Automatic Identification of Bioprostheses on X-ray Angiographic Sequences of Transcatheter Aortic Valve Implantation Procedures Using Deep Learning
Abstract
:1. Introduction
2. Background
2.1. Angiographic Imaging in TAVI
2.2. U-Net
2.3. Evaluation Metrics
3. Materials and Methods
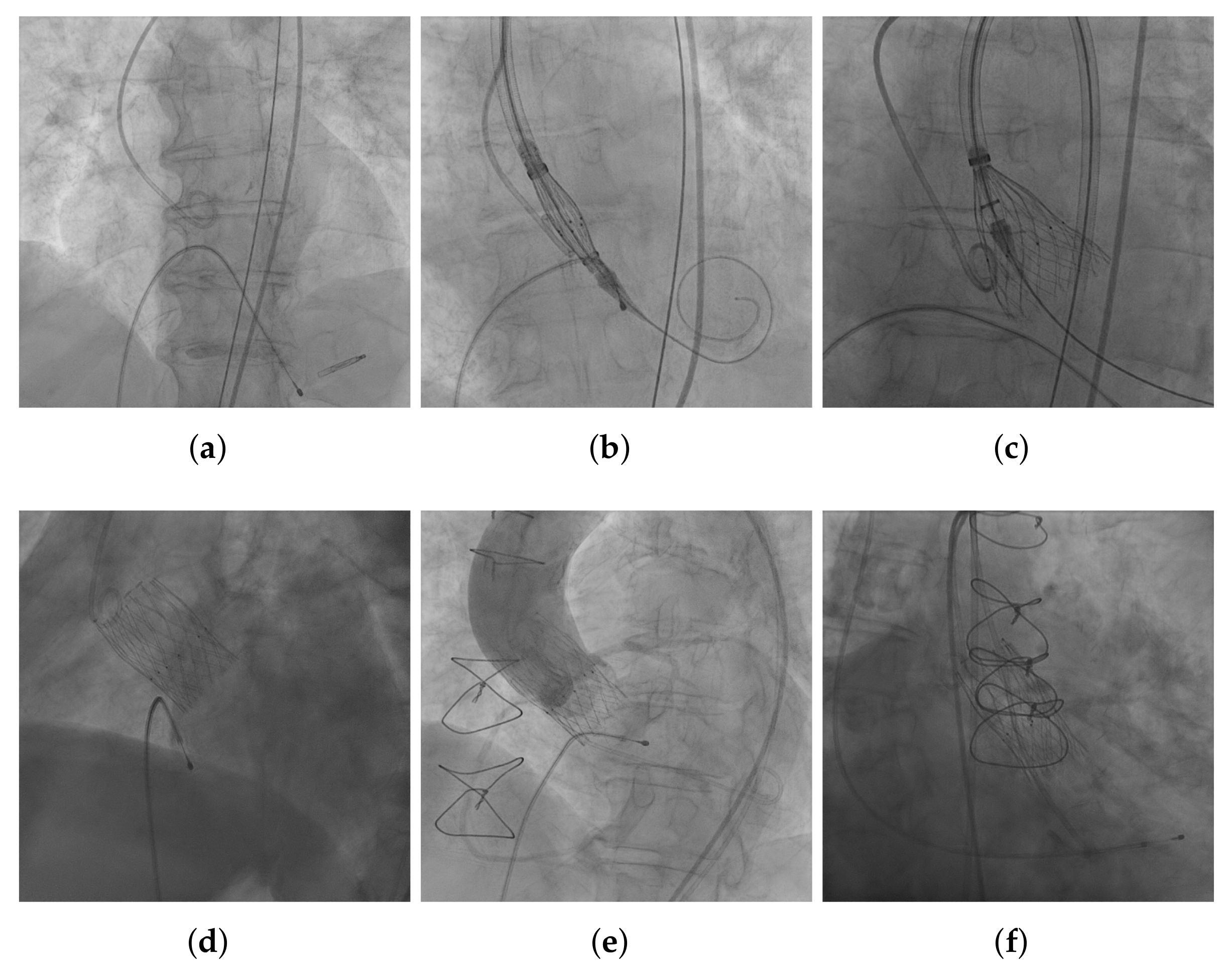
3.1. Image Acquisition and Annotation
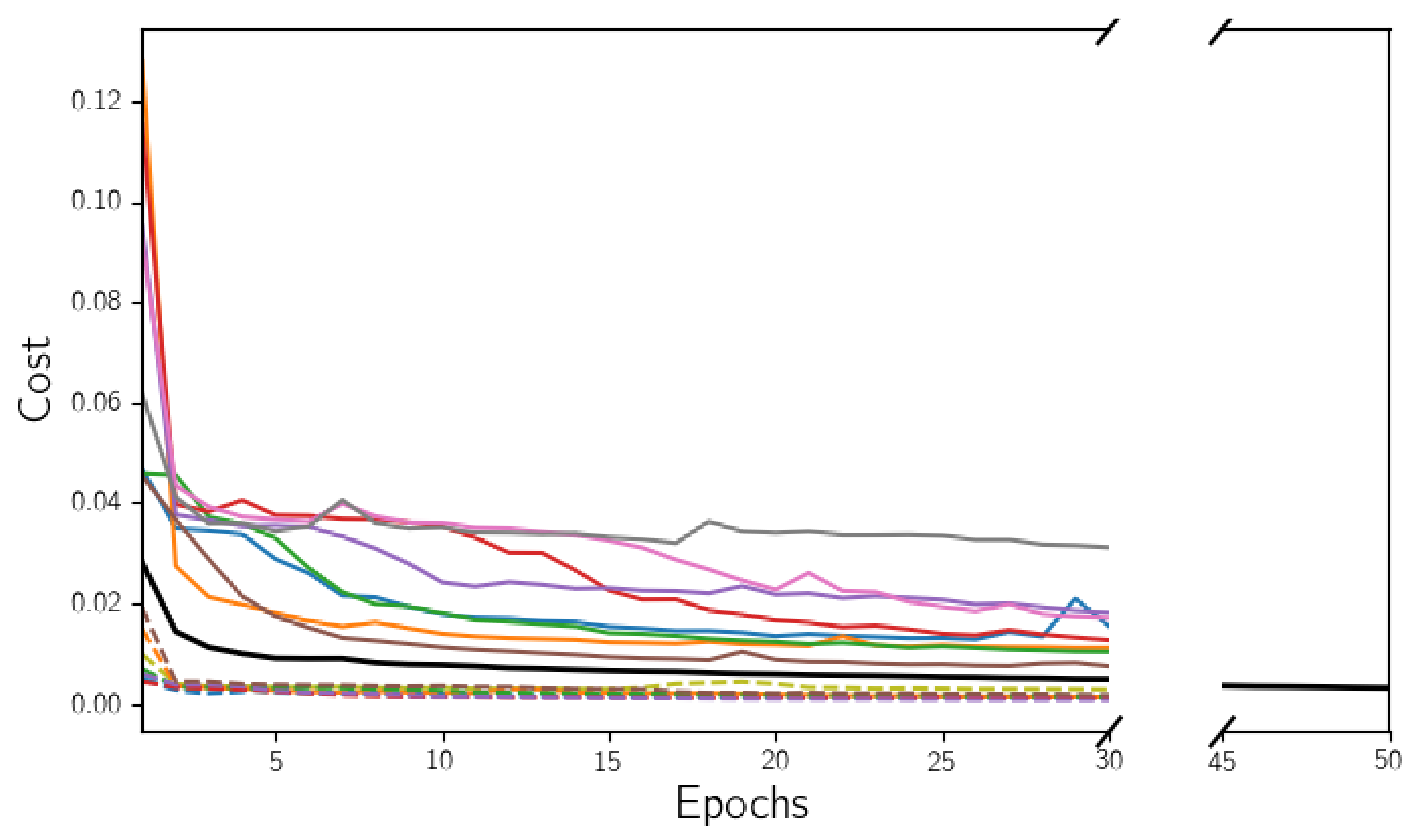
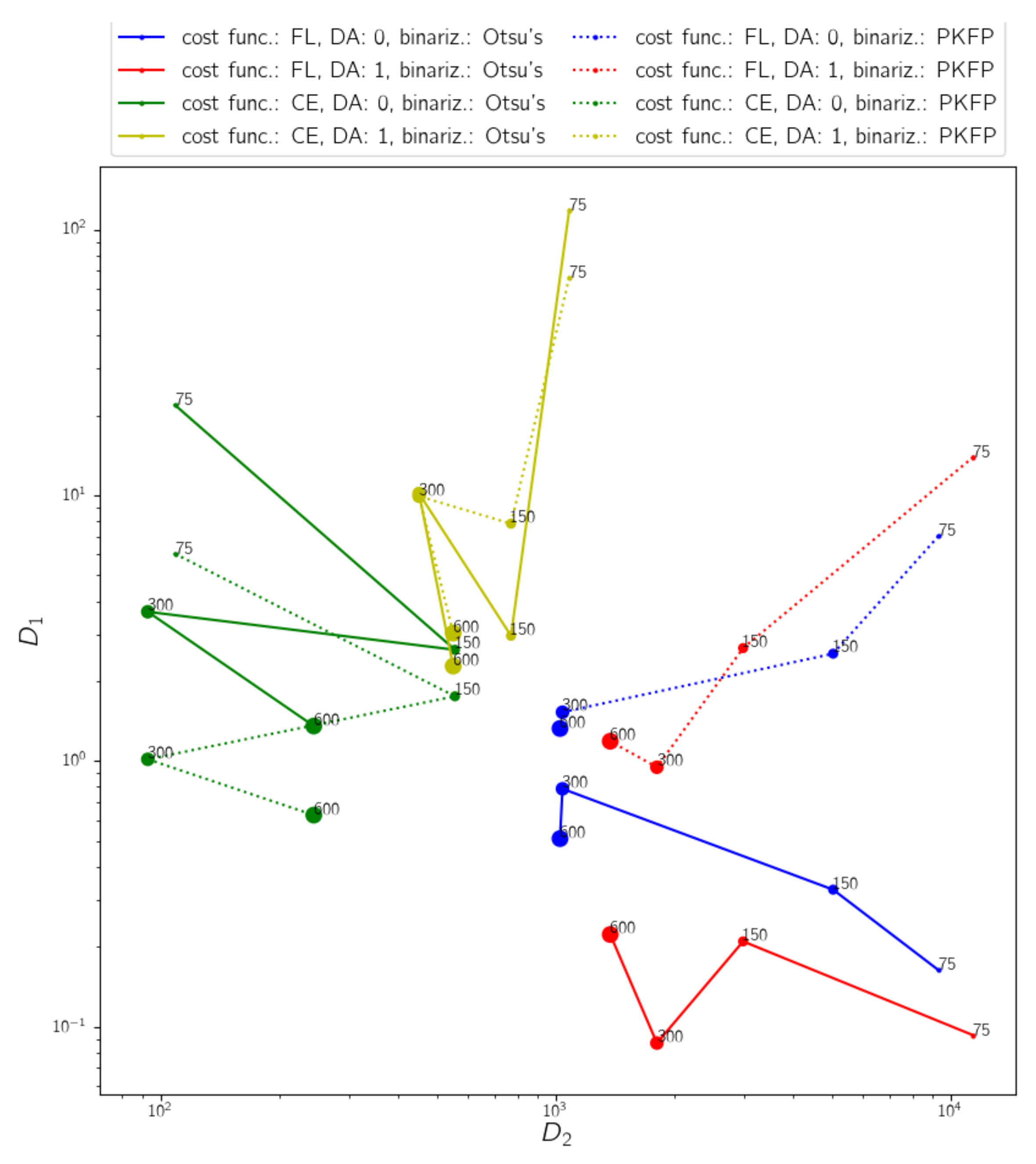
3.2. Hyperparameter Tuning
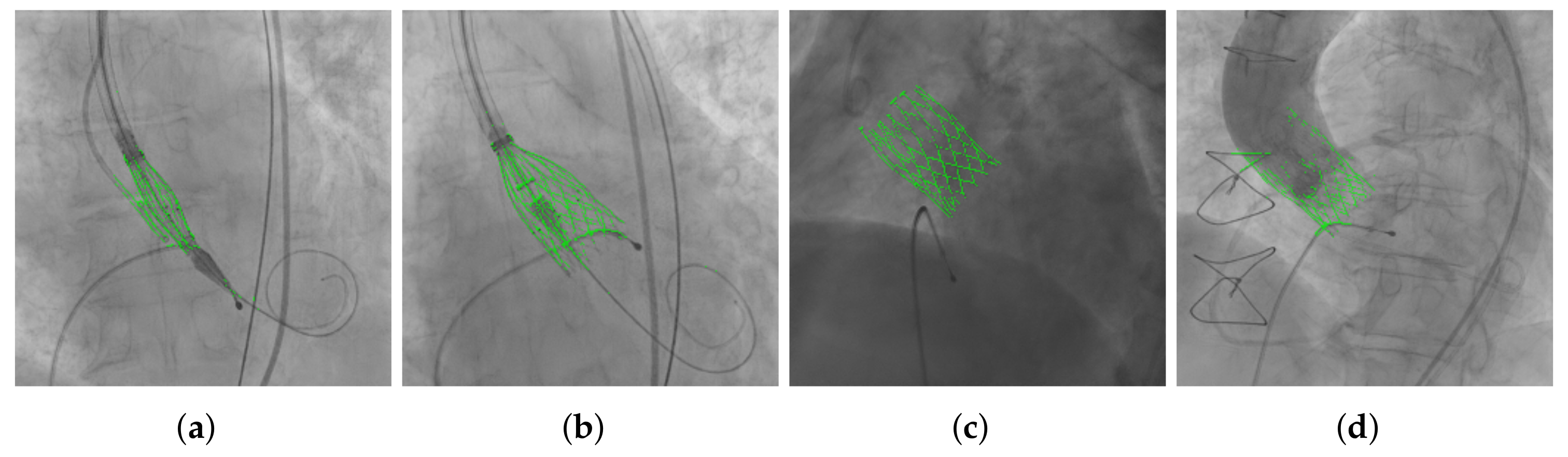
3.3. THV Segmentation
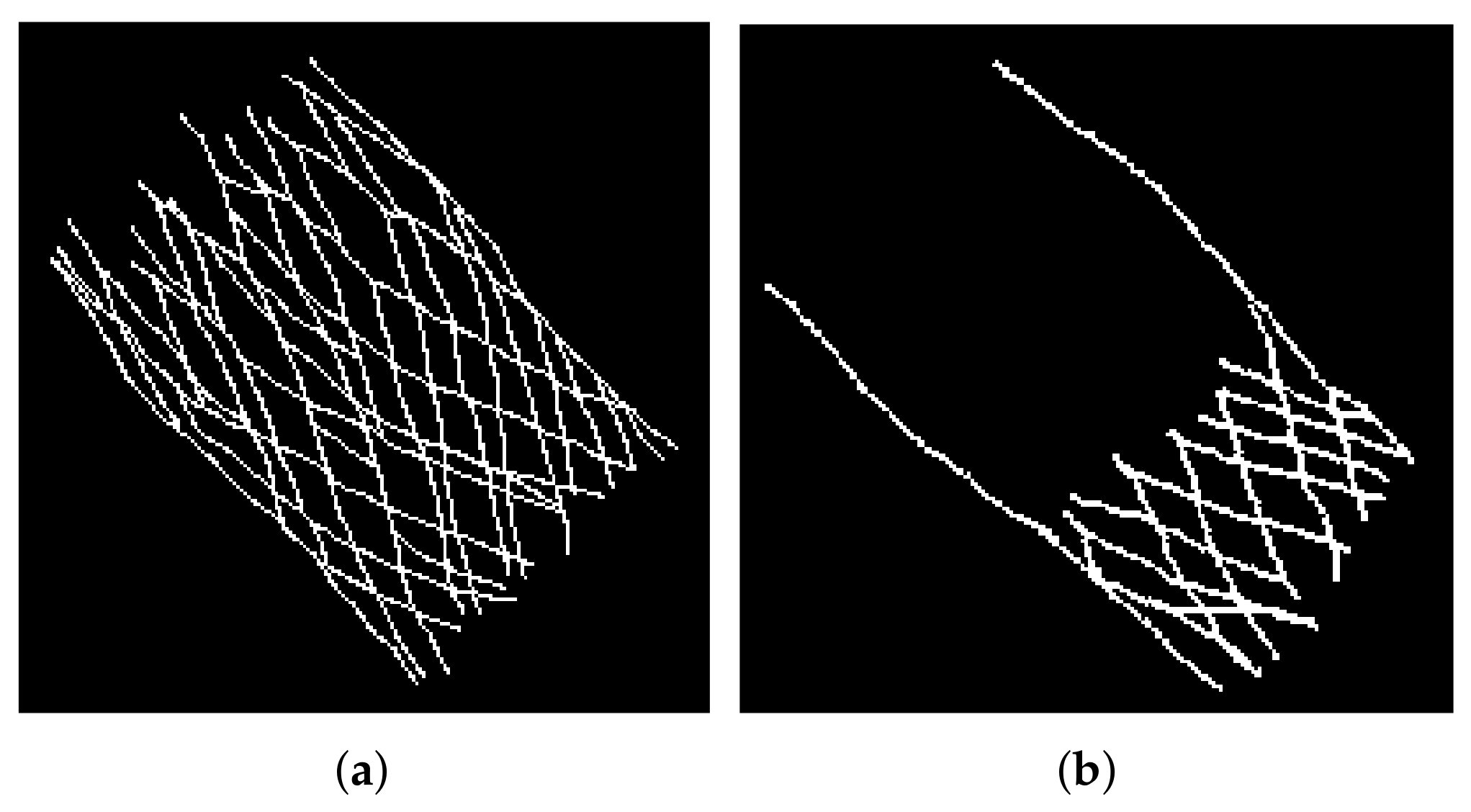
3.4. Evaluation of the Segmentation Results
4. Experiments and Results
4.1. Hyperparameter Tuning for the Model Selection
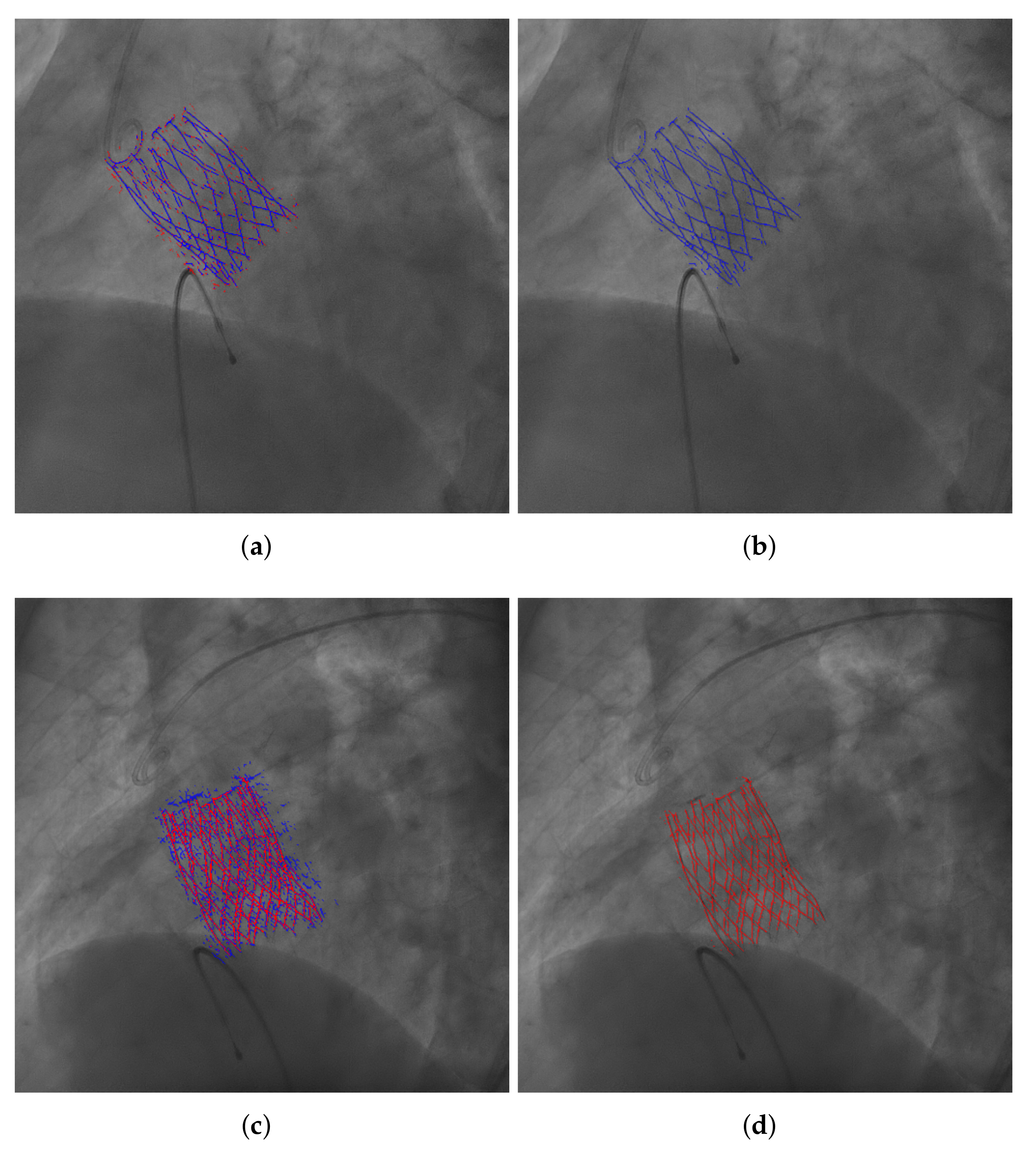
4.2. Evaluation of the Results of the Selected Model
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, D.R.; Mack, M.J.; Kaul, S.; Agnihotri, A.; Alexander, K.P.; Bailey, S.R.; Calhoon, J.H.; Carabello, B.A.; Desai, M.Y.; Edwards, F.H.; et al. 2012 ACCF/AATS/SCAI/STS Expert Consensus Document on Transcatheter Aortic Valve Replacement. J. Am. Coll. Cardiol. 