Identification of Six microRNAs as Potential Biomarkers for Pemphigus Vulgaris: From Diagnosis to Pathogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Ethics
2.2. Diagnostic Criteria
2.3. Inclusion and Exclusion Criteria
2.4. Serum Sample Collection and Processing
2.5. RNA Extraction, Reverse Transcription, and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.6. Target Gene Enrichment Analysis
2.7. Statistical Analysis
3. Results
3.1. Demographic and Clinical Characteristics
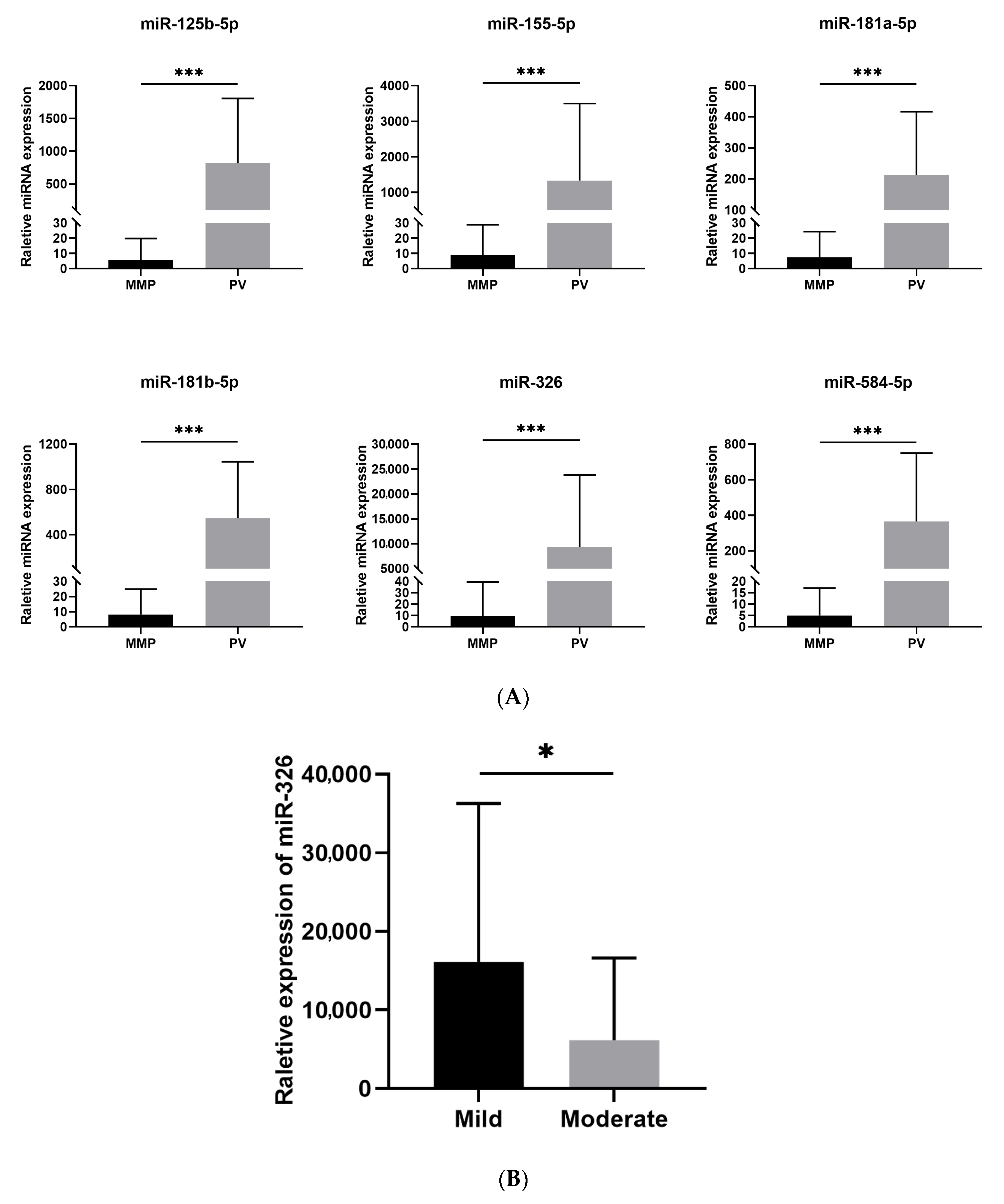
3.2. MiRNAs Expression Profiles
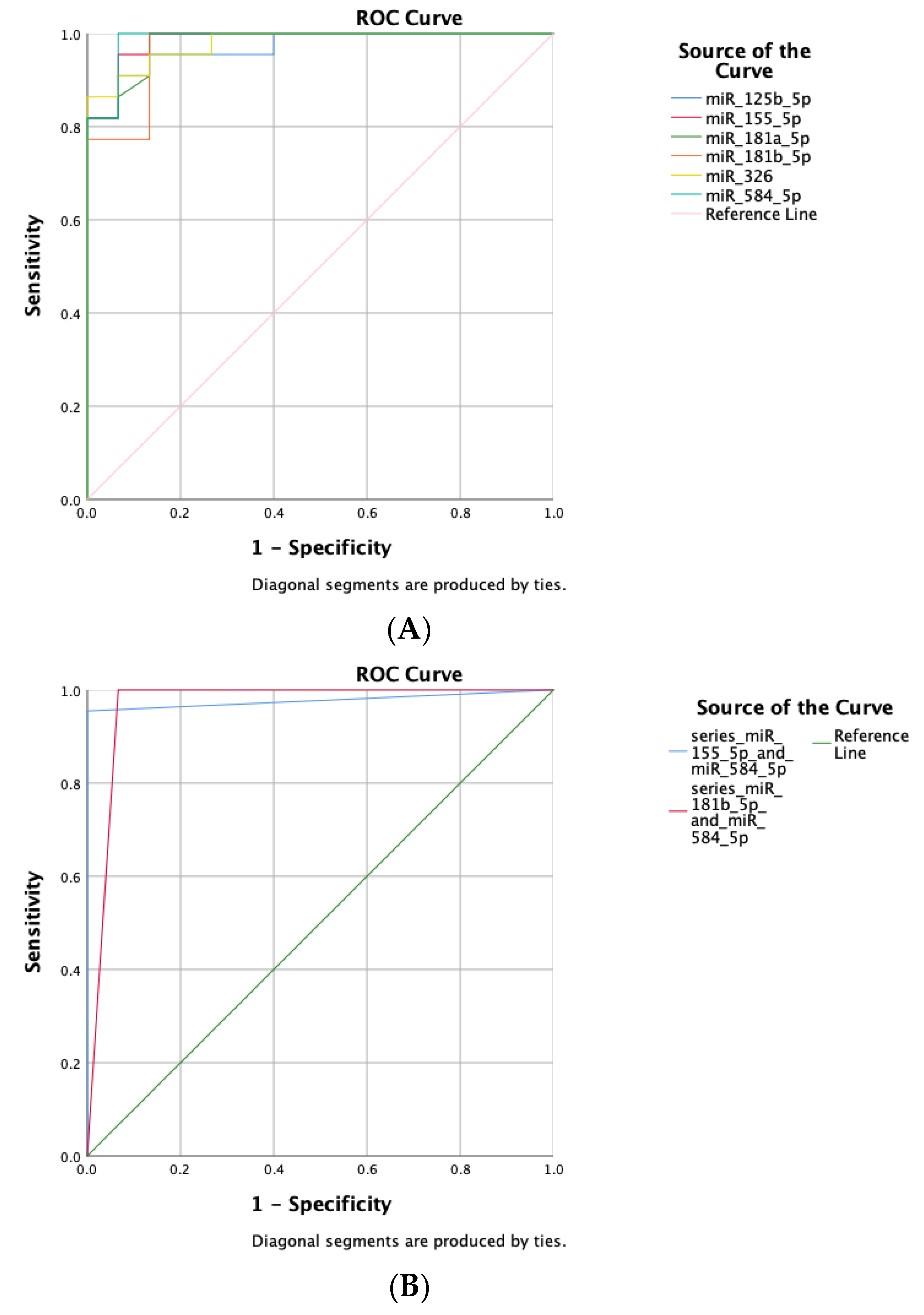
3.3. Diagnostic Efficacy
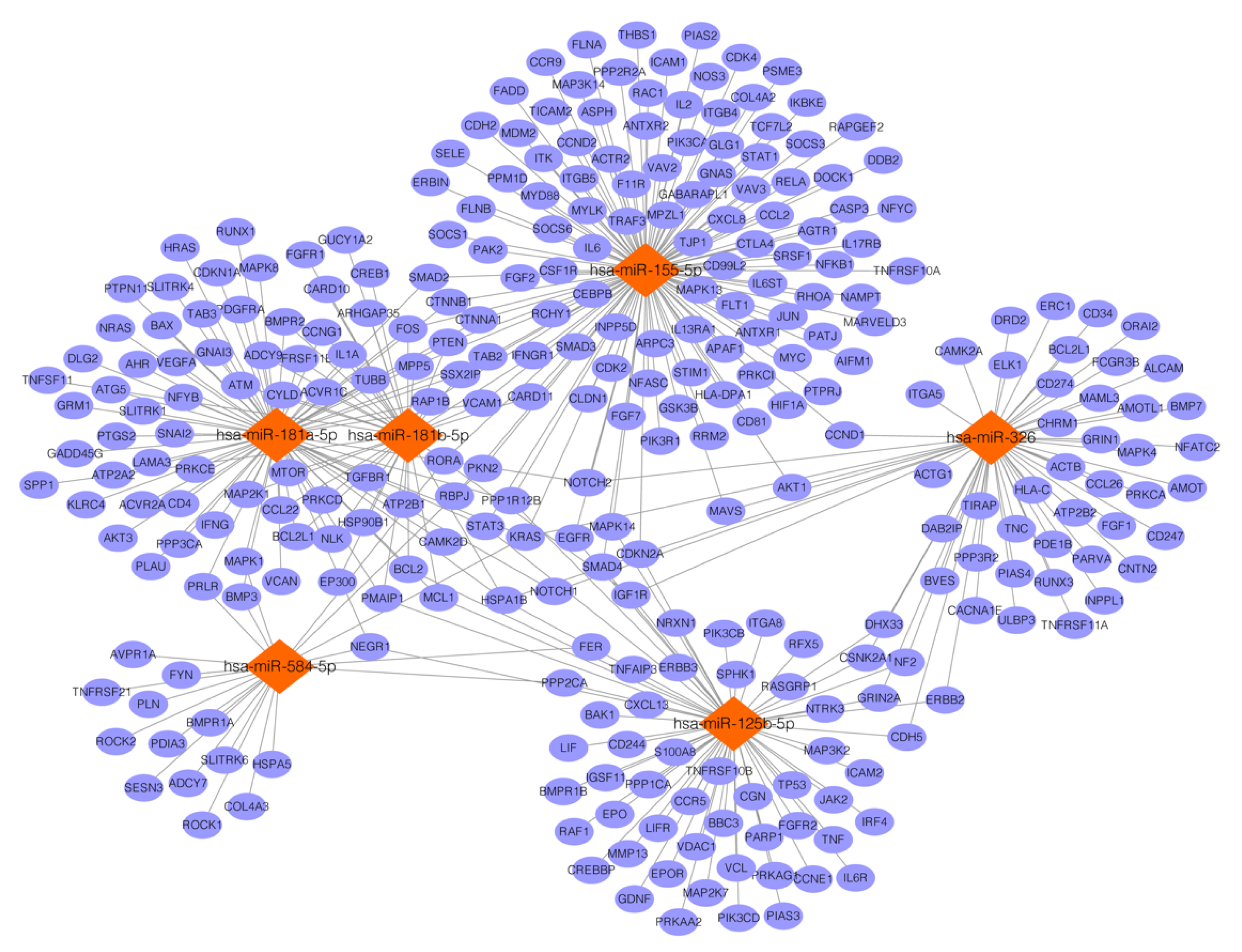
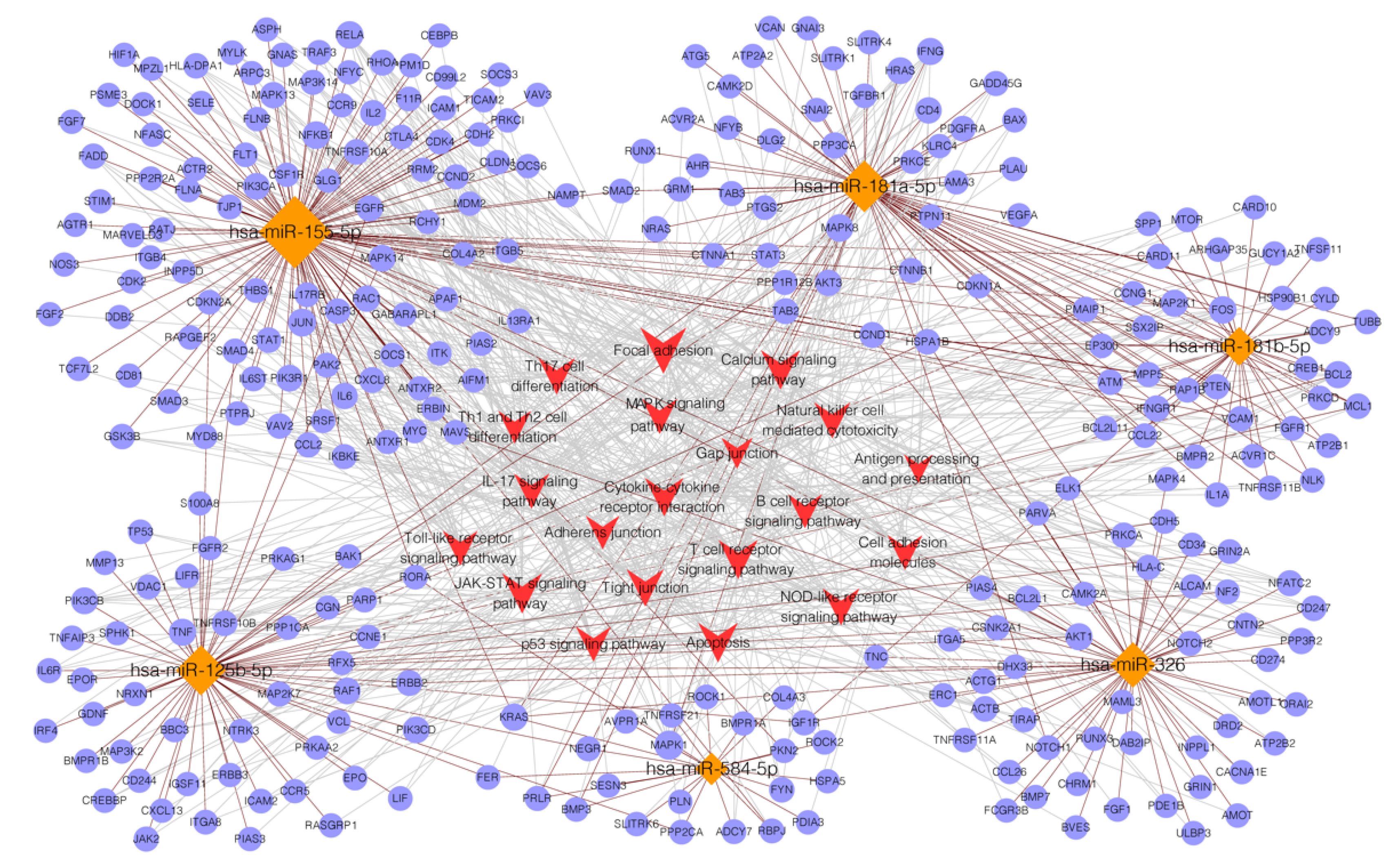
3.4. Targets Prediction for miRNAs
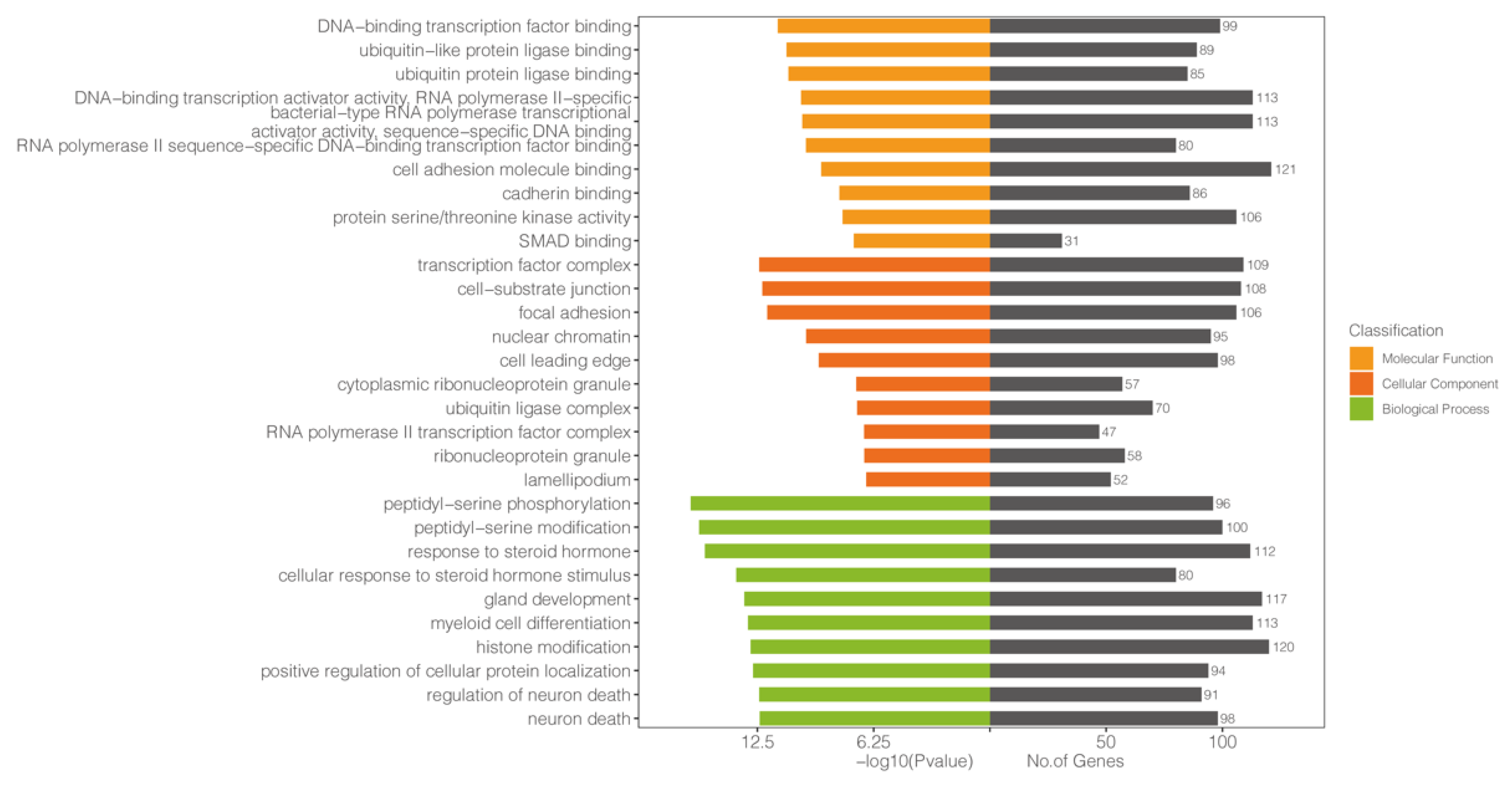
3.5. GO Analysis
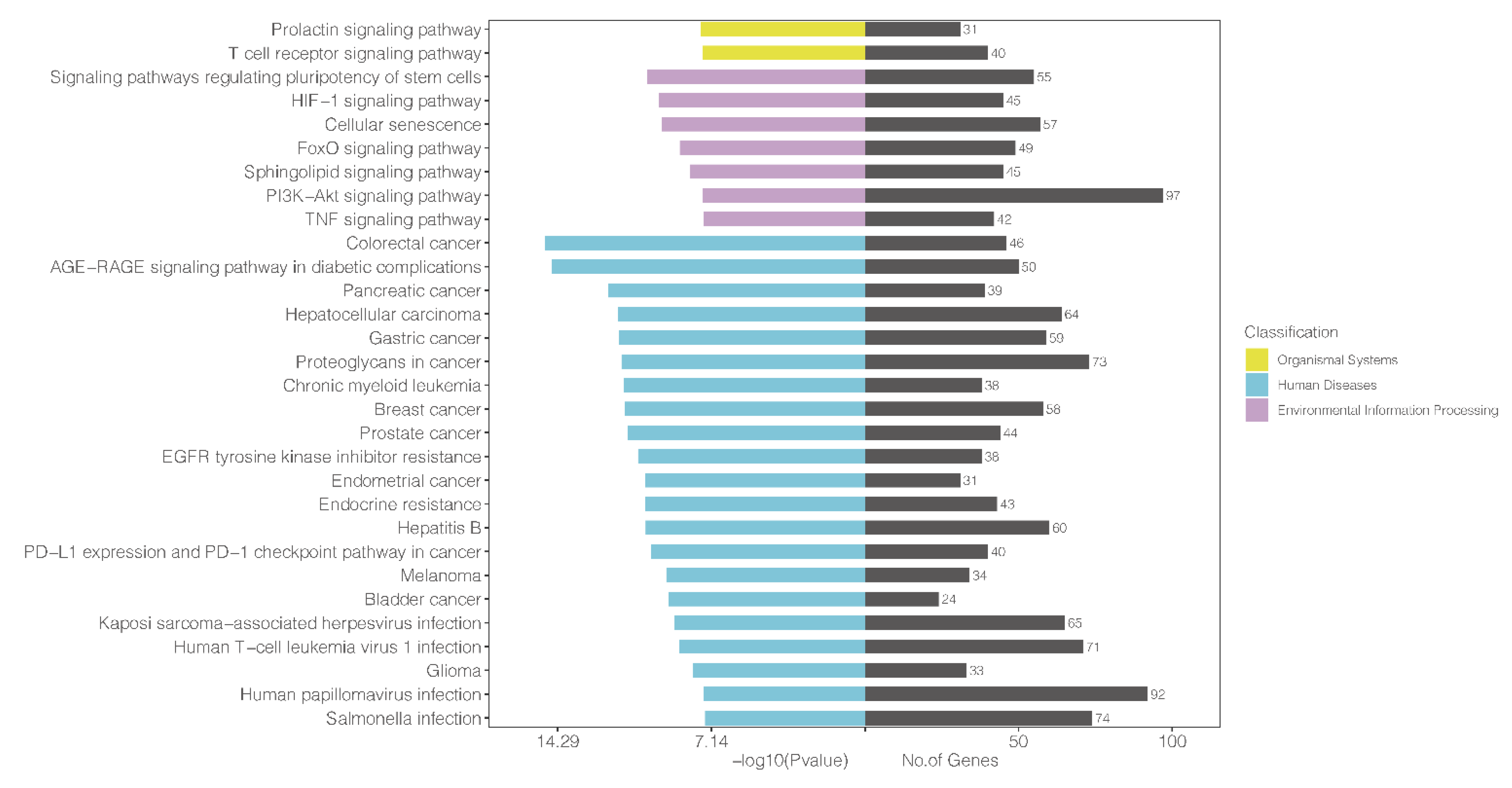
3.6. KEGG Analysis
3.7. Adverse Events
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Amagai, M.; Tanikawa, A.; Shimizu, T.; Hashimoto, T.; Ikeda, S.; Kurosawa, M.; Niizeki, H.; Aoyama, Y.; Iwatsuki, K.; Kitajima, Y. Japanese guidelines for the management of pemphigus. J. Dermatol. 2014, 41, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Hübner, F.; Recke, A.; Zillikens, D.; Linder, R.; Schmidt, E. Prevalence and age distribution of pemphigus and pemphigoid diseases in Germany. J. Investig. Dermatol. 2016, 136, 2495–2498. [Google Scholar] [CrossRef]
- Wertenteil, S.; Garg, A.; Strunk, A.; Alloo, A. Prevalence estimates for pemphigus in the United States: A sex- and age-adjusted population analysis. JAMA Dermatol. 2019, 155, 627–629. