Analysis of Early Biomarkers Associated with the Development of Critical Respiratory Failure in Coronavirus Disease 2019 (COVID-19)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Endpoints
2.3. Biomarker Analysis
2.4. Statistical Analysis
3. Results
3.1. Patient Characteristics and Outcomes
3.2. Comparison of Biomarkers between Patients with and without MW
3.3. Cox Proportional Hazards Regression Analysis for Detecting Biomarkers Associated with Time-to-MV
3.4. ROC Curve Analysis for Determining the Association between Biomarkers and the Start of MV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Rabi, F.A.; Al Zoubi, M.S.; Kasasbeh, G.A.; Salameh, D.M.; Al-Nasser, A.D. SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far. Pathogens 2020, 9, 231. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Hasan, S.S.; Capstick, T.; Ahmed, R.; Kow, C.S.; Mazhar, F.; Merchant, H.A.; Zaidi, S.T.R. Mortality in COVID-19 patients with acute respiratory distress syndrome and corticosteroids use: A systematic review and meta-analysis. Expert Rev. Respir. Med. 2020, 14, 1149–1163. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [Green Version]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef]
- Słomka, A.; Kowalewski, M.; Żekanowska, E. Coronavirus Disease 2019 (COVID-19): A Short Review on Hematological Manifestations. Pathogens 2020, 9, 493. [Google Scholar] [CrossRef]
- Chen, R.; Huang, Y.; Quan, J.; Liu, J.; Wang, H.; Billiar, T.R.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon 2020, 6, e05672. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Laguna-Goya, R.; Utrero-Rico, A.; Talayero, P.; Lasa-Lazaro, M.; Ramirez-Fernandez, A.; Naranjo, L.; Segura-Tudela, A.; Cabrera-Marante, O.; de Frias, E.R.; Garcia-Garcia, R.; et al. IL-6–based mortality risk model for hospitalized patients with COVID-19. J. Allergy Clin. Immunol. 2020, 146, 799–807.e9. [Google Scholar] [CrossRef]
- Popadic, V.; Klasnja, S.; Milic, N.; Rajovic, N.; Aleksic, A.; Milenkovic, M.; Crnokrak, B.; Balint, B.; Todorovic-Balint, M.; Mrda, D.; et al. Predictors of Mortality in Critically Ill COVID-19 Patients Demanding High Oxygen Flow: A Thin Line between Inflammation, Cytokine Storm, and Coagulopathy. Oxidative Med. Cell. Longev. 2021, 2021, 6648199. [Google Scholar] [CrossRef]
- Herold, T.; Jurinovic, V.; Arnreich, C.; Lipworth, B.J.; Hellmuth, J.C.; von Bergwelt-Baildon, M.; Klein, M.; Weinberger, T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020, 146, 128–136.e4. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, X.; Fan, Q.; Liu, H.; Liu, X.; Liu, Z.; Zhang, Z. d-dimer levels on admission to predict in-hospital mortality in patients with COVID-19. J. Thromb. Haemost. 2020, 18, 1324–1329. [Google Scholar] [CrossRef]
- Ayanian, S.; Reyes, J.; Lynn, L.; Teufel, K. The association between biomarkers and clinical outcomes in novel coronavirus pneumonia in a US cohort. Biomark. Med. 2020, 14, 1091–1097. [Google Scholar] [CrossRef]
- Li, Y.; Deng, Y.; Ye, L.; Sun, H.; Du, S.; Huang, H.; Zeng, F.; Chen, X.; Deng, G. Clinical Significance of Plasma d-Dimer in COVID-19 Mortality. Front. Med. 2021, 8, 638097. [Google Scholar] [CrossRef]
- Zhan, H.; Chen, H.; Liu, C.; Cheng, L.; Yan, S.; Li, H.; Li, Y. Diagnostic Value of d-Dimer in COVID-19: A Meta-Analysis and Meta-Regression. Clin. Appl. Thromb. Hemost. 2021, 27, 10760296211010976. [Google Scholar] [CrossRef]
- Knight, S.R.; Ho, A.; Pius, R.; Buchan, I.; Carson, G.; Drake, T.M.; Dunning, J.; Fairfield, C.J.; Gamble, C.; Green, C.A.; et al. Risk stratification of patients admitted to hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: Development and validation of the 4C Mortality Score. BMJ 2020, 370, m3339. [Google Scholar] [CrossRef]
- Liang, W.; Liang, H.; Ou, L.; Chen, B.; Chen, A.; Li, C.; Li, Y.; Guan, W.; Sang, L.; Lu, J.; et al. Development and Validation of a Clinical Risk Score to Predict the Occurrence of Critical Illness in Hospitalized Patients With COVID-19. JAMA Intern. Med. 2020, 180, 1081–1089. [Google Scholar] [CrossRef]
- National Institutes of Health Website. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/management-of-covid-19/ (accessed on 13 May 2020).