2012, 59, 1200–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leon, M.B.; Smith, C.R.; Mack, M.J.; Makkar, R.R.; Svensson, L.G.; Kodali, S.K.; Thourani, V.H.; Tuzcu, E.M.; Miller, D.C.; Herrmann, H.C.; et al. Transcatheter or surgical aortic-valve replacement in intermediate-risk patients. N. Engl. J. Med. 2016, 374, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, G.S.; Gillam, L.D.; Hahn, R.T.; Kapadia, S.; Leipsic, J.; Lerakis, S.; Tuzcu, M.; Douglas, P.S. A practical guide to multimodality imaging of transcatheter aortic valve replacement. JACC Cardiovasc. Imaging 2012, 5, 441–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanke, P.; Weir-McCall, J.R.; Achenbach, S.; Delgado, V.; Hausleiter, J.; Jilaihawi, H.; Marwan, M.; Nørgaard, B.L.; Piazza, N.; Schoenhagen, P.; et al. Computed tomography imaging in the context of transcatheter aortic valve implantation (TAVI)/transcatheter aortic valve replacement (TAVR) an expert consensus document of the Society of Cardiovascular Computed Tomography. JACC Cardiovasc. Imaging 2019, 12, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kurra, V.; Kapadia, S.R.; Tuzcu, E.M.; Halliburton, S.S.; Svensson, L.; Roselli, E.E.; Schoenhagen, P. Pre-Procedural Imaging of Aortic Root Orientation and Dimensions: Comparison Between X-Ray Angiographic Planar Imaging and 3-Dimensional Multidetector Row Computed Tomography. JACC Cardiovasc. Interv. 