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.; Kasperkiewicz, M.; Joly, P. Pemphigus. Lancet 2019, 394, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Murrell, D.F.; Peña, S.; Joly, P.; Marinovic, B.; Hashimoto, T.; Diaz, L.A.; Sinha, A.A.; Payne, A.S.; Daneshpazhooh, M.; Eming, R.; et al. Diagnosis and management of pemphigus: Recommendations of an international panel of experts. J. Am. Acad. Dermatol. 2020, 82, 575–585.e571. [Google Scholar] [CrossRef] [PubMed]
- Joly, P.; Horvath, B.; Patsatsi, A.; Uzun, S.; Bech, R.; Beissert, S.; Bergman, R.; Bernard, P.; Borradori, L.; Caproni, M.; et al. Updated S2K guidelines on the management of pemphigus vulgaris and foliaceus initiated by the european academy of dermatology and venereology (EADV). J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Harman, K.E.; Brown, D.; Exton, L.S.; Groves, R.W.; Hampton, P.J.; Mohd, M.M.F.; Setterfield, J.F.; Yesudian, P.D. British Association of Dermatologists’ guidelines for the management of pemphigus vulgaris 2017. Br. J. Derm. 2017, 177, 1170–1201. [Google Scholar] [CrossRef] [PubMed]
- Nosrati, A.; Hodak, E.; Mimouni, T.; Oren-Shabtai, M.; Levi, A.; Leshem, Y.A.; Mimouni, D. Treatment of Pemphigus with Rituximab: Real-Life Experience in a Cohort of 117 Patients in Israel. Dermatology 2021, 237, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell. Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Long, H.; Wang, X.; Chen, Y.; Wang, L.; Zhao, M.; Lu, Q. Dysregulation of microRNAs in autoimmune diseases: Pathogenesis, biomarkers and potential therapeutic targets. Cancer Lett. 2018, 428, 90–103. [Google Scholar] [CrossRef]
- Miao, C.; Wang, X.; Zhou, W.; Huang, J. The emerging roles of exosomes in autoimmune diseases, with special emphasis on microRNAs in exosomes. Pharm. Res. 2021, 169, 105680. [Google Scholar] [CrossRef] [PubMed]
- Evangelatos, G.; Fragoulis, G.E.; Koulouri, V.; Lambrou, G.I. MicroRNAs in rheumatoid arthritis: From pathogenesis to clinical impact. Autoimmun. Rev. 2019, 18, 102391. [Google Scholar] [CrossRef] [PubMed]