- Pisano, A.; Yavorovskiy, A.; Verniero, L.; Landoni, G. Indications for Tracheal Intubation in Patients With Coronavirus Disease 2019 (COVID-19). J. Cardiothorac. Vasc. Anesth. 2021, 35, 1276–1280. [Google Scholar] [CrossRef]
- Hoshino, T.; Okamoto, M.; Sakazaki, Y.; Kato, S.; Young, H.A.; Aizawa, H. Role of Proinflammatory Cytokines IL-18 and IL-1β in Bleomycin-Induced Lung Injury in Humans and Mice. Am. J. Respir. Cell Mol. Biol. 2009, 41, 661–670. [Google Scholar] [CrossRef]
- Stilund, M.; Gjelstrup, M.C.; Petersen, T.; Møller, H.J.; Rasmussen, P.V.; Christensen, T. Biomarkers of Inflammation and Axonal Degeneration/Damage in Patients with Newly Diagnosed Multiple Sclerosis: Contributions of the Soluble CD163 CSF/Serum Ratio to a Biomarker Panel. PLoS ONE 2015, 10, e0119681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajiri, M.; Okamoto, M.; Fujimoto, K.; Johkoh, T.; Ono, J.; Tominaga, M.; Azuma, K.; Kawayama, T.; Ohta, S.; Izuhara, K.; et al. Serum level of periostin can predict long-term outcome of idiopathic pulmonary fibrosis. Respir. Investig. 2015, 53, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Andersson, U.; Tracey, K.J. HMGB1 Is a Therapeutic Target for Sterile Inflammation and Infection. Annu. Rev. Immunol. 2011, 29, 139–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, R.; Chen, R.; Zhang, Q.; Hou, W.; Wu, S.; Cao, L.; Huang, J.; Yu, Y.; Fan, X.-G.; Yan, Z.; et al. HMGB1 in health and disease. Mol. Asp. Med. 2014, 40, 1–116. [Google Scholar] [CrossRef] [Green Version]
- Andersson, U.; Yang, H.; Harris, H. High-mobility group box 1 protein (HMGB1) operates as an alarmin outside as well as inside cells. Semin. Immunol. 2018, 38, 40–48. [Google Scholar] [CrossRef]
- Park, J.S.; Svetkauskaite, D.; He, Q.; Kim, J.Y.; Strassheim, D.; Ishizaka, A.; Abraham, E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 2004, 279, 7370–7377. [Google Scholar] [CrossRef] [Green Version]
- Andersson, U.; Ottestad, W.; Tracey, K.J. Extracellular HMGB1: A therapeutic target in severe pulmonary inflammation including COVID-19? Mol. Med. 2020, 26, 42. [Google Scholar] [CrossRef]
- Qu, L.; Chen, C.; Chen, Y.; Li, Y.; Tang, F.; Huang, H.; He, W.; Zhang, R.; Shen, L. High-Mobility Group Box 1 (HMGB1) and Autophagy in Acute Lung Injury (ALI): A Review. Med. Sci. Monit. 2019, 25, 1828–1837. [Google Scholar] [CrossRef]
- Tseng, C.-C.; Fang, W.-F.; Leung, S.Y.; Chen, H.-C.; Chang, Y.-C.; Wang, C.-C.; Chang, H.-C.; Lin, M.-C. Impact of Serum Biomarkers and Clinical Factors on Intensive Care Unit Mortality and 6-Month Outcome in Relatively Healthy Patients with Severe Pneumonia and Acute Respiratory Distress Syndrome. Dis. Markers 2014, 2014, 804654. [Google Scholar] [CrossRef] [Green Version]
