An Ensemble of Transfer Learning Models for the Prediction of Skin Cancers with Conditional Generative Adversarial Networks
Abstract
1. Introduction
- Exploration of image augmentation methods, such as flip, affine, linear contrast, multiply, and Gaussian blur (image transformation methods) to balance the dataset.
- Exploration of the Conditional GAN architecture for generating skin cancer images.
- Performance analysis of the fine-tuned pre-trained models VGG16, ResNet50, and ResNet101 on both balanced and unbalanced datasets.
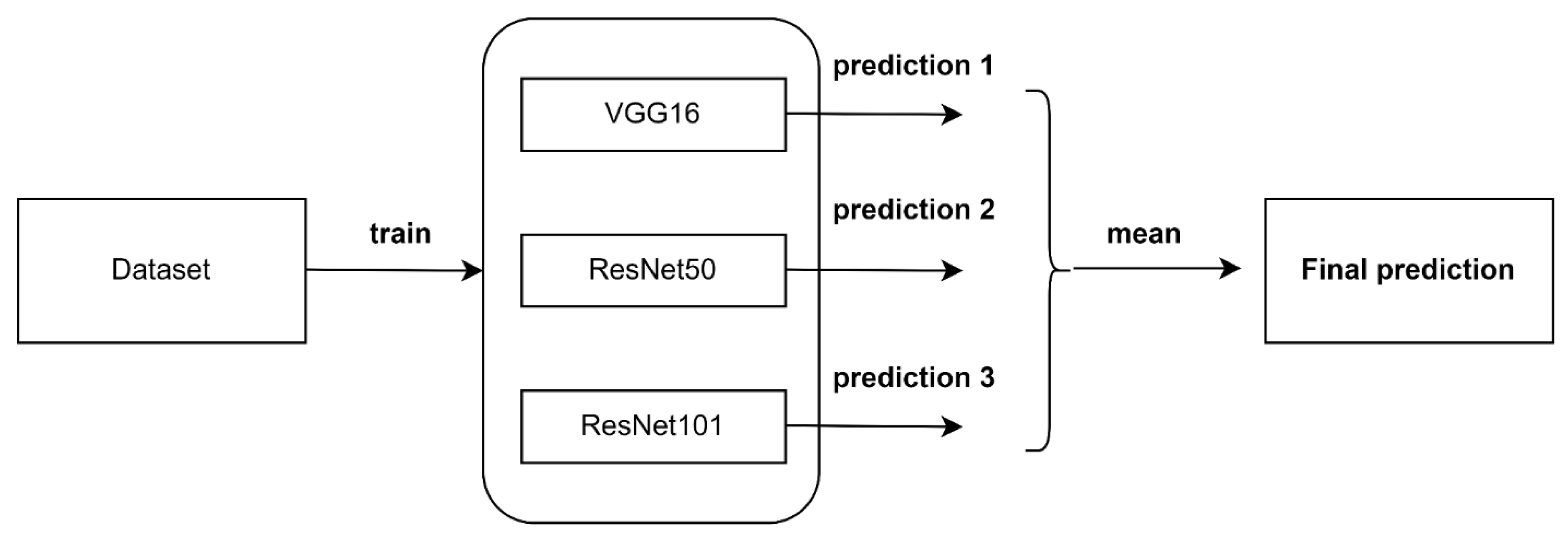
- An ensemble algorithm by combining the predictions of the three fine-tuned models to improve the performance obtained by deep individual models.
