Automated Classification of Idiopathic Pulmonary Fibrosis in Pathological Images Using Convolutional Neural Network and Generative Adversarial Networks
Abstract
:1. Introduction
1.1. Background
1.2. Related Works
1.3. Objective and Contributions
- Image generation using GANs is introduced for use as input data for classification. By using GAN with progressive growing mechanism, high-resolution images are generated in a stable manner. Generated images can improve the classification accuracy of IIPs.
- The CNN model used for classification is trained in two steps: a rough pretraining using generated pathological images, and fine tuning using real images to obtain high accuracy.
2. Method
2.1. Outline
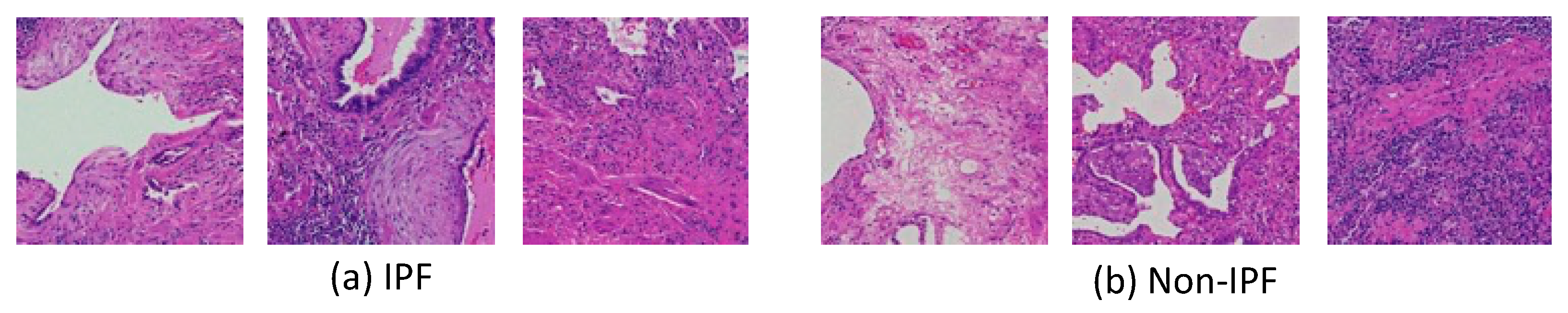
2.2. Image Dataset
2.3. Image Preprocessing
2.4. Data Augmentation by Generative Adversarial Networks
2.5. Two-Step Image Classification
2.6. Visualization
2.7. Evaluation Metrics
3. Results
3.1. Synthesized Patch Images Using PGGAN
3.2. Image Classification Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Travis, W.D.; Costabel, U.; Hansell, D.M.; King, T.E.; Lynch, D.A.; Nicholson, A.G.; Ryerson, C.J.; Ryu, J.H.; Selman, M.; Wells, A.U.; et al. An official American Thoracic Society/European Respiratory Society statement: Update of the international multidisciplinary classification of the idiopathic interstitial pneumonias. Am. J. Respir. Crit. Care Med. 2013, 188, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Hanna, M.G.; Parwani, A.; Sirintrapun, S.J. Whole slide imaging: Technology and applications. Adv. Anat. Pathol. 2020, 27, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Teramoto, A.; Fujita, H.; Yamamuro, O.; Tamaki, T. Automated detection of pulmonary nodules in PET/CT images: Ensemble false-positive reduction using a convolutional neural network technique. Med. Phys. 2016, 43, 2821–2827. [Google Scholar] [CrossRef]
- Teramoto, A.; Tsukamoto, T.; Kiriyama, Y.; Fujita, H. Automated classification of lung cancer types from cytological images using deep convolutional neural networks. BioMed Res. Int. 2017, 2017, 4067832. [Google Scholar] [CrossRef] [Green Version]
- Teramoto, A.; Yamada, A.; Kiriyama, Y.; Tsukamoto, T.; Yan, K.; Zhang, L.; Imaizumi, K.; Saito, K.; Fujita, H. Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network. Inform. Med. Unlocked 2019, 16, 100205. [Google Scholar] [CrossRef]
- Teramoto, A.; Kiriyama, Y.; Tsukamoto, T.; Sakurai, E.; Michiba, A.; Imaizumi, K.; Saito, K.; Fujita, H. Weakly supervised learning for classification of lung cytological images using attention-based multiple instance learning. Sci. Rep. 2021, 11, 20317. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Jiao, Z.; Choi, J.W.; Halsey, K.; Tran, T.M.L.; Hsieh, B.; Wang, D.; Eweje, F.; Wang, R.; Chang, K.; Wu, J.; et al. Prognostication of patients with COVID-19 using artificial intelligence based on chest X-rays and clinical data: A retrospective study. Lancet Digit. Healthy 2021, 3, e286–e294. [Google Scholar] [CrossRef]