- Hamada, N.; Maeyama, T.; Kawaguchi, T.; Yoshimi, M.; Fukumoto, J.; Yamada, M.; Yamada, S.; Kuwano, K.; Nakanishi, Y. The Role of High Mobility Group Box1 in Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2008, 39, 440–447. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Iwamoto, H.; Sakamoto, S.; Horimasu, Y.; Masuda, T.; Miyamoto, S.; Nakashima, T.; Ohshimo, S.; Fujitaka, K.; Hamada, H.; et al. Serum high-mobility group box 1 is associated with the onset and severity of acute exacerbation of idiopathic pulmonary fibrosis. Respirology 2020, 25, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Sakamoto, S.; Isshiki, T.; Furuya, K.; Kurosaki, A.; Homma, S. Association of serum high-mobility group box protein 1 level with outcomes of acute exacerbation of idiopathic pulmonary fibrosis and fibrosing nonspecific interstitial pneumonia. PLoS ONE 2018, 13, e0196558. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Long, X.; Xu, Q.; Tan, J.; Wang, G.; Cao, Y.; Wei, J.; Luo, H.; Zhu, H.; Huang, L.; et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 992–994. [Google Scholar] [CrossRef] [PubMed]
- Andersson, U.; Tracey, K.J. HMGB1 in Sepsis. Scand. J. Infect. Dis. 2003, 35, 577–584. [Google Scholar] [CrossRef]
- Kohno, N.; Awaya, Y.; Oyama, T.; Yamakido, M.; Akiyama, M.; Inoue, Y.; Yokoyama, A.; Hamada, H.; Fujioka, S.; Hiwada, K. KL-6, a Mucin-like Glycoprotein, in Bronchoalveolar Lavage Fluid from Patients with Interstitial Lung Disease. Am. Rev. Respir. Dis. 1993, 148, 637–642. [Google Scholar] [CrossRef]
- Yokoyama, A.; Kondo, K.; Nakajima, M.; Matsushima, T.; Takahashi, T.; Nishimura, M.; Bando, M.; Sugiyama, Y.; Totani, Y.; Ishizaki, T.; et al. Prognostic value of circulating KL-6 in idiopathic pulmonary fibrosis. Respirology 2006, 11, 164–168. [Google Scholar] [CrossRef]
- Ishii, H.; Mukae, H.; Kadota, J.; Kaida, H.; Nagata, T.; Abe, K.; Matsukura, S.; Kohno, S. High serum concentrations of surfactant protein A in usual interstitial pneumonia compared with non-specific interstitial pneumonia. Thorax 2003, 58, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, N.; Hattori, N.; Yokoyama, A.; Kohno, N. Utility of KL-6/MUC1 in the clinical management of interstitial lung diseases. Respir. Investig. 2012, 50, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Kondo, T.; Hattori, N.; Ishikawa, N.; Murai, H.; Haruta, Y.; Hirohashi, N.; Tanigawa, K.; Kohno, N. KL-6 concentration in pulmonary epithelial lining fluid is a useful prognostic indicator in patients with acute respiratory distress syndrome. Respir. Res. 2011, 12, 32. [Google Scholar] [CrossRef] [Green Version]