- Papara, C.; Zillikens, D.; Sadik, C.D.; Baican, A. MicroRNAs in pemphigus and pemphigoid diseases. Autoimmun. Rev. 2021, 20, 102852. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatsonis, C.A.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; de Vet, H.C.W.; et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015, 351, h5527. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.; Lamberts, A.; Diercks, G.F.H.; Pas, H.H.; Meijer, J.M.; Bolling, M.C.; Horváth, B. Oral Lesions in Autoimmune Bullous Diseases: An Overview of Clinical Characteristics and Diagnostic Algorithm. Am. J. Clin. Dermatol. 2019, 20, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- The Gene Ontology Resource. Available online: http://geneontology.org (accessed on 3 November 2022).
- GenomeNet Database Resources. Available online: http://www.genome.jp (accessed on 3 November 2022).
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Shimizu, T.; Takebayashi, T.; Sato, Y.; Niizeki, H.; Aoyama, Y.; Kitajima, Y.; Iwatsuki, K.; Hashimoto, T.; Yamagami, J.; Werth, V.P.; et al. Grading criteria for disease severity by pemphigus disease area index. J. Derm. 2014, 41, 969–973. [Google Scholar] [CrossRef]
- Xiao, C.; Nemazee, D.; Gonzalez-Martin, A. MicroRNA control of B cell tolerance, autoimmunity and cancer. Semin. Cancer Biol. 2020, 64, 102–107. [Google Scholar] [CrossRef]
- Pauley, K.M.; Cha, S.; Chan, E.K. MicroRNA in autoimmunity and autoimmune diseases. J. Autoimmun. 2009, 32, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Bystryn, J.C.; Steinman, N.M. The adjuvant therapy of pemphigus. An update. Arch. Dermatol. 1996, 132, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Kuo, C.-F.; Chen, Y.-H.; Yang, Y.