2. Literature Review
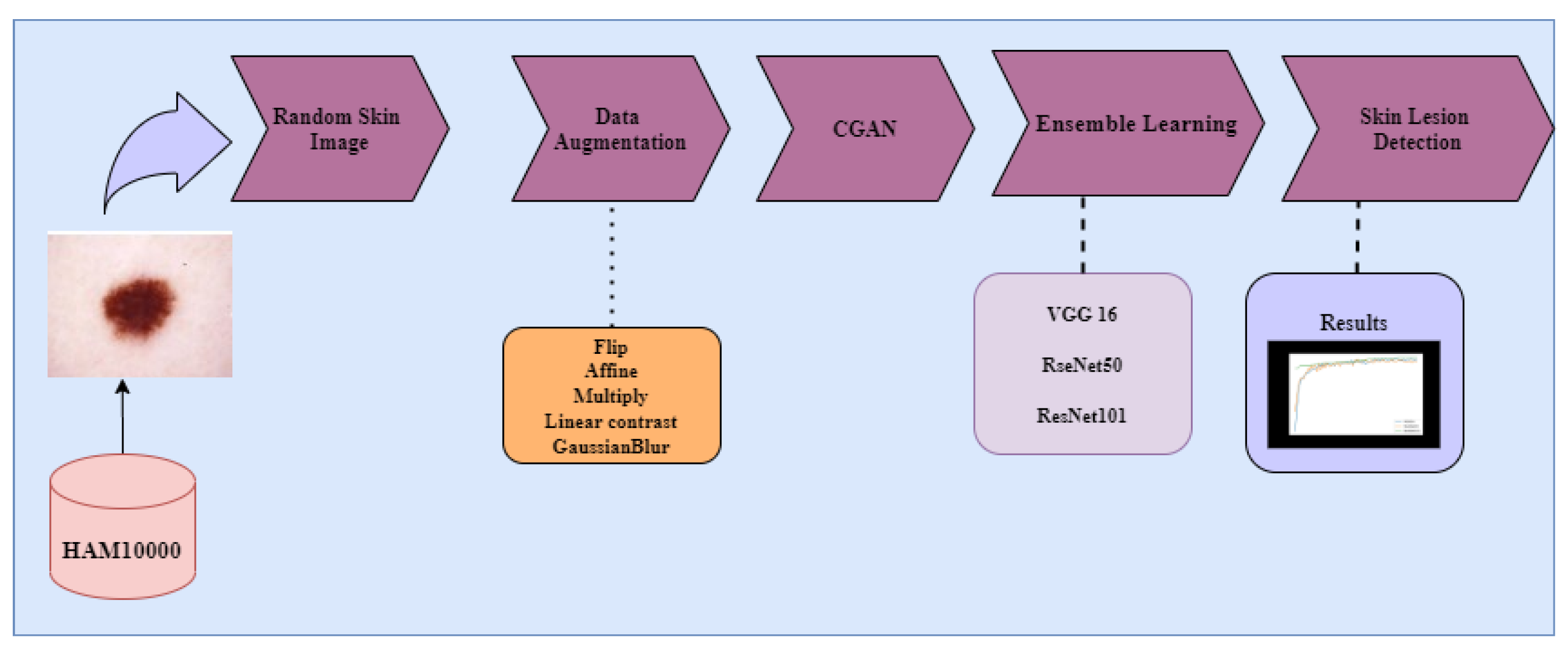
3. Materials and Methods
3.1. Dataset
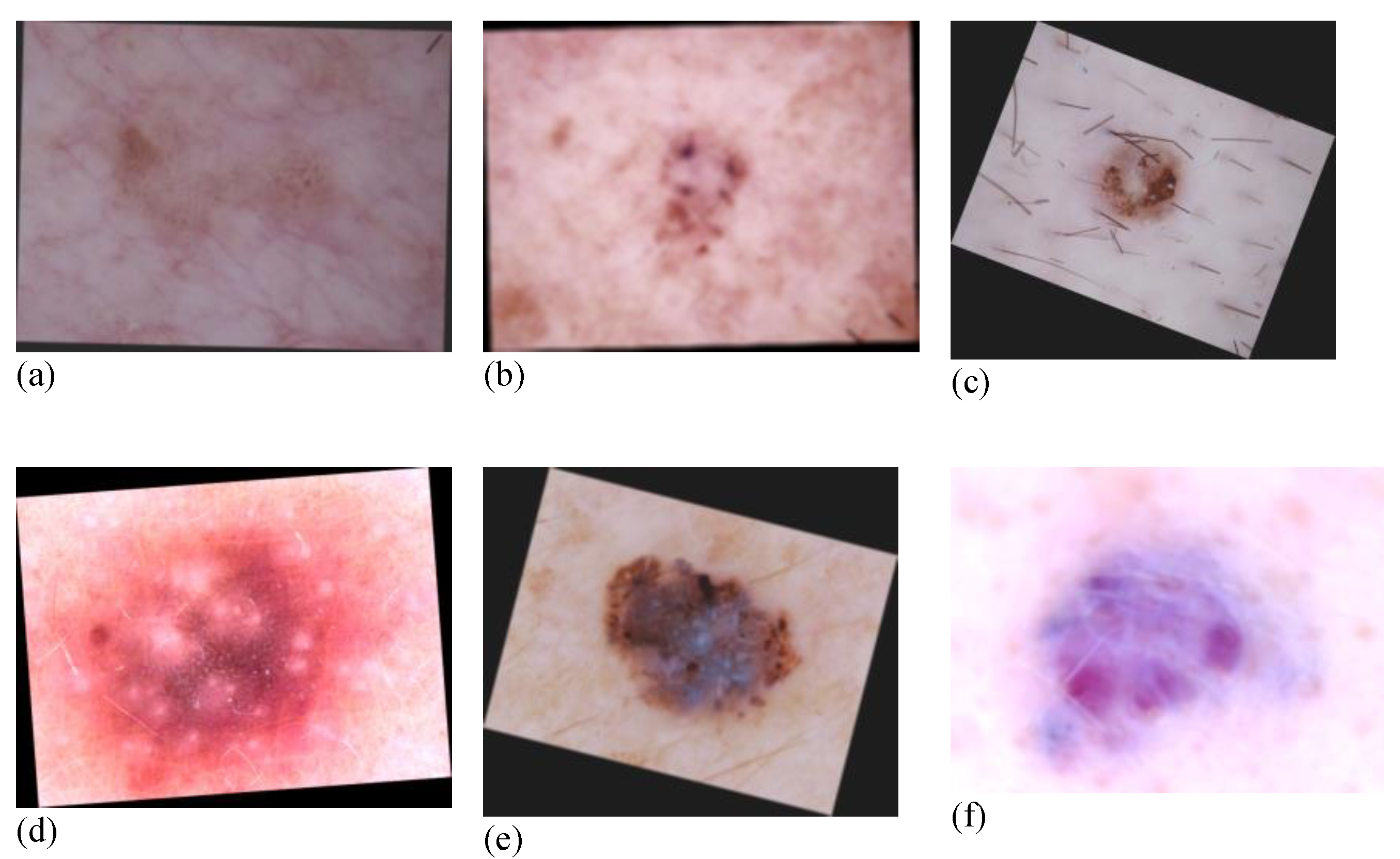
3.2. Data Augmentation
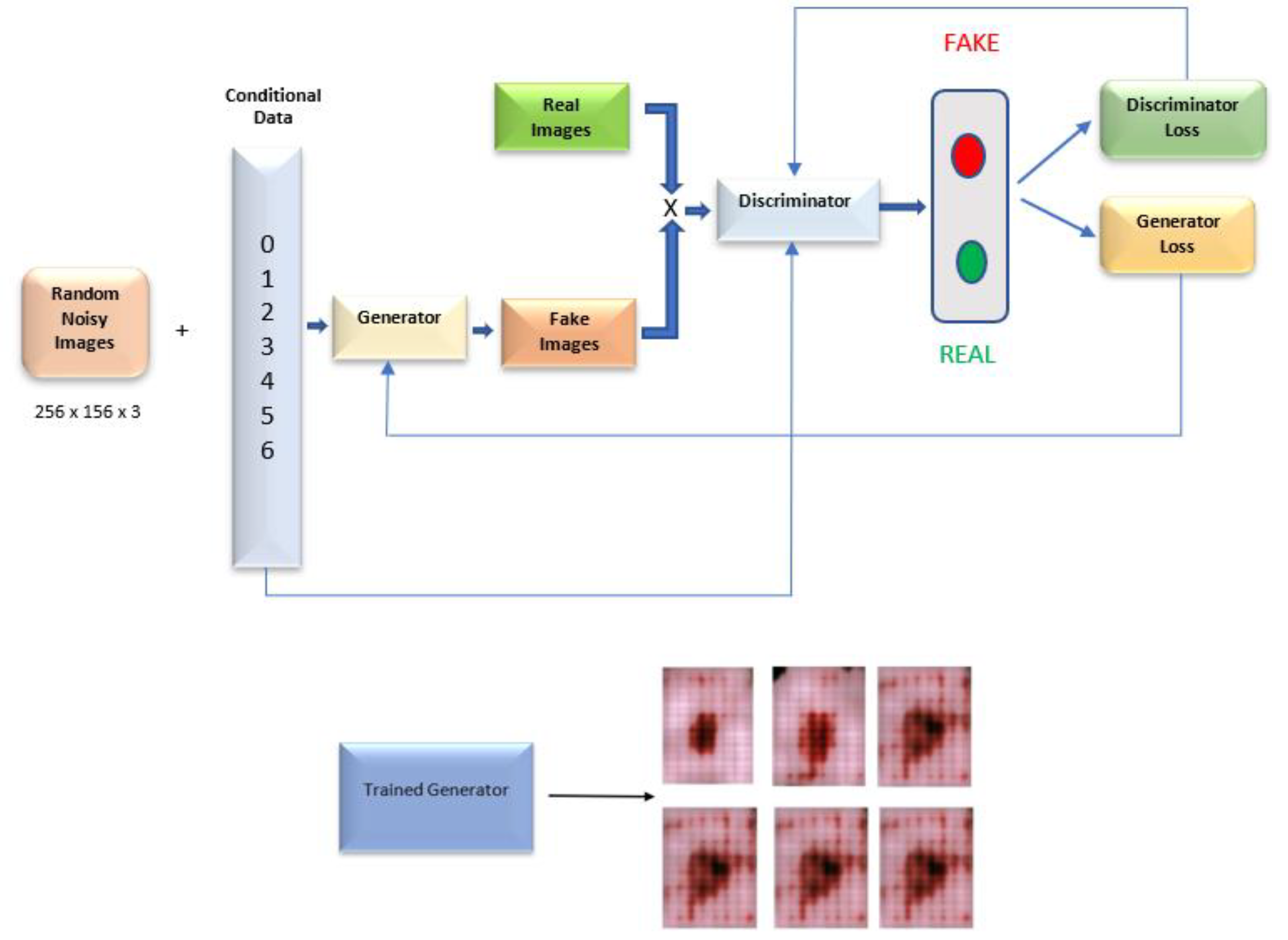
3.3. Image Generation Conditional Generative Adversarial Networks (CGANs)
3.4. Classification Model Development
3.5. Experimental Setup
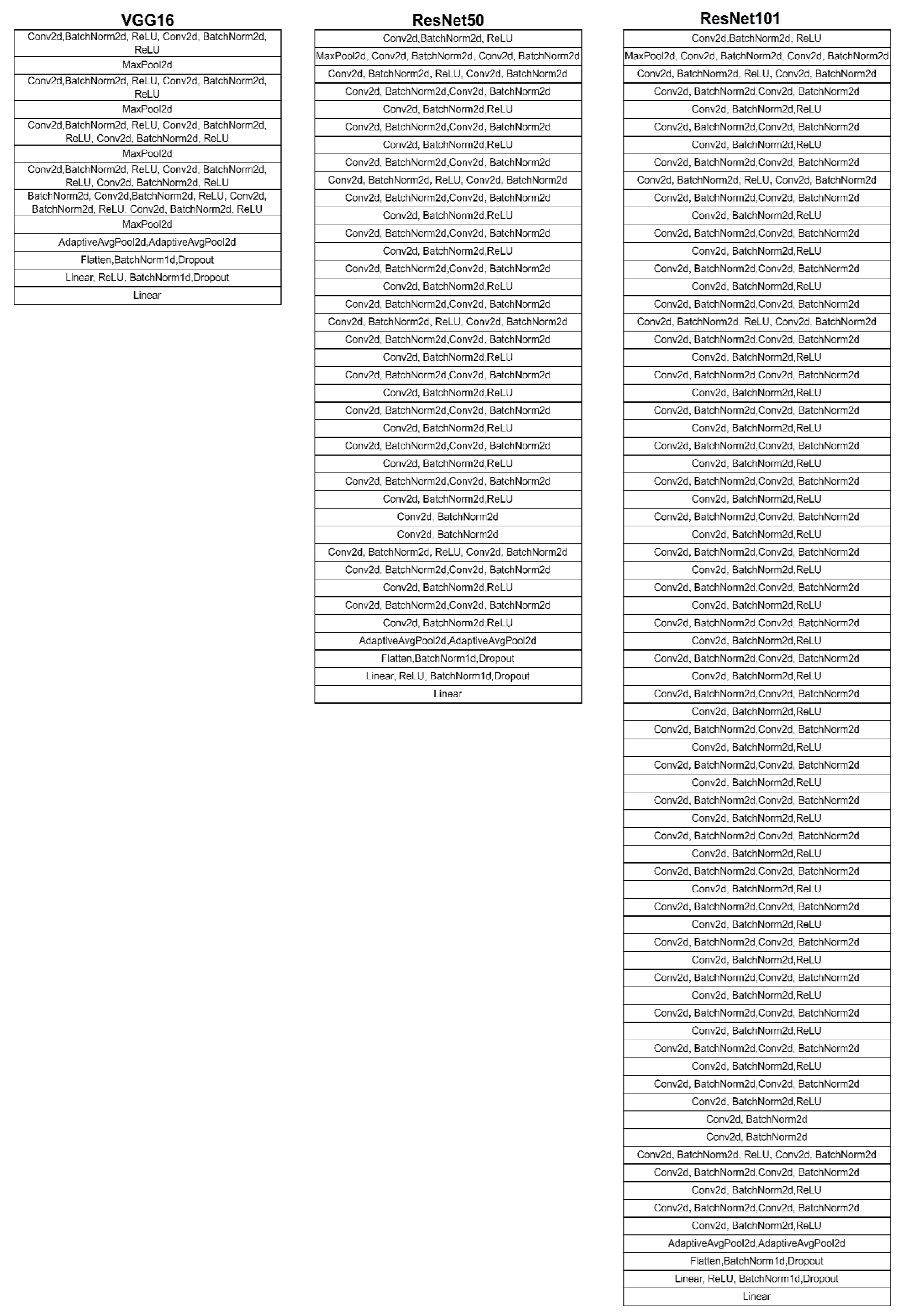
3.6. VGG16
3.7. ResNet50
3.8. ResNet101
3.9. Ensemble Algorithm
3.10. Performance Evaluation
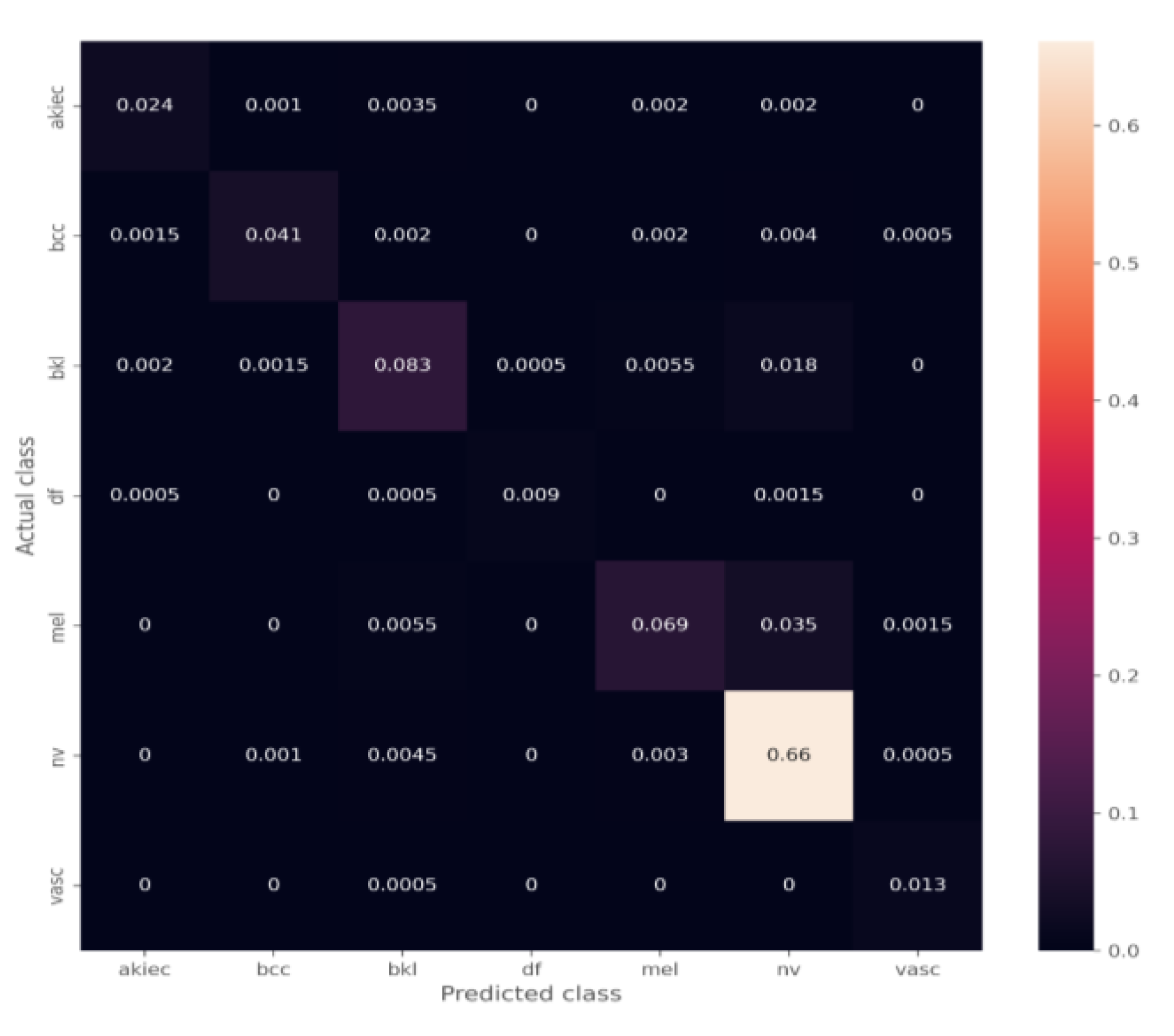
4. Results
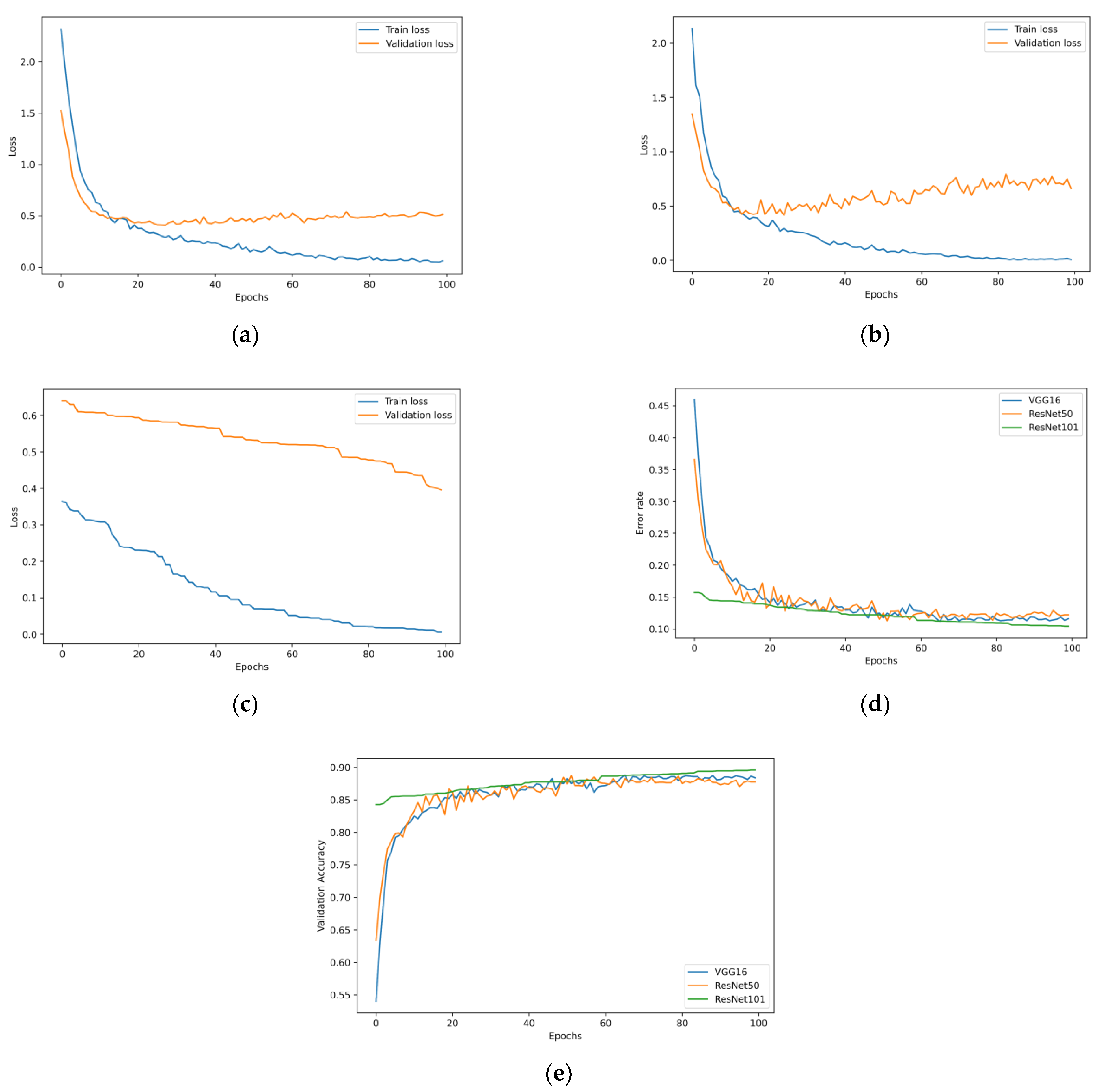