- Ishizaka, A.; Matsuda, T.; Albertine, K.H.; Koh, H.; Tasaka, S.; Hasegawa, N.; Kohno, N.; Kotani, T.; Morisaki, H.; Takeda, J.; et al. Elevation of KL-6, a lung epithelial cell marker, in plasma and epithelial lining fluid in acute respiratory distress syndrome. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 286, L1088–L1094. [Google Scholar] [CrossRef]
- Sato, H.; Callister, M.E.J.; Mumby, S.; Quinlan, G.J.; Welsh, K.I.; Dubois, R.M.; Evans, T.W. KL-6 levels are elevated in plasma from patients with acute respiratory distress syndrome. Eur. Respir. J. 2004, 23, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Matthay, M.A.; Zemans, R.L.; Zimmerman, G.A.; Arabi, Y.M.; Beitler, J.R.; Mercat, A.; Herridge, M.; Randolph, A.G.; Calfee, C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers 2019, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- d’Alessandro, M.; Cameli, P.; Refini, R.M.; Bergantini, L.; Alonzi, V.; Lanzarone, N.; Bennett, D.; Rana, G.D.; Montagnani, F.; Scolletta, S.; et al. Serum KL-6 concentrations as a novel biomarker of severe COVID-19. J. Med. Virol. 2020, 92, 2216–2220. [Google Scholar] [CrossRef] [PubMed]
- Awano, N.; Inomata, M.; Kuse, N.; Tone, M.; Takada, K.; Muto, Y.; Fujimoto, K.; Akagi, Y.; Mawatari, M.; Ueda, A.; et al. Serum KL-6 level is a useful biomarker for evaluating the severity of coronavirus disease 2019. Respir. Investig. 2020, 58, 440–447. [Google Scholar] [CrossRef]
- Deng, K.; Fan, Q.; Yang, Y.; Deng, X.; He, R.; Tan, Y.; Lan, Y.; Deng, X.; Pan, Y.; Wang, Y.; et al. Prognostic roles of KL-6 in disease severity and lung injury in COVID-19 patients: A longitudinal retrospective analysis. J. Med. Virol. 2021, 93, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, M.; Bergantini, L.; Cameli, P.; Curatola, G.; Remediani, L.; Sestini, P.; Bargagli, E.; Siena COVID Unit. Peripheral biomarkers’ panel for severe COVID-19 patients. J. Med. Virol. 2021, 93, 1230–1232. [Google Scholar] [CrossRef]
- Crisan-Dabija, R.; Covic, A.; Brinza, C.; Popa, I.V.; Burlacu, A. Involvement of KL-6 Biomarker in Interstitial Lung Disease Induced by SARS-CoV-2 Infection: A Systematic Review. Appl. Sci. 2021, 11, 3482. [Google Scholar] [CrossRef]
- D’Alessandro, M.; Bergantini, L.; Cameli, P.; Curatola, G.; Remediani, L.; Bennett, D.; Bianchi, F.; Perillo, F.; Volterrani, L.; Mazzei, M.A.; et al. Serial KL-6 measurements in COVID-19 patients. Intern. Emerg. Med. 2021, 16, 1541–1545. [Google Scholar] [CrossRef]
- Nishida, S.; Fukazawa, R.; Imai, T.; Takeda, S.; Hayakawa, J.; Takeuchi, H.; Shimizu, K.; Kawakami, Y.; Takase, M. Serum KL-6 and surfactant protein D in children with 2009 pandemic H1N1 influenza infection. Pediatr. Int. 2011, 53, 910–914. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, Y.; Chi, H.; Chen, S.; Peng, M.; Luo, L.; Chen, L.; Li, J.; Shen, B.; Wang, D. The hemocyte counts as a potential biomarker for predicting disease progression in COVID-19: A retrospective study. Clin. Chem. Lab. Med. 2020, 58, 1106–1115. [Google Scholar] [CrossRef]
