methionine PET assessment: Patterns of integration in therapy planning. Eur. J. Nucl. Med. Mol. Imaging 2012, 39, 771–781. [Google Scholar] [CrossRef]


| Method | Accuracy | Specificity | Sensitivity | AUC |
|---|---|---|---|---|
| Snapshot 1 | 63% | 39% | 87% | 0.63 |
| Snapshot 2 | 67% | 74% | 61% | 0.67 |
| Snapshot 3 | 70% | 74% | 65% | 0.70 |
| Snapshot 4 | 61% | 74% | 48% | 0.61 |
| Snapshot 5 | 59% | 52% | 65% | 0.59 |
| Ensemble | 70% | 70% | 70% | 0.70 |
| Method | Accuracy | Specificity | Sensitivity | AUC |
|---|---|---|---|---|
| CNN 1 | 63% | 48% | 78% | 0.63 |
| CNN 2 | 54% | 48% | 61% | 0.54 |
| CNN 3 | 61% | 78% | 43% | 0.61 |
| CNN 4 | 61% | 65% | 57% | 0.61 |
| CNN 5 | 57% | 43% | 70% | 0.57 |
| Ensemble | 67% | 70% | 65% | 0.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Ahmed, K.; Hall, L.O.; Goldgof, D.B.; Gatenby, R. Ensembles of Convolutional Neural Networks for Survival Time Estimation of High-Grade Glioma Patients from Multimodal MRI. Diagnostics 2022, 12, 345. https://doi.org/10.3390/diagnostics12020345
Ben Ahmed K, Hall LO, Goldgof DB, Gatenby R. Ensembles of Convolutional Neural Networks for Survival Time Estimation of High-Grade Glioma Patients from Multimodal MRI. Diagnostics. 2022; 12(2):345. https://doi.org/10.3390/diagnostics12020345
Chicago/Turabian StyleBen Ahmed, Kaoutar, Lawrence O. Hall, Dmitry B. Goldgof, and Robert Gatenby. 2022. "Ensembles of Convolutional Neural Networks for Survival Time Estimation of High-Grade Glioma Patients from Multimodal MRI" Diagnostics 12, no. 2: 345. https://doi.org/10.3390/diagnostics12020345
APA StyleBen Ahmed, K., Hall, L. O., Goldgof, D. B., & Gatenby, R. (2022). Ensembles of Convolutional Neural Networks for Survival Time Estimation of High-Grade Glioma Patients from Multimodal MRI. Diagnostics, 12(2), 345. https://doi.org/10.3390/diagnostics12020345