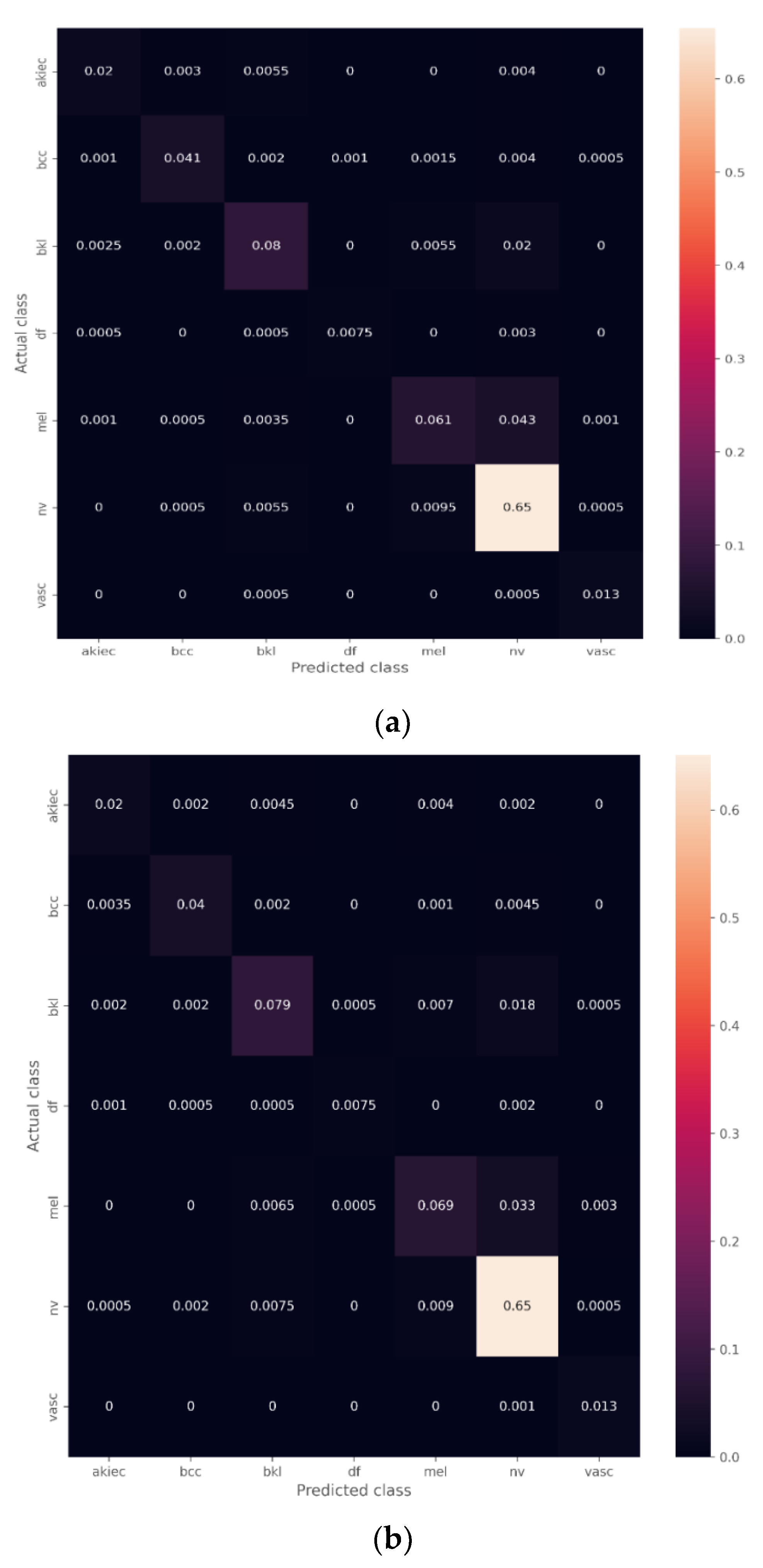
4.1. Transfer Learning Model with the Unbalanced Dataset
The Effect of the Ensemble Algorithm
4.2. Transfer Learning Model on the Balanced Data Obtained by Data Augmentation
4.3. The Effect of the Ensemble Algorithm
5. Discussion
Performance Comparison with Previous Works
6. Limitations
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Armstrong, B.K.; Kricker, A. Skin cancer. Dermatol. Clin. 1995, 13, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Simões, M.C.F.; Sousa, J.J.S.; Pais, A.A.C.C. Skin cancer and new treatment perspectives: A review. Cancer Lett. 2015, 357, 8–42. [Google Scholar] [CrossRef]