2010, 3, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Binder, R.; Leipsic, J.; Wood, D.; Moore, T.; Toggweiler, S.; Willson, A.; Gurvitch, R.; Freeman, M.; Webb, J. Prediction of Optimal Deployment Projection for Transcatheter Aortic Valve Replacement Angiographic 3-Dimensional Reconstruction of the Aortic Root Versus Multidetector Computed Tomography. Circ. Cardiovasc. Interv. 2012, 5, 247–252. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.; Sun, W. Transcatheter valve underexpansion limits leaflet durability: Implications for valve-in-valve procedures. Ann. Biomed. Eng. 2017, 45, 394–404. [Google Scholar] [CrossRef] [Green Version]
- Wagner, M.G.; Hatt, C.R.; Dunkerley, D.A.P.; Bodart, L.E.; Raval, A.N.; Speidel, M.A. A dynamic model-based approach to motion and deformation tracking of prosthetic valves from biplane x-ray images. Med. Phys. 2018, 45, 2583–2594. [Google Scholar] [CrossRef]
- Arsalan, M.; Owais, M.; Mahmood, T.; Choi, J.; Park, K.R. Artificial intelligence-based diagnosis of cardiac and related diseases. J. Clin. Med. 2020, 9, 871. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, F.; Niu, Y. A Convolutional Neural Network Combining Discriminative Dictionary Learning and Sequence Tracking for Left Ventricular Detection. Sensors 2021, 21, 3693. [Google Scholar] [CrossRef]
- Galea, R.R.; Diosan, L.; Andreica, A.; Popa, L.; Manole, S.; Bálint, Z. Region-of-Interest-Based Cardiac Image Segmentation with Deep Learning. Appl. Sci. 2021, 11, 1965. [Google Scholar] [CrossRef]
- Dey, D.; Slomka, P.J.; Leeson, P.; Comaniciu, D.; Shrestha, S.; Sengupta, P.P.; Marwick, T.H. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019, 73, 1317–1335. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Kamnitsas, K.; Glocker, B. Analyzing Overfitting Under Class Imbalance in Neural Networks for Image Segmentation. IEEE Trans. Med. Imaging 2021, 40, 1065–1077. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Lin, T.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar] [CrossRef] [Green Version]