- Shi, X.; Wang, L.; Li, Y.; Wu, J.; Huang, H. GCLDNet: Gastric cancer lesion detection network combining level feature aggregation and attention feature fusion. Front. Oncol. 2021, 12, 901475. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shi, X.; Yang, L.; Pu, C.; Tan, Q.; Yang, Z.; Huang, H. MC-GAT: Multi-layer collaborative generative adversarial transformer for cholangiocarcinoma classification from hyperspectral pathological images. Biomed. Opt. Express 2022, 13, 5794–5812. [Google Scholar] [CrossRef]
- Takeuchi, N.; Teramoto, A.; Imaizumi, K.; Saito, K.; Fujita, H. Analysis of idiopathic interstitial pneumonia in CT images using 3D U-net. Med. Image Inf. Sci. 2021, 38, 26–131. [Google Scholar] [CrossRef]
- Uegami, W.; Bychkov, A.; Ozasa, M.; Uehara, K.; Kataoka, K.; Johkoh, T.; Kondoh, Y.; Sakanashi, H.; Fukuoka, J. MIXTURE of human expertise and deep learning—Developing an explainable model for predicting pathological diagnosis and survival in patients with interstitial lung disease. Mod. Pathol. 2022, 35, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Toda, R.; Teramoto, A.; Tsujimoto, M.; Toyama, H.; Imaizumi, K.; Saito, K.; Fujita, H. Synthetic CT image generation of shape-controlled lung cancer using semi-conditional InfoGAN and its applicability for type classification. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 241–251. [Google Scholar] [CrossRef]
- Teramoto, A.; Tsukamoto, T.; Yamada, A.; Kiriyama, Y.; Imaizumi, K.; Saito, K.; Fujita, H. Deep learning approach to classification of lung cytological images: Two-step training using actual and synthesized images by progressive growing of generative adversarial networks. PLoS ONE 2020, 15, e0229951. [Google Scholar] [CrossRef]
- Yoshida, M.; Teramoto, A.; Kudo, K.; Matsumoto, S.; Saito, K.; Fujita, H. Automated extraction of cerebral infarction region in Head MR image using pseudo cerebral infarction image by CycleGAN. Appl. Sci. 2022, 12, 489. [Google Scholar] [CrossRef]
- Onishi, Y.; Teramoto, A.; Tsujimoto, M.; Tsukamoto, T.; Saito, K.; Toyama, H.; Imaizumi, K.; Fujita, H. Multiplanar analysis for pulmonary nodule classification in CT images using deep convolutional neural network and generative adversarial networks. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 173–178. [Google Scholar] [CrossRef]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive growing of GANs for improved quality, stability, and variation. arXiv 2017, arXiv:1710.10196. [Google Scholar]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein GAN. arXiv 2017, arXiv:1701.07875. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2016, arXiv:1511.06434v2. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. IEEE Conf. Comput. Vis. Pattern Recognit. CVPR 2015, 2015, 1–9. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. IEEE Conf. Comput. Vis. Pattern Recognit. CVPR 2016, 2016, 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. IEEE Conf. Comput. Vis. Pattern Recognit. CVPR 2017, 2017, 2261–2269. [Google Scholar] [CrossRef] [Green Version]
- Efron, B. Estimating the error rate of a prediction rule: Improvement on cross-validation. J. Am. Stat. Assoc. 1983, 78, 316–331. [Google Scholar] [CrossRef]
- Walsh, S.L.; Maher, T.M.; Kolb, M.; Poletti, V.; Nusser, R.; Richeldi, L.; Vancheri, C.; Wilsher, M.L.; Antoniou, K.M.; Behr, J.; et al. Diagnostic accuracy of a clinical diagnosis of idiopathic pulmonary fibrosis: An international case–cohort study. Eur. Respir. J. 2017, 50, 1700936. [Google Scholar] [CrossRef]








| (a) Image-Based Classification | |||||
| CNN Model | Data Augmentation | Sensitivity | Specificity | Accuracy | AUC |
| VGG-16 | w/o GAN DA | 0.601 ± 0.062 | 0.547 ± 0.031 | 0.588 ± 0.050 | 0.615 ± 0.039 |
| w GAN DA | 0.582 ± 0.040 | 0.587 ± 0.047 | 0.583 ± 0.019 | 0.618 ± 0.009 | |
| VGG-19 | w/o GAN DA | 0.609 ± 0.027 | 0.592 ± 0.015 | 0.605 ± 0.021 | 0.641 ± 0.021 |
| w GAN DA | 0.607 ± 0.006 | 0.563 ± 0.014 | 0.596 ± 0.005 | 0.618 ± 0.007 | |
| InceptionV3 | w/o GAN DA | 0.625 ± 0.020 | 0.556 ± 0.023 | 0.608 ± 0.019 | 0.618 ± 0.032 |
| w GAN DA | 0.595 ± 0.011 | 0.527 ± 0.004 | 0.578 ± 0.008 | 0.592 ± 0.006 | |
| ResNet-50 | w/o GAN DA | 0.614 ± 0.021 | 0.578 ± 0.011 | 0.605 ± 0.014 | 0.619 ± 0.019 |
| w GAN DA | 0.540 ± 0.089 | 0.483 ± 0.090 | 0.526 ± 0.044 | 0.533 ± 0.009 | |
| DenseNet-121 | w/o GAN DA | 0.628 ± 0.018 | 0.568 ± 0.019 | 0.613 ± 0.009 | 0.628 ± 0.009 |
| w GAN DA | 0.683 ± 0.008 | 0.521 ± 0.016 | 0.642 ± 0.002 | 0.644 ± 0.006 | |
| DenseNet-169 | w/o GAN DA | 0.658 ± 0.019 | 0.554 ± 0.007 | 0.632 ± 0.013 | 0.639 ± 0.022 |
| w GAN DA | 0.691 ± 0.010 | 0.522 ± 0.015 | 0.649 ± 0.004 | 0.649 ± 0.004 | |
| DenseNet-201 | w/o GAN DA | 0.652 ± 0.036 | 0.557 ± 0.058 | 0.628 ± 0.013 | 0.632 ± 0.023 |
| w GAN DA | 0.663 ± 0.013 | 0.548 ± 0.012 | 0.634 ± 0.007 | 0.646 ± 0.006 | |
| (b) Case-Based Classification | |||||
| CNN Model | Data Augmentation | Sensitivity | Specificity | Accuracy | AUC |
| VGG-16 | w/o GAN DA | 0.806 ± 0.127 | 0.583 ± 0.083 | 0.694 ± 0.087 | 0.701 ± 0.064 |
| w GAN DA | 0.861 ± 0.048 | 0.611 ± 0.048 | 0.736 ± 0.024 | 0.811 ± 0.039 | |