-W. Incidence, mortality, and causes of death of patients with pemphigus in Taiwan: A nationwide population-based study. J. Investig. Dermatol. 2012, 132, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Langan, S.M.; Smeeth, L.; Hubbard, R.; Fleming, K.M.; Smith, C.J.P.; West, J. Bullous pemphigoid and pemphigus vulgaris--incidence and mortality in the UK: Population based cohort study. BMJ 2008, 337, a180. [Google Scholar] [CrossRef] [PubMed]
- Frampton, J.E. Rituximab: A Review in Pemphigus Vulgaris. Am. J. Clin. Derm. 2020, 21, 149–156. [Google Scholar] [CrossRef]
- Yang, A.; Xuan, R.; Melbourne, W.; Tran, K.; Murrell, D.F. Validation of the BIOCHIP test for the diagnosis of bullous pemphigoid, pemphigus vulgaris and pemphigus foliaceus. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 153–160. [Google Scholar] [CrossRef]
- Cialfi, S.; Oliviero, C.; Ceccarelli, S.; Marchese, C.; Barbieri, L.; Biolcati, G.; Uccelletti, D.; Palleschi, C.; Barboni, L.; De Bernardo, C.; et al. Complex multipathways alterations and oxidative stress are associated with Hailey-Hailey disease. Br. J. Derm. 2010, 162, 518–526. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Ding, J.; Li, L.; Wang, Z. Identification of ATP2C1 mutations in the patients of Hailey-Hailey disease. BMC Med. Genet. 2020, 21, 120. [Google Scholar] [CrossRef]
- Shah, A.A.; Dey-Rao, R.; Seiffert-Sinha, K.; Sinha, A.A. Increased oxidative stress in pemphigus vulgaris is related to disease activity and HLA-association. Autoimmunity 2016, 49, 248–257. [Google Scholar] [CrossRef]
- Lin, X.; Chen, M.; Li, X.; Wang, H.; Bao, Y. Low SOCS3 expression in CD4(+) T cells from pemphigus vulgaris patients enhanced Th1- and Th17-cell differentiation and exacerbated acantholysis via STAT activation. Mol. Immunol. 2022, 150, 114–125. [Google Scholar] [CrossRef]