- Moccia, S.; De Momi, E.; El Hadji, S.; Mattos, L.S. Blood vessel segmentation algorithms—Review of methods, datasets and evaluation metrics. Comput. Methods Programs Biomed. 2018, 158, 71–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLong, E.; DeLong, D.; Clarke-Pearson, D. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Jaccard, P. The distribution of the flora in the alpine zone. 1. New Phytol. 1912, 11, 37–50. [Google Scholar] [CrossRef]
- Rockafellar, R.T.; Wets, R.J.B. Variational Analysis; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; Volume 317. [Google Scholar]
- Chollet, F. Keras. Available online: https://keras.io (accessed on 22 December 2021).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: tensorflow.org (accessed on 22 December 2021).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Luque, A.; Carrasco, A.; Martín, A.; de las Heras, A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recognit. 2019, 91, 216–231. [Google Scholar] [CrossRef]
| Training Set Size | Data Aug. () | Loss Function | Best Epoch | Best Loss Value | (Otsu’s Binarization) | (PKFP Binarization) | |
|---|---|---|---|---|---|---|---|
| 75 | 0 | CE | 30 | 0.0171 | 21.87 ± 26.43 | 6.03 ± 3.79 | 108.79 ± 81.54 |
| FL | 6 | 0.0028 | 0.16 ± 0.55 | 7.02 ± 3.75 | 9314.98 ± 3149.51 | ||