- Cassano, R.; Cuconato, M.; Calviello, G.; Serini, S.; Trombino, S. Recent advances in nanotechnology for the treatment of melanoma. Molecules 2021, 26, 785. [Google Scholar] [CrossRef] [PubMed]
- Buljan, M.; Bulat, V.; Šitum, M.; Lugović Mihić, L.; Stanić-Duktaj, S. Variations in clinical presentation of basal cell carcinoma. Acta Clin. Croat. 2008, 47, 30. [Google Scholar]
- Kanavy, H.E.; Gerstenblith, M.R. Ultraviolet radiation and melanoma. Semin. Cutan. Med. Surg. 2011, 30, 222–228. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Shorfuzzaman, M. An explainable stacked ensemble of deep learning models for improved melanoma skin cancer detection. Multimed Syst. 2022, 28, 1309–1323. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.U.; Aslam, N.; Anwar, T.; Aljameel, S.S.; Ullah, M.; Khan, R.; Rehman, A.; Akhtar, N. Remote Diagnosis and Triaging Model for Skin Cancer Using EfficientNet and Extreme Gradient Boosting. Complexity 2021, 2021, 1–13. [Google Scholar] [CrossRef]
- Wolff, K.; Johnson, R.C.; Saavedra, A.; Roh, E.K. Fitzpatrick’s Color Atlas and Synopsis of Clinical Dermatology; McGraw Hill Professional, 2017. McGraw-Hill Medical Pub. Division, 2005. Available online: https://accessmedicine.mhmedical.com/book.aspx?bookID=2043 (accessed on 6 December 2022).