| All Cases | Asymptomatic or Mild at Admission | Moderate at Admission | Severe at Admission | p Value | |
|---|---|---|---|---|---|
| N | 135 | 42 | 66 | 27 | |
| Age (years) | 50.0 (35.0–70.0) | 32.5 (26.0–49.0) | 53.5 (39.0–72.3) | 67.0 (51.0–72.0) | <0.00010 * |
| Gender, Male | 69 (51%) | 8 (19.1%) | 40 (60.6%) | 21 (77.8%) | <0.00010 * |
| Smoker | 56 (47.5%) | 19 (48.7%) | 28 (46.7%) | 9 (47.4%) | 1.0 |
| Complication | |||||
| Diabetes mellitus | 22 (16.3%) | 1 (2.4%) | 11 (16.7%) | 10 (37.0%) | 0.014 * |
| Hypertension | 33 (24.4%) | 4 (9.5%) | 16 (24.2%) | 13 (48.2%) | 0.0014 * |
| Malignant disease | 9 (6.7%) | 1 (2.4%) | 5 (7.6%) | 3 (11.1%) | 0.39 |
| Data at admission | |||||
| Neutrophil counts (/µL) | 3737.0 (2664.0–5883.0) | 3278.2 (2303.2–4670.6) | 3465.5 (2556.9–5032.9) | 5893.0 (4012.0–8333.4) | <0.0001 * |
| Lymphocyte counts (/µL) | 1360.1 (952.0–1757.5) | 1635.1 (1038.0–1823.8) | 1211.8 (922.1–1610.8) | 1354.5 (844.9–1925.7) | 0.048 * |
| Platelet counts (×104/µL) | 18.5 (15.6–24.0) | 20.4 (16.9–24.5) | 18.7 (15.4–25.5) | 18.0 (13.6–20.5) | 0.18 |
| CRP (mg/dL) | 1.5 (0.22–4.7) | 0.14 (0.05–0.59) | 2.3 (1.1–4.6) | 6.9 (3.0–10.7) | <0.00010 * |
| Lactate dehydrogenase (IU/L) | 219.0 (177.0–296.0) | 174.5 (156.8–201.3) | 220.5 (188.5–264.3) | 390.0 (298.0–485.0) | <0.00010 * |
| Ferritin (ng/mL) | 337.0 (114.0–743.1) | 99.5 (33.8–136.0) | 371.8 (251.6–668.6) | 1116.4 (643.0–1698.9) | <0.00010 * |
| Interleukin-6 (pg/mL) | 9.1 (2.4–20.8) | 1.8 (1.3–4.5) | 11.4 (5.8–21.2) | 20.4 (8.4–58.9) | <0.00010 * |
| Interleukin-18 (pg/mL) | 295.0 (207.5–406.0) | 227.5 (175.0–310.0) | 290.0 (210.0–406.0) | 502.5 (353.5–678.8) | <0.00010 * |
| KL-6 (IU/mL) | 220.5 (185.0–294.3) | 204.5 (168.8–264.3) | 224.0 (184.0–289.0) | 312.0 (210.0–410.0) | 0.0010 * |
| HMGB-1 (ng/mL) | 6.5 (4.1–9.4) | 7.3 (5.8–11.0) | 5.4 (3.2–8.8) | 6.6 (5.1–11.9) | 0.13 |
| Soluble CD163 (ng/mL) | 558.5 (459.8–724.0) | 513.0 (435.0–694.0) | 529.5 (440.5–631.5) | 748.0 (501.0–886.0) | 0.1 |
| d-dimer (µg/mL) | 0.50 (0.30–1.1) | 0.40 (0.20–0.55) | 0.40 (0.20–1.1) | 1.1 (0.60–1.7) | <0.00010 * |
| Data at day 3 | <0.00010 * | ||||
| Neutrophil counts (/µL) | 3011.4 (2073.0–4951.0) | 2521.1 (1740.6–3844.9) | 2808.0 (2007.8–4048.9) | 6706.0 (3063.7–9683.2) | <0.00010 * |
| Lymphocyte counts (/µL) | 1278.9 (900.6–1847.3) | 1818.0 (1332.4–2118.0) | 1171.8 (792.0–1729.0) | 1040.7 (599.8–1321.3) | <0.00010 * |
| Platelet counts (×104/µL) | 20.5 (16.3–26.4) | 20.7 (17.0–26.1) | 19.8 (15.1–27.2) | 22.0 (17.6–28.3) | 0.70 |