| 1 | CE | 30 | 0.0312 | 118.38 ± 218.96 | 65.96 ± 56.89 | 1082.49 ± 666.88 | |
| FL | 30 | 0.0028 | 0.093 ± 0.40 | 13.95 ± 8.07 | 11415.52 ± 3787.17 | ||
| 150 | 0 | CE | 30 | 0.0128 | 2.62 ± 3.69 | 1.75 ± 2.11 | 556.16 ± 232.48 |
| FL | 30 | 0.0015 | 0.33 ± 0.56 | 2.54 ± 1.45 | 5041.36 ± 1334.57 | ||
| 1 | CE | 30 | 0.0182 | 2.96 ± 4.87 | 7.82 ± 8.99 | 771.07 ± 313.18 | |
| FL | 30 | 0.0016 | 0.21 ± 0.42 | 2.66 ± 1.73 | 2983.14 ± 1011.65 | ||
| 300 | 0 | CE | 30 | 0.0085 | 3.65 ± 3.15 | 1.02 ± 1.07 | 92.81 ± 62.13 |
| FL | 29 | 0.0010 | 0.79 ± 2.03 | 1.53 ± 2.49 | 1040.91 ± 220.49 | ||
| 1 | CE | 26 | 0.0129 | 10.16 ± 17.12 | 9.92 ± 13.54 | 451.25 ± 161.92 | |
| FL | 30 | 0.0012 | 0.09 ± 0.18 | 0.95 ± 0.97 | 1804.71 ± 353.82 | ||
| 600 | 0 | CE | 30 | 0.0075 | 1.36 ± 1.56 | 0.63 ± 0.73 | 244.23 ± 90.97 |
| FL | 30 | 0.0009 | 0.51 ± 3.40 | 1.33 ± 4.63 | 1025.85 ± 170.54 | ||
| 1 | CE | 29 | 0.0112 | 2.28 ± 4.82 | 3.03 ± 5.02 | 550.40 ± 176.58 | |
| FL | 30 | 0.0012 | 0.22 ± 0.37 | 1.19 ± 1.14 | 1374.78 ± 233.04 |
| Metric | Bin. Method | Exp. 1 (460 Frames) | Exp. 2 (65 Frames) | Exp. 3 (3 Frames) | Exp. 4 (6 Frames) |
|---|---|---|---|---|---|