- Rajput, G.; Agrawal, S.; Raut, G.; Vishvakarma, S.K. An accurate and noninvasive skin cancer screening based on imaging technique. Int. J. Imaging Syst. Technol. 2022, 32, 354–368. [Google Scholar] [CrossRef]
- Argenziano, G.; Soyer, H.P. Dermoscopy of pigmented skin lesions--a valuable tool for early. Lancet Oncol. 2001, 2, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Kittler, H.; Pehamberger, H.; Wolff, K.; Binder, M. Diagnostic accuracy of dermoscopy. Lancet Oncol. 2002, 3, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Fabbrocini, G.; De Vita, V.; Pastore, F.; D’Arco, V.; Mazzella, C.; Annunziata, M.C.; Cacciapuoti, S.; Mauriello, M.C.; Monfrecola, A. Teledermatology: From prevention to diagnosis of nonmelanoma and melanoma skin cancer. Int. J. Telemed. Appl. 2011, 2011, 1–5. [Google Scholar] [CrossRef]
- Ali, A.-R.A.; Deserno, T.M. A systematic review of automated melanoma detection in dermatoscopic images and its ground truth data. Med. Imaging 2012 Image Percept. Obs. Perform. Technol. Assess. 2012, 8318, 421–431. [Google Scholar]
- Sinz, C.; Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.-Y.; Kreusch, J.; et al. Accuracy of dermatoscopy for the diagnosis of nonpigmented cancers of the skin. J. Am. Acad. Dermatol. 2017, 77, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep Learning, Sparse Coding, and SVM for Melanoma Recognition in Dermoscopy Images; Springer: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Srinivas, C.; Nandini Prasad, K.S.; Zakariah, M.; Alothaibi, Y.A.; Shaukat, K.; Partibane, B.; Awal, H. Deep Transfer Learning Approaches in Performance Analysis of Brain Tumor Classification Using MRI Images. J. Healthc. Eng. 2022, 2022, 3264367. [Google Scholar] [CrossRef]
- Shaukat, K.; Suhuai, L.; Vijay, V.; Hameed, I.A.; Chen, S.; Liu, D.; Li, J. Performance comparison and current challenges of using machine learning techniques in cybersecurity. Energies 2020, 13, 2509. [Google Scholar] [CrossRef]
- Shaukat, K.; Luo, S.; Vijay, V. A novel method for improving the robustness of deep learning-based malware detectors against adversarial attacks. Eng. Appl. Artif. Intell. 2022, 116, 105461. [Google Scholar] [CrossRef]
- Shaukat, K.; Luo, S.; Chen, S.; Liu, D. Cyber threat detection using machine learning techniques: A performance evaluation perspective. In Proceedings of the 2020 International Conference on Cyber Warfare and Security (ICCWS), Norfolk, VI, USA, 12–13 March 2020; pp. 1–6. [Google Scholar]
- Shaukat, K.; Luo, S.; Vijay, V.; Hameed, I.A.; Xu, M. A Survey on Machine Learning Techniques for Cyber Security in the Last Decade. IEEE Access 2020, 8, 222310–222354. [Google Scholar] [CrossRef]
- Kousis, I.; Perikos, I.; Hatzilygeroudis, I.; Virvou, M. Deep Learning Methods for Accurate Skin Cancer Recognition and Mobile Application. Electronics 2022, 11, 1294. [Google Scholar] [CrossRef]
- Khan, N.H.; Mir, M.; Qian, L.; Baloch, M.; Khan, M.F.A.; Rehman, A.-U.; Ngowi, E.E.; Wu, D.-D.; Ji, X.-Y. Skin cancer biology and barriers to treatment: Recent applications of polymeric micro/nanostructures. J. Adv. Res. 2021, 36, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Tajjour, S.; Garg, S.; Chandel, S.S.; Sharma, D. A novel hybrid artificial neural network technique for the early skin cancer diagnosis using color space conversions of original images. Int. J. Imaging Syst. Technol. 2022. [Google Scholar] [CrossRef]
- Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183. [Google Scholar] [CrossRef] [PubMed]
- Barata, A.C.F.; Celebi, M.E.; Marques, J.S. A survey of feature extraction in dermoscopy image analysis of skin cancer. IEEE J. Biomed. Health Inform. 2018, 23, 1096–1109. [Google Scholar] [CrossRef]
- Celebi, M.E.; Kingravi, H.A.; Uddin, B.; Iyatomi, H.; Aslandogan, Y.A.; Stoecker, W.V.; Moss, R.H. A methodological approach to the classification of dermoscopy images. Comput. Med. imaging Graph. 2007, 31, 362–373. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imaging 2016, 36, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef]
- Yap, M.H.; Goyal, M.; Osman, F.; Ahmad, E.; Martí, R.; Denton, E.; Juette, A.; Zwiggelaar, R. End-to-end breast ultrasound lesions recognition with a deep learning approach. In Medical imaging 2018: Biomedical applications in Molecular, Structural, and Functional Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10578, p. 1057819. [Google Scholar]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Skin cancer classification using deep learning and transfer learning. In Proceedings of the 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), Cairo, Egypt, 20–22 December 2018; pp. 90–93. [Google Scholar]