| CRP (mg/dL) | 1.4 (0.26–4.6) | 0.070 (0.050–0.32) | 2.1 (0.77–7.1) | 5.0 (1.8–9.4) | <0.00010 * |
| Lactate dehydrogenase (IU/L) | 214.0 (170.0–306.8) | 164.0 (138.0–195.0) | 226.0 (181.0–275.0) | 371.0 (310.5–491.8) | <0.00010 * |
| Ferritin (ng/mL) | 467.8 (142.3–978.5) | 85.1 (21.0–132.2) | 469.8 (228.0–964.5) | 869.2 (666.6–1681.8) | <0.00010 * |
| Interleukin-6 (pg/mL) | 7.7 (2.4–23.1) | 1.8 (1.2–2.7) | 10.0 (4.2–20.0) | 20.2 (5.8–37.9) | <0.00010 * |
| HMGB-1 (ng/mL) | 8.5 (5.1–14.7) | 6.3 (4.3–9.5) | 7.0 (5.0–11.0) | 15.2 (8.7–21.4) | 0.00010 * |
| Soluble CD163 (ng/mL) | 617.0 (459.5–886.0) | 463.0 (342.8–630.5) | 597.0 (459.3–808.8) | 878.0 (650.5–1000.0) | <0.00010 * |
| d-dimer (µg/mL) | 0.60 (0.40–1.3) | 0.40 (0.20–0.60) | 0.60 (0.40–1.4) | 1.0 (0.50–4.8) | 0.0016 * |
| All Cases at Admission | Asymptomatic or Mild at Admission | Moderate at Admission | Severe at Admission | p Value | |
|---|---|---|---|---|---|
| N | 135 | 42 | 66 | 27 | |
| Started MV or died | 11 (8.1%) | 0 | 6 (9.1%) | 5 (18.5%) | 0.015 * |
| Started MV and survived | 6 (4.4%) | 0 | 4 (6.1%) | 2 (7.4%) | 0.21 |
| Started MV and died | 4 (3.0%) | 0 | 1 (1.5%) | 3 (11.1%) | 0.034 * |
| Survived without MV | 124 (91.9%) | 42 (100%) | 60 (90.9%) | 22 (81.5%) | 0.015 * |
| Died without MV | 1 (0.74%) | 0 | 1 (1.5%) | 0 | 1 |
| Duration from admission to start of MV | 2.0 (0–8.0) | - | 8.0 (1.5–8.5) | 0 (0–2.5) | 0.084 |
| Started MV | |||
|---|---|---|---|
| Yes | No | p Value | |
| N | 10 (7.4%) | 125 (92.6%) | |
| Data at admission | |||
| Neutrophil counts (/µL) | 6104.1 (4162.6–8076.9) | 3570.0 (2642.4–5302.8) | 0.017 * |
| Lymphocyte counts (/µL) | 1007.0 (749.0–1384.2) | 1419.6 (958.0–1765.1) | 0.084 * |
| Platelet counts (×104/µL) | 14.4 (12.3–25.9) | 18.9 (16.2–23.9) | 0.070 |
| CRP (mg/dL) | 9.2 (3.7–17.9) | 1.4 (0.19–4.2) | 0.00030 * |
| Lactate dehydrogenase (IU/L) | 381.0 (229.8–525.0) | 213.0 (174.5–277.5) | 0.0017 * |
| Ferritin (ng/mL) | 1243.3 (627.1–2095.0) | 294.9 (102.5–635.6) | 0.00020 * |
| Interleukin-6 (pg/mL) | 315.5 (215.8–1027.0) | 213.0 (183.0–285.0) | 0.052 |
| Interleukin-18 (pg/mL) | 5.0 (3.6–19.1) | 6.6 (4.1–9.4) | 0.67 |
| KL-6 (IU/mL) | 827.5 (541.0–971.5) | 539.5 (446.8–695.5) | 0.094 |
| HMGB-1 (ng/mL) | 7.0 (4.7–11.2) | 6.3 (4.0–9.5) | 0.67 |
| Soluble CD163 (ng/mL) | 717.0 (465.0–886.0) | 541.0 (435.0–696.0) | 0.19 |
| d-dimer (µg/mL) | 1.9 (0.60–3.9) | 0.40 (0.20–0.95) | 0.00070 * |
| Data at day 3 | |||
| Neutrophil counts (/µL) | 6532.0 (3443.0–9246.2) | 2953.0 (2044.5–4524.6) | 0.012 * |
| Lymphocyte counts (/µL) | 568.3 (440.6–885.5) | 1344.0 (983.5–1866.4) | 0.00070 * |
| Platelet counts (×104/µL) | 15.3 (13.3–21.8) | 20.8 (16.9–26.9) | 0.033 * |
| CRP (mg/dL) | 6.0 (2.0–25.2) | 1.2 (0.19–4.0) | 0.0031 * |