- Mizushima, T.; Arakawa, S.; Sanada, Y.; Yoshino, I.; Miyazaki, D.; Urushima, H.; Tsujimoto, Y.; Ito, T.; Shimizu, S. Inhibition of epithelial cell death by Bcl-2 improved chronic colitis in IL-10 KO mice. Am. J. Pathol. 2013, 183, 1936–1944. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Brégégère, F.; Frusić-Zlotkin, M.; Feinmesser, M.; Michel, B.; Milner, Y. Possible apoptotic mechanism in epidermal cell acantholysis induced by pemphigus vulgaris autoimmunoglobulins. Apoptosis 2004, 9, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Amber, K.T.; Valdebran, M.; Grando, S.A. Non-Desmoglein Antibodies in Patients With Pemphigus Vulgaris. Front. Immunol. 2018, 9, 1190. [Google Scholar] [CrossRef] [PubMed]
- Kurzen, H.; Brenner, S. Significance of autoimmunity to non-desmoglein targets in pemphigus. Autoimmunity 2006, 39, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Chernyavsky, A.I.; Arredondo, J.; Kitajima, Y.; Sato-Nagai, M.; Grando, S.A. Desmoglein versus non-desmoglein signaling in pemphigus acantholysis: Characterization of novel signaling pathways downstream of pemphigus vulgaris antigens. J. Biol. Chem. 2007, 282, 13804–13812. [Google Scholar] [CrossRef]
- Fang, H.; Li, Q.; Wang, G. The role of T cells in pemphigus vulgaris and bullous pemphigoid. Autoimmun. Rev. 2020, 19, 102661. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chen, Y.; Zhou, Y.; Wang, Y.; Zheng, J.; Pan, M. Cognate Th2-B cell interaction is essential for the autoantibody production in pemphigus vulgaris. J. Clin. Immunol. 2012, 32, 114–123. [Google Scholar] [CrossRef]
- Rizzo, C.; Fotino, M.; Zhang, Y.; Chow, S.; Spizuoco, A.; Sinha, A.A. Direct characterization of human T cells in pemphigus vulgaris reveals elevated autoantigen-specific Th2 activity in association with active disease. Clin. Exp. Derm. 2005, 30, 535–540. [Google Scholar] [CrossRef]