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.S.; Rozeira, J. PH 2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Mahbod, A.; Schaefer, G.; Ellinger, I.; Ecker, R.; Pitiot, A.; Wang, C. Fusing fine-tuned deep features for skin lesion classification. Comput. Med. Imaging Graph. 2019, 71, 19–29. [Google Scholar] [CrossRef]
- Gessert, N.; Sentker, T.; Madesta, F.; Schmitz, R.; Kniep, H.; Baltruschat, I.; Werner, R.; Schlaefer, A. Skin lesion classification using cnns with patch-based attention and diagnosis-guided loss weighting. IEEE Trans. Biomed. Eng. 2019, 67, 495–503. [Google Scholar] [CrossRef]
- Liu, L.; Mou, L.; Zhu, X.X.; Mandal, M. Automatic skin lesion classification based on mid-level feature learning. Comput. Med. Imaging Graph. 2020, 84, 101765. [Google Scholar] [CrossRef]
- Aburaed, N.; Panthakkan, A.; Al-Saad, M.; Amin, S.A.; Mansoor, W. Deep Convolutional Neural Network (DCNN) for Skin Cancer Classification. In Proceedings of the 2020 27th IEEE International Conference on Electronics, Circuits and Systems (ICECS), Glasgow, Scotland, 23–25 November 2020; pp. 1–4. [Google Scholar]
- Huynh, A.T.; Hoang, V.-D.; Vu, S.; Le, T.T.; Nguyen, H.D. Skin Cancer Classification Using Different Backbones of Convolutional Neural Networks. In Proceedings of the International Conference on Industrial, Engineering and Other Applications of Applied Intelligent Systems, Kitakyushu, Japan, 19–22 July 2022; pp. 160–172. [Google Scholar]
- Garg, R.; Maheshwari, S.; Shukla, A. Decision support system for detection and classification of skin cancer using CNN. In Innovations in Computational Intelligence and Computer Vision; Springer: New York, NY, USA, 2021; pp. 578–586. [Google Scholar]
- Khan, M.H.-M.; Sahib-Kaudeer, N.G.; Dayalen, M.; Mahomedaly, F.; Sinha, G.R.; Nagwanshi, K.K.; Taylor, A. Multi-Class Skin Problem Classification Using Deep Generative Adversarial Network (DGAN). Comput. Intell. Neurosci. 2022, 2022, 1–13. [Google Scholar]
- Alam, T.M.; Shaukat, K.; Khan, W.A.; Hameed, I.A.; Almuqren, L.A.; Raza, M.A.; Aslam, M.; Luo, S. An Efficient Deep Learning-Based Skin Cancer Classifier for an Imbalanced Dataset. Diagnostics 2022, 12, 2115. [Google Scholar] [CrossRef]
- Yang, X.; Khushi, M.; Shaukat, K. Biomarker CA125 feature engineering and class imbalance learning improves ovarian cancer prediction. In Proceedings of the 2020 IEEE Asia-Pacific Conference on Computer Science and Data Engineering (CSDE), Gold Coast, Australia, 16–18 December 2020; pp. 1–6. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. Data descriptor: The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Rashid, H.; Tanveer, M.A.; Khan, H.A. Skin lesion classification using GAN based data augmentation. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 916–919. [Google Scholar]
- Diz, J.; Marreiros, G.; Freitas, A. Applying data mining techniques to improve breast cancer diagnosis. J. Med. Syst. 2016, 40, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Advances in Neural Information Processing Systems; Universitéde Montréal: Montreal, QC, Canada, 2014. [Google Scholar]
- Lisa, T.; Jude, S. Transfer Learning Handbook of Research on Machine Learning Applications. IGI Glob. 2009, 242–264. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv1409.1556. [Google Scholar]
- Mohakud, R.; Rajashree, D. Designing a grey wolf optimization based hyper-parameter optimized convolutional neural network classifier for skin cancer detection. J. King Saud Univ.Comput. Inf. Sci. 2022, 34, 6280–6291. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, J.; Niu, J.; Zhang, Y.; Wang, W. Roadside units deployment for content downloading in vehicular networks. In Proceedings of the IEEE International Conference on Communications (ICC), Budapest, Hungary, 9–13 June 2013; pp. 6365–6370. [Google Scholar] [CrossRef]
- Chaturvedi, S.S.; Gupta, K.; Prasad, P.S. Skin lesion analyser: An efficient seven-way multi-class skin cancer classification using MobileNet. In Proceedings of the International Conference on Advanced Machine Learning Technologies and Applications, Jaipur, India, 13–15 February 2020; pp. 165–176. [Google Scholar]
- Khan, M.A.; Javed, M.Y.; Sharif, M.; Saba, T.; Rehman, A. Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification. In Proceedings of the 2019 international conference on computer and information sciences (ICCIS), Aljouf, Saudi Arabia, 3–4 April 2019; pp. 1–7. [Google Scholar]
- Sae-Lim, W.; Wettayaprasit, W.; Aiyarak, P. Convolutional neural networks using MobileNet for skin lesion classification. In Proceedings of the 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), Pataya, Chonburi, Thailand, 10–12 July 2019; pp. 242–247. [Google Scholar]