| Lactate dehydrogenase (IU/L) | 326.5 (292.8–446.3) | 207.0 (165.8–284.3) | 0.0020 * |
| Ferritin (ng/mL) | 1104.4 (640.7–2199.9) | 420.5 (126.1–865.9) | 0.0022 * |
| Interleukin-6 (pg/mL) | 56.4 (27.9–164.0) | 6.7 (2.3–18.6) | 0.0011 * |
| HMGB-1 (ng/mL) | 16.4 (14.8–32.9) | 7.9 (5.0–11.5) | 0.0037 * |
| Soluble CD163 (ng/mL) | 766.0 (502.5–1000.0) | 610.5 (459.3–825.8) | 0.32 |
| d-dimer (µg/mL) | 4.8 (0.55–60.1) | 0.50 (0.30–1.0) | 0.0071 * |
| A. Analysis of biomarkers at admission | |||
| RR | 95%CI | pValue | |
| Neutrophil (µL) | 54.7 | 3.4–489.5 | 0.0077 * |
| Lymphocyte (µL) | 0.022 | 0.00013–1.6 | 0.086 |
| Platelet (×104/µL) | 0.14 | 0.0014–7.5 | 0.35 |
| CRP (mg/dL) | 51.4 | 8.7–318.4 | <0.00010 * |
| Lactate dehydrogenase (IU/L) | 188.5 | 14.6–2152.1 | 0.00020 * |
| Ferritin (ng/mL) | 108.7 | 10.3–845.6 | 0.00070 * |
| Interleukin-6 (pg/mL) | 26.6 | 3.2–139.2 | 0.0053 * |
| Interleukin-18 (pg/mL) | 7.2 | 0.057–106.5 | 0.35 |
| KL-6 (IU/mL) | 89.4 | 8.6–654.1 | 0.0012 * |
| HMGB-1 (ng/mL) | 1.9 | 0.0015–55.9 | 0.81 |
| Soluble CD163 (ng/mL) | 10.8 | 0.34–340.9 | 0.17 |
| d-dimer (µg/mL) | 91.6 | 9.5–697.5 | 0.00070 * |
| B. Analysis of biomarkers at 3 days after admission | |||
| RR | 95%CI | pValue | |
| Neutrophil (µL) | 41.1 | 2.9–344.9 | 0.0090 * |
| Lymphocyte (µL) | 0.038 | 0.00057–0.0018 § | 0.33 |
| Platelet (×104/µL) | 0.013 | 0.00012–0.87 | 0.043 * |
| CRP (mg/dL) | 53.2 | 7.6–338.6 | 0.00020 * |
| Lactate dehydrogenase (IU/L) | 25.8 | 2.5–180.0 | 0.0087 * |
| Ferritin (ng/mL) | 20.5 | 2.0–117.0 | 0.015 * |
| Interleukin-6 (pg/mL) | 45.1 | 4.7–293.2 | 0.0031 * |
| HMGB-1 (ng/mL) | 2219.8 | 33.0–600,284.4 | 0.00050 * |
| Soluble CD163 (ng/mL) | 3.7 | 0.29–54.6 | 0.31 |
| d-dimer (µg/mL) | 123.3 | 10.7–1659.5 | 0.00070 * |
| A. Analysis of biomarkers at admission | |||
| RR | 95%CI | pValue | |
| CRP (mg/dL) | 33.1 | 3.2–372.6 | 0.0028 * |
| Interleukin-6 (pg/mL) | 14.5 | 0.67–142.7 | 0.041 * |
| KL-6 (IU/mL) | 64.7 | 3.6–804.4 | 0.0013 * |
| B. Analysis of biomarkers at 3 days after admission | |||
| RR | 95%CI | pValue | |
| Platelet (×104/µL) | −0.37 § | −0.72–−0.14 § | 0.00060 * |
| CRP (mg/dL) | −0.33 § | −0.96–−0.044 § | 0.021 * |
| HMGB-1 (ng/mL) | 0.22 § | 0.054–0.46 § | 0.011 * |
| d-dimer (µg/mL) | 0.93 § | 0.083–2.1 § | 0.00037 * |
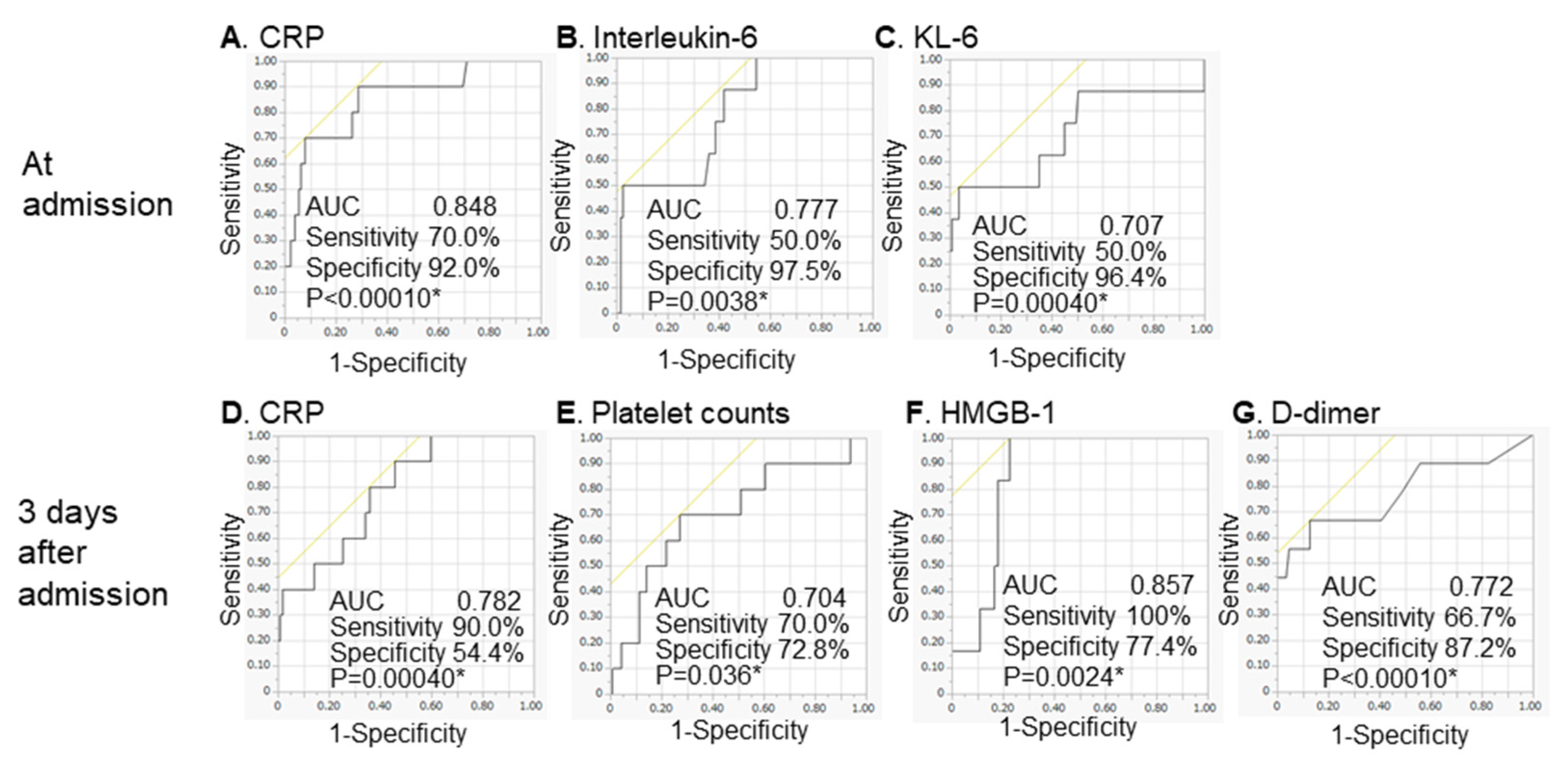
| A. Analysis of biomarkers at admission | |||||
| Cut-Off | AUC | Sensitivity | Specificity | pValue | |
| Neutrophil (/µL) | 4672.6 | 0.727 | 80.0% | 69.6% | 0.0089 * |
| Lymphocyte (/µL) | 1333.5 | 0.665 | 80.0% | 54,4% | 0.083 |
| Platelet (×104/µL) | 14.9 | 0.672 | 70.0% | 84.8% | 0.33 |
| CRP (mg/dL) | 8.0 | 0.848 | 70.0% | 92.0% | <0.0001 * |
| Lactate dehydrogenase (IU/L) | 372.0 | 0.798 | 60.0% | 89.6% | 0.0003 * |