| Otsu’s | 0.91 ± 0.86 | – | 67.95 ± 75.11 | 10.21 ± 8.73 | |
| PKFP | 0.30 ± 0.38 | – | 52.94 ± 62.72 | 3.92 ± 4.16 | |
| – | 138.48 ± 75.76 | 2.40 ± 9.21 | 204.45 ± 19.83 | 369.83 ± 48.50 | |
| Accuracy | Otsu’s | 0.99 ± 0.01 | 0.99 ± 0.01 | 0.99 ± 0.01 | 0.99 ± 0.01 |
| PKFP | 0.99 ± 0.01 | 0.99 ± 0.01 | 0.99 ± 0.01 | 0.99 ± 0.01 | |
| Precision | Otsu’s | 0.73 ± 0.05 | 0 | 0.51 ± 0.06 | 0.46 ± 0.06 |
| PKFP | 0.63 ± 0.07 | 0 | 0.43 ± 0.06 | 0.34 ± 0.04 | |
| Recall | Otsu’s | 0.87 ± 0.04 | 0 | 0.70 ± 0.09 | 0.75 ± 0.04 |
| PKFP | 0.97 ± 0.02 | 0 | 0.80 ± 0.09 | 0.93 ± 0.03 | |
| Dice Coefficient | Otsu’s | 0.15 ± 0.03 | 0 | 0.29 ± 0.05 | 0.33 ± 0.04 |
| PKFP | 0.22 ± 0.06 | 0 | 0.36 ± 0.05 | 0.47 ± 0.06 | |
| Jaccard Index | Otsu’s | 0.74 ± 0.04 | 0 | 0.55 ± 0.06 | 0.50 ± 0.05 |
| PKFP | 0.64 ± 0.07 | 0 | 0.47 ± 0.06 | 0.36 ± 0.03 | |
| AUROC | Otsu’s | 0.94 ± 0.02 | – | 0.85 ± 0.05 | 0.89 ± 0.02 |
| PKFP | 0.98 ± 0.01 | – | 0.90 ± 0.04 | 0.96 ± 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busto, L.; Veiga, C.; González-Nóvoa, J.A.; Loureiro-Ga, M.; Jiménez, V.; Baz, J.A.; Íñiguez, A. Automatic Identification of Bioprostheses on X-ray Angiographic Sequences of Transcatheter Aortic Valve Implantation Procedures Using Deep Learning. Diagnostics 2022, 12, 334. https://doi.org/10.3390/diagnostics12020334
Busto L, Veiga C, González-Nóvoa JA, Loureiro-Ga M, Jiménez V, Baz JA, Íñiguez A. Automatic Identification of Bioprostheses on X-ray Angiographic Sequences of Transcatheter Aortic Valve Implantation Procedures Using Deep Learning. Diagnostics. 2022; 12(2):334. https://doi.org/10.3390/diagnostics12020334
Chicago/Turabian StyleBusto, Laura, César Veiga, José A. González-Nóvoa, Marcos Loureiro-Ga, Víctor Jiménez, José Antonio Baz, and Andrés Íñiguez. 2022. "Automatic Identification of Bioprostheses on X-ray Angiographic Sequences of Transcatheter Aortic Valve Implantation Procedures Using Deep Learning" Diagnostics 12, no. 2: 334. https://doi.org/10.3390/diagnostics12020334
APA StyleBusto, L., Veiga, C., González-Nóvoa, J. A., Loureiro-Ga, M., Jiménez, V., Baz, J. A., & Íñiguez, A. (2022). Automatic Identification of Bioprostheses on X-ray Angiographic Sequences of Transcatheter Aortic Valve Implantation Procedures Using Deep Learning. Diagnostics, 12(2), 334. https://doi.org/10.3390/diagnostics12020334