- Asothai, R.; Anand, V.; Das, D.; Antil, P.S.; Khandpur, S.; Sharma, V.K.; Sharma, A. Distinctive Treg associated CCR4-CCL22 expression profile with altered frequency of Th17/Treg cell in the immunopathogenesis of Pemphigus Vulgaris. Immunobiology 2015, 220, 1129–1135. [Google Scholar] [CrossRef]
- Xu, R.C.; Zhu, H.Q.; Li, W.P.; Zhao, X.Q.; Yuan, H.J.; Zheng, J.; Pan, M. The imbalance of Th17 and regulatory T cells in pemphigus patients. Eur. J. Derm. 2013, 23, 795–802. [Google Scholar] [CrossRef]
- Arakawa, M.; Dainichi, T.; Yasumoto, S.; Hashimoto, T. Lesional Th17 cells in pemphigus vulgaris and pemphigus foliaceus. J. Derm. Sci. 2009, 53, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhou, S.; Liu, Z.; Cong, W.; Fei, X.; Zeng, W.; Zhu, H.; Xu, R.; Wang, Y.; Zheng, J.; et al. Pivotal Role of Lesional and Perilesional T/B Lymphocytes in Pemphigus Pathogenesis. J. Investig. Derm. 2017, 137, 2362–2370. [Google Scholar] [CrossRef]
- Kowalski, E.H.; Kneibner, D.; Kridin, K.; Amber, K.T. Serum and blister fluid levels of cytokines and chemokines in pemphigus and bullous pemphigoid. Autoimmun. Rev. 2019, 18, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Vielmuth, F.; Walter, E.; Fuchs, M.; Radeva, M.Y.; Buechau, F.; Magin, T.M.; Spindler, V.; Waschke, J. Keratins Regulate p38MAPK-Dependent Desmoglein Binding Properties in Pemphigus. Front. Immunol. 2018, 9, 528. [Google Scholar] [CrossRef] [PubMed]
- Kasperkiewicz, M.; Ellebrecht, C.T.; Takahashi, H.; Yamagami, J.; Zillikens, D.; Payne, A.S.; Amagai, M. Pemphigus. Nat. Rev. Dis. Primers 2017, 3, 17026. [Google Scholar] [CrossRef] [PubMed]
- Bilgic Temel, A.; Murrell, D.F. Pharmacological advances in pemphigus. Curr. Opin Pharm. 2019, 46, 44–49. [Google Scholar] [CrossRef]