- Huang, H.-W.; Hsu, B.W.-Y.; Lee, C.-H.; Tseng, V.S. Development of a light-weight deep learning model for cloud applications and remote diagnosis of skin cancers. J. Dermatol. 2021, 48, 310–316. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Sharif, M.; Kadry, S.; Nam, Y. Computer decision support system for skin cancer localization and classification. Comput. Mater. Contin. 2021, 68, 1041–1064. [Google Scholar]
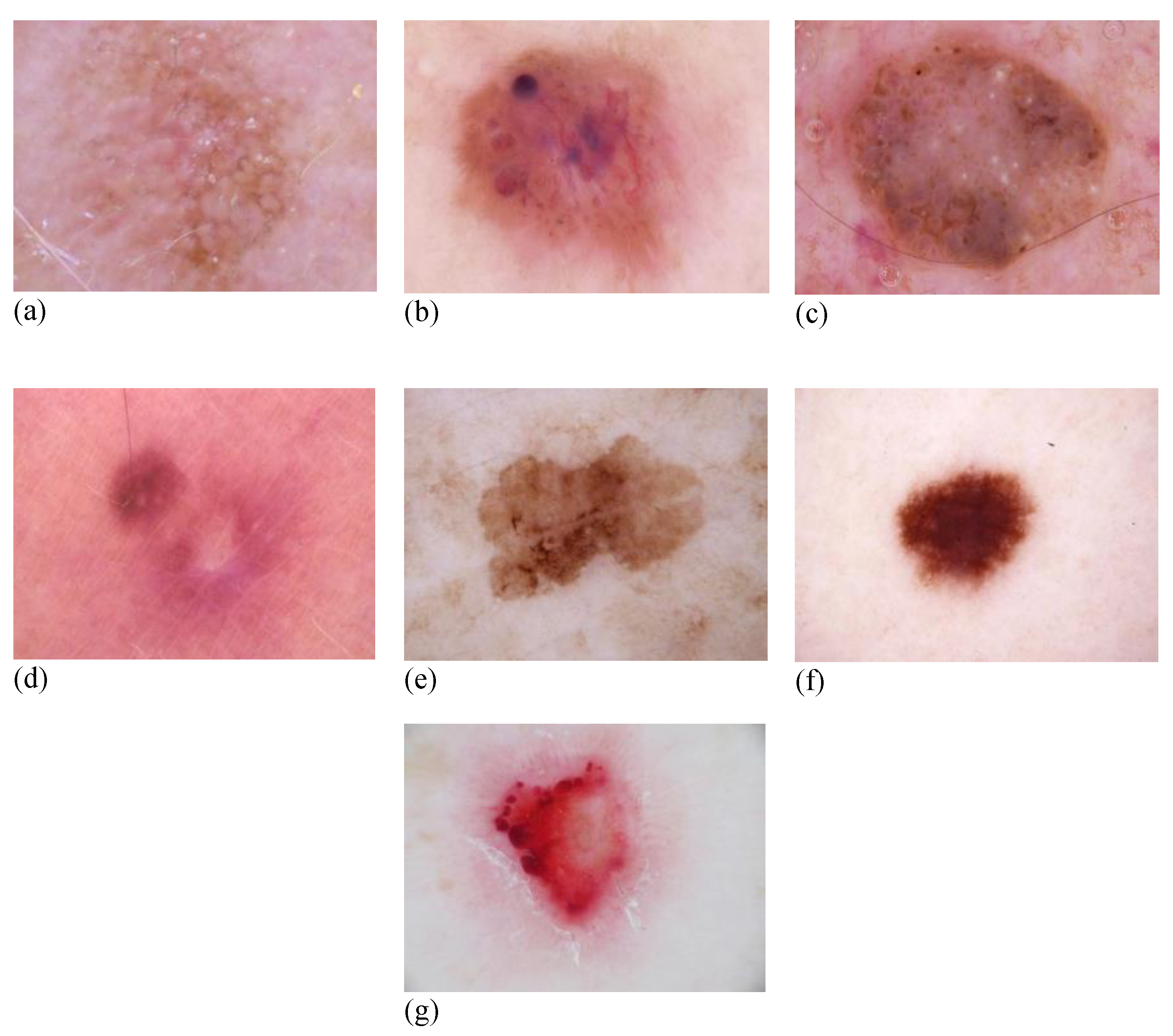


| Class | Train | Validation | Test | Total | Benign/Malignant |
|---|---|---|---|---|---|
| Actinic Keratosis (AKIEC) | 236 | 26 | 65 | 327 | Benign or Malignant |
| Basal Cell Carcinoma (BCC) | 371 | 41 | 102 | 514 | Malignant |
| Benign Keratosis (BKL) | 792 | 88 | 219 | 1099 | Benign |
| Dermatofibroma (DF) | 83 | 9 | 23 | 115 | Benign |
| Melanoma (MEL) | 802 | 89 | 222 | 1113 | Malignant |
| Nevus (NV) | 4828 | 536 | 1341 | 6705 | Benign |
| Vascular Cancer (VASC) | 103 | 11 | 28 | 142 | Benign or Malignant |
| flip | 50% of horizontal and vertical flip on all images. |
| Affine | Translation: Move each image −20 to +20% per axis. Rotation: Rotate each image by −30 to 30 degrees. Scaling: Zoom in each image by 0.5 to 1.5 times. |
| Multiply | Multiplication of each image by a random value sampled from [0.8, 1.2]. |
| Linear contrast | Change contrast by equation. 127 + alpha × (v-127). V: Pixel value. Alpha: Samples from [0.6, 1.4]. |
| Gaussian Blur | Blur the images using Gaussian kernel with standard deviation sampled from the interval [0.0, 3.0]. |
| Model | Accuracy (%) | Recall (%) | Precision (%) | F1 Score (%) |
|---|---|---|---|---|
| VGG16 | 87.7 | 75.08 | 84.66 | 79.06 |
| ResNet50 | 87.9 | 75.57 | 81.69 | 78.01 |
| ResNet101 | 88.15 | 75.96 | 84.48 | 79.57 |
| Ensemble model | 90 | 80.66 | 88.06 | 83.77 |
| Ensemble Models | Unbalanced Dataset | Balanced Dataset | ||||
|---|---|---|---|---|---|---|
| Class of Skin Cancer | Recall (%) | Precision (%) | F1 Score (%) | Recall (%) | Precision (%) | F1 Score (%) |
| AKIEC | 73.84 | 85.71 | 79.33 | 84.61 | 94.82 | 89.43 |
| BCC | 80.39 | 92.13 | 85.86 | 90.19 | 94.84 | 92.46 |
| BKL | 75.34 | 83.33 | 79.13 | 84.93 | 90.73 | 87.73 |
| DF | 78.26 | 94.73 | 85.71 | 95.65 | 95.65 | 95.65 |
| MEL | 61.71 | 84.56 | 71.35 | 72.07 | 92.48 | 81.01 |
| NV | 98.65 | 91.62 | 95.00 | 99.03 | 93.91 | 96.40 |
| VASC | 96.42 | 84.37 | 90 | 96.42 | 90 | 93.10 |
| Model | Accuracy (%) | Recall (%) | Precision (%) | F1 Score (%) |
|---|---|---|---|---|
| VGG16 | 92 | 85.07 | 91.84 | 88.07 |
| ResNet50 | 92.1 | 84.65 | 88.65 | 86.26 |
| ResNet101 | 92.25 | 85.40 | 90.63 | 87.79 |
| Ensemble model | 93.5 | 88.98 | 93.20 | 90.82 |
| S. No. | Year | Dataset | References | Models | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|---|---|---|---|
| 1. | 2020 | HAM10000 | [54] | Darknet-53 + NasNet Mobile | 83.1% | 92.61% | - | 88.03% |
| 2. | 2019 | HAM10000 | [55] | ResNet50 + ResNet101 | 89.8% | - | - | - |
| 3. | 2019 | HAM10000 | [56] | MobileNet | 83.23% | - | - | - |
| 4. | 2021 | KCGMH and HAM10000 | [57] | VGG16 | 85.8% | - | - | - |
| 5. | 2021 | HAM10000 | [58] | 90.67% | 91.2% | 90.3% | 89.99% | |
| 6. | HAM10000 | Proposed work (augmentation + ensemble model) | VGG16 + ResNet50, + ResNet101 | 93.5% | 93.20% | 88.98% | 90.82% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Rasheed, A.; Ksibi, A.; Ayadi, M.; Alzahrani, A.I.A.; Zakariah, M.; Ali Hakami, N. An Ensemble of Transfer Learning Models for the Prediction of Skin Cancers with Conditional Generative Adversarial Networks. Diagnostics 2022, 12, 3145. https://doi.org/10.3390/diagnostics12123145
Al-Rasheed A, Ksibi A, Ayadi M, Alzahrani AIA, Zakariah M, Ali Hakami N. An Ensemble of Transfer Learning Models for the Prediction of Skin Cancers with Conditional Generative Adversarial Networks. Diagnostics. 2022; 12(12):3145. https://doi.org/10.3390/diagnostics12123145
Chicago/Turabian StyleAl-Rasheed, Amal, Amel Ksibi, Manel Ayadi, Abdullah I. A. Alzahrani, Mohammed Zakariah, and Nada Ali Hakami. 2022. "An Ensemble of Transfer Learning Models for the Prediction of Skin Cancers with Conditional Generative Adversarial Networks" Diagnostics 12, no. 12: 3145. https://doi.org/10.3390/diagnostics12123145
APA StyleAl-Rasheed, A., Ksibi, A., Ayadi, M., Alzahrani, A. I. A., Zakariah, M., & Ali Hakami, N. (2022). An Ensemble of Transfer Learning Models for the Prediction of Skin Cancers with Conditional Generative Adversarial Networks. Diagnostics, 12(12), 3145. https://doi.org/10.3390/diagnostics12123145