| Ferritin (ng/mL) | 706.5 | 0.855 | 80.0% | 78.3% | 0.0004 * |
| Interleukin-6 (pg/mL) | 133.0 | 0.777 | 50.0% | 97.5% | 0.0038 * |
| Interleukin-18 (pg/mL) | 281.0 | 0.655 | 85.7% | 50.0% | 0.35 |
| KL-6 (IU/mL) | 382.0 | 0.707 | 50.0% | 96.4% | 0.0004 * |
| HMGB-1 (ng/mL) | 7.0 | 0.547 | 57.1% | 39.8% | 0.82 |
| Soluble CD163 (ng/mL) | 675.0 | 0.648 | 71.4% | 72.8% | 0.19 |
| d-dimer (µg/mL) | 1.2 | 0.789 | 70.0% | 81.3% | 0.0006 * |
| B. Analysis of biomarkers at 3 days after admission | |||||
| Cut-Off | AUC | Sensitivity | Specificity | pValue | |
| Neutrophil (µL) | 5765.5 | 0.752 | 66.7% | 83.0% | 0.011 * |
| Lymphocyte (µL) | 790.0 | 0.826 | 80.0% | 85.8% | 0.35 |
| Platelet (×104/µL) | 17.2 | 0.704 | 70.0% | 72.8% | 0.036 * |
| CRP (mg/dL) | 1.4 | 0.782 | 90.0% | 54.4% | 0.0004 * |
| Lactate dehydrogenase (IU/L) | 308.0 | 0.795 | 80.0% | 79.2% | 0.0078 * |
| Ferritin (ng/mL) | 434.2 | 0.798 | 100.0% | 50.6% | 0.0092 * |
| Interleukin-6 (pg/mL) | 33.8 | 0.899 | 83.3% | 91.9% | 0.0004 * |
| HMGB-1 (ng/mL) | 13.4 | 0.857 | 100.0% | 77.4% | 0.0024 * |
| Soluble CD163 (ng/mL) | 712.0 | 0.621 | 66.7% | 66.7% | 0.32 |
| d-dimer (µg/mL) | 1.9 | 0.772 | 66.7% | 87.2% | <0.0001 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, H.; Okamoto, M.; Nagasaki, Y.; Yoshio, S.; Nouno, T.; Yano, C.; Tanaka, T.; Watanabe, F.; Shibata, N.; Arimizu, Y.; et al. Analysis of Early Biomarkers Associated with the Development of Critical Respiratory Failure in Coronavirus Disease 2019 (COVID-19). Diagnostics 2022, 12, 339. https://doi.org/10.3390/diagnostics12020339
Yamada H, Okamoto M, Nagasaki Y, Yoshio S, Nouno T, Yano C, Tanaka T, Watanabe F, Shibata N, Arimizu Y, et al. Analysis of Early Biomarkers Associated with the Development of Critical Respiratory Failure in Coronavirus Disease 2019 (COVID-19). Diagnostics. 2022; 12(2):339. https://doi.org/10.3390/diagnostics12020339
Chicago/Turabian StyleYamada, Hiroyoshi, Masaki Okamoto, Yoji Nagasaki, Suzuyo Yoshio, Takashi Nouno, Chiyo Yano, Tomohiro Tanaka, Fumi Watanabe, Natsuko Shibata, Yoko Arimizu, and et al. 2022. "Analysis of Early Biomarkers Associated with the Development of Critical Respiratory Failure in Coronavirus Disease 2019 (COVID-19)" Diagnostics 12, no. 2: 339. https://doi.org/10.3390/diagnostics12020339
APA StyleYamada, H., Okamoto, M., Nagasaki, Y., Yoshio, S., Nouno, T., Yano, C., Tanaka, T., Watanabe, F., Shibata, N., Arimizu, Y., Fukamachi, Y., Zaizen, Y., Hamada, N., Kawaguchi, A., Hoshino, T., & Morita, S. (2022). Analysis of Early Biomarkers Associated with the Development of Critical Respiratory Failure in Coronavirus Disease 2019 (COVID-19). Diagnostics, 12(2), 339. https://doi.org/10.3390/diagnostics12020339