- Tavakolpour, S. Current and future treatment options for pemphigus: Is it time to move towards more effective treatments? Int. Immunopharmacol. 2017, 53, 133–142. [Google Scholar] [CrossRef]

| miRNAs | Forward Primer Sequence (5′-3′) |
|---|---|
| hsa-miR-125b-5p | CTGAGACCCTAACTTGTGAAA |
| hsa-miR-146a-5p | AGAACTGAATTCCATGGGTTAA |
| hsa-miR-148a-3p | AGTGCACTACAGAACTTTGTAA |
| hsa-miR-150-5p | CCCAACCCTTGTACCAGTG |
| hsa-miR-155-5p | TTAATGCTAATCGTGATAGGGG |
| hsa-miR-181a-5p | TTCAACGCTGTCGGTGAGTAA |
| hsa-miR181b-5p | ATTCATTGCTGTCGGTGGGT |
| hsa-miR-326 | TGGGCCCTTCCTCCAGAAA |
| hsa-miR-338-3p | CAGCATCAGTGATTTTGTTGAA |
| hsa-miR-423-5p | GGGCAGAGAGCGAGACTTT |
| hsa-miR-424-5p | CAGCAGCAATTCATGTTTTGAA |
| hsa-miR-584-5p | GTTTGCCTGGGACTGAGAAA |
| 5S | GGAGACCGCCTGGGAATA |
| Group | Sex (Female/Male) | Age (Mean ± SD) | ELISA (U/mL) | PDAI Score (Mean ± SD) | Disease Severity | ||
|---|---|---|---|---|---|---|---|
| Dsg3 (Mean ± SD) | Dsg1 (Mean ± SD) | Mild (n) | Moderate (n) | ||||
| PV group (n = 22) | 12/10 | 53.23 ± 8.74 | 148.39 ± 33.84 | 40.29 ± 42.71 | 11.09 ± 6.21 | 7 | 15 |
| MMP group (n = 15) | 10/5 | 58.80 ± 10.65 | - | - | - | - | - |
| NC group (n = 10) | 6/4 | 53.50 ± 9.09 | - | - | - | - | - |
| miRNAs | AUC | Cutoff | Youden Index | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Area | SE | 95% CI | |||||||
| miR-125b-5p | 0.970 | 0.024 | 0.923–1.000 | 8.575 | 0.822 | 95.5 | 86.7 | 91.3 | 92.9 |
| miR-155-5p | 0.985 | 0.015 | 0.955–1.000 | 39.025 | 0.888 | 95.5 | 93.3 | 95.5 | 93.3 |
| miR-181a-5p | 0.980 | 0.018 | 0.946–1.000 | 67.150 | 0.818 | 81.8 | 100 | 100 | 78.9 |
| miR181b-5p | 0.970 | 0.025 | 0.922–1.000 | 15.835 | 0.867 | 100 | 86.7 | 91.7 | 100 |
| miR-326 | 0.979 | 0.018 | 0.943–1.000 | 178.440 | 0.864 | 86.4 | 100 | 100 | 83.3 |