| VGG-19 | w/o GAN DA | 0.806 ± 0.048 | 0.639 ± 0.048 | 0.722 ± 0.024 | 0.765 ± 0.022 |
| w GAN DA | 0.889 ± 0.048 | 0.639 ± 0.048 | 0.764 ± 0.024 | 0.843 ± 0.019 | |
| InceptionV3 | w/o GAN DA | 0.944 ± 0.048 | 0.583 ± 0.000 | 0.764 ± 0.024 | 0.757 ± 0.030 |
| w GAN DA | 0.806 ± 0.096 | 0.667 ± 0.000 | 0.736 ± 0.048 | 0.744 ± 0.040 | |
| ResNet-50 | w/o GAN DA | 0.889 ± 0.048 | 0.611 ± 0.048 | 0.750 ± 0.042 | 0.722 ± 0.047 |
| w GAN DA | 0.611 ± 0.048 | 0.444 ± 0.048 | 0.528 ± 0.024 | 0.522 ± 0.028 | |
| DenseNet-121 | w/o GAN DA | 0.861 ± 0.127 | 0.583 ± 0.000 | 0.722 ± 0.064 | 0.734 ± 0.033 |
| w GAN DA | 0.972 ± 0.048 | 0.694 ± 0.048 | 0.833 ± 0.000 | 0.843 ± 0.005 | |
| DenseNet-169 | w/o GAN DA | 0.972 ± 0.048 | 0.583 ± 0.000 | 0.778 ± 0.024 | 0.786 ± 0.013 |
| w GAN DA | 0.944 ± 0.048 | 0.639 ± 0.048 | 0.792 ± 0.042 | 0.826 ± 0.045 | |
| DenseNet-201 | w/o GAN DA | 0.833 ± 0.220 | 0.583 ± 0.000 | 0.708 ± 0.110 | 0.726 ± 0.074 |
| w GAN DA | 0.833 ± 0.083 | 0.667 ± 0.083 | 0.750 ± 0.083 | 0.809 ± 0.024 | |
| (a) Classification without PGGAN (DenseNet-169) | |||
| Predicted Class | |||
| Non-IPF | IPF | ||
| Actual class | Non-IPF | 7 | 5 |
| IPF | 0 | 12 | |
| (b) Classification Using PGGAN (DenseNet-121) | |||
| Predicted Class | |||
| Non-IPF | IPF | ||
| Actual class | Non-IPF | 9 | 3 |
| IPF | 1 | 11 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teramoto, A.; Tsukamoto, T.; Michiba, A.; Kiriyama, Y.; Sakurai, E.; Imaizumi, K.; Saito, K.; Fujita, H. Automated Classification of Idiopathic Pulmonary Fibrosis in Pathological Images Using Convolutional Neural Network and Generative Adversarial Networks. Diagnostics 2022, 12, 3195. https://doi.org/10.3390/diagnostics12123195
Teramoto A, Tsukamoto T, Michiba A, Kiriyama Y, Sakurai E, Imaizumi K, Saito K, Fujita H. Automated Classification of Idiopathic Pulmonary Fibrosis in Pathological Images Using Convolutional Neural Network and Generative Adversarial Networks. Diagnostics. 2022; 12(12):3195. https://doi.org/10.3390/diagnostics12123195
Chicago/Turabian StyleTeramoto, Atsushi, Tetsuya Tsukamoto, Ayano Michiba, Yuka Kiriyama, Eiko Sakurai, Kazuyoshi Imaizumi, Kuniaki Saito, and Hiroshi Fujita. 2022. "Automated Classification of Idiopathic Pulmonary Fibrosis in Pathological Images Using Convolutional Neural Network and Generative Adversarial Networks" Diagnostics 12, no. 12: 3195. https://doi.org/10.3390/diagnostics12123195
APA StyleTeramoto, A., Tsukamoto, T., Michiba, A., Kiriyama, Y., Sakurai, E., Imaizumi, K., Saito, K., & Fujita, H. (2022). Automated Classification of Idiopathic Pulmonary Fibrosis in Pathological Images Using Convolutional Neural Network and Generative Adversarial Networks. Diagnostics, 12(12), 3195. https://doi.org/10.3390/diagnostics12123195