| miR-584-5p | 0.988 | 0.014 | 0.960–1.000 | 10.455 | 0.933 | 100 | 93.3 | 95.7 | 100 |
| miR-155-5p + miR-584-5p | 0.977 | 0.026 | 0.925–1.000 | - | 0.955 | 95.5 | 100 | 93.8 | 100 |
| miR-181b-5p + miR-584-5p | 0.967 | 0.038 | 0.892–1.000 | - | 0.933 | 100 | 93.3 | 100 | 95.7 |
| miRNAs | miRDB | miRTarBase | miRWalk | TargetScan | Candidate Target Genes |
|---|---|---|---|---|---|
| miR-125b-5p | 925 | 432 | 6148 | 1736 | 622 |
| miR-155-5p | 701 | 904 | 2842 | 3296 | 1030 |
| miR-181a-5p | 1408 | 553 | 3106 | 3363 | 727 |
| miR181b-5p | 1408 | 375 | 5335 | 290 | 408 |
| miR-326 | 759 | 138 | 7930 | 3317 | 391 |
| miR-584-5p | 402 | 67 | 7670 | 2667 | 233 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, W.; Xing, Y.; Li, C.; Zhou, P.; Hu, X.; Hua, H.; Wei, P. Identification of Six microRNAs as Potential Biomarkers for Pemphigus Vulgaris: From Diagnosis to Pathogenesis. Diagnostics 2022, 12, 3058. https://doi.org/10.3390/diagnostics12123058
He W, Xing Y, Li C, Zhou P, Hu X, Hua H, Wei P. Identification of Six microRNAs as Potential Biomarkers for Pemphigus Vulgaris: From Diagnosis to Pathogenesis. Diagnostics. 2022; 12(12):3058. https://doi.org/10.3390/diagnostics12123058
Chicago/Turabian StyleHe, Wenxiu, Yixiao Xing, Chunlei Li, Peiru Zhou, Xiaosheng Hu, Hong Hua, and Pan Wei. 2022. "Identification of Six microRNAs as Potential Biomarkers for Pemphigus Vulgaris: From Diagnosis to Pathogenesis" Diagnostics 12, no. 12: 3058. https://doi.org/10.3390/diagnostics12123058
APA StyleHe, W., Xing, Y., Li, C., Zhou, P., Hu, X., Hua, H., & Wei, P. (2022). Identification of Six microRNAs as Potential Biomarkers for Pemphigus Vulgaris: From Diagnosis to Pathogenesis. Diagnostics, 12(12), 3058. https://doi.org/10.3390/diagnostics12123